Abstract
The TP53 gene product, p53, is a pleiotropic transcription factor induced by stress, which functions to promote cell cycle arrest, apoptosis and senescence. Genome-wide profiling has revealed an extensive system of long noncoding RNAs (lncRNAs) that is integral to the p53 signalling network. As a research tool, we implemented a public access database called TP53LNC-DB that annotates currently available information relating lncRNAs to p53 signalling in humans.
Introduction
Long noncoding RNAs (lncRNAs) are broadly defined as RNA transcripts of >200 nucleotides with no protein-coding activity (1). Apart from translation, the biosynthesis and regulation of lncRNAs occur similarly to coding gene transcription with RNA polymerase II–mediated transcription, capping and polyadenylation, along with alternative splicing (2, 3). Notably, current estimates suggest that lncRNA genes may be nearly as abundant, if not more so, as their mRNA cousins (4). Indeed, there is growing interest in the function of specific lncRNAs in both physiological and pathological processes (5–7).
Among the body of work involving lncRNAs, there has been a strong association with the tumor suppressor p53 (8, 9). P53 is the protein product of the TP53 gene, and among a large group of genes defined as either tumor suppressors or oncogenes, TP53 is arguably the most notorious gene linked with the aetiology of cancer. This reputation comes from the observation that TP53 is rendered ineffective by mutation in ~50% of all cancer cases and possibly inactivated by a range of indirect mechanisms in others (10, 11). It has become well known that p53 directly or indirectly regulates a diverse array of cellular pathways (12, 13). Most notably, the p53 signalling network is essential for normal cell growth and genomic stability, working to impose cell cycle arrest or apoptosis in response to cellular stress, including but not limited to DNA damage and environmental challenges, together with responding to oncogenic changes in cells (14).
The cellular levels of p53 are tightly controlled, and these have been shown to be regulated by an array of transcriptional and post-transcriptional mechanisms (15, 16). For example, p53 levels are classically regulated by proteasomal degradation mediated through the E3 ubiquitin ligases MDM2, COP1 and Pirh2, among others (16–19). However, there is a growing body of evidence to establish links between lncRNAs and the p53 signalling network. In particular, a large number of lncRNAs appear to be implicated in cellular signalling pathways alongside of p53, functioning as regulators or effectors in the execution of downstream functions of p53 (20). For example, p53 transcriptionally upregulates the lncRNA TRINGS (Tp53-regulated inhibitor of necrosis under glucose starvation) to protect cancer cells from necrosis under conditions of glucose starvation (21). Similarly, the lncRNA PANDA is also upregulated by p53, working to regulate cell death genes downstream of p53 (22). Other lncRNAs have been shown to exert control over p53, accomplishing this through a variety of mechanisms. The lncRNA DINO binds directly to the p53 protein and promotes its stabilization (23), while MEG3 also impacts p53 gene expression through effects of MDM2 that prevent p53 degradation (24). Conversely, the lncRNA RoR represses p53 in response to DNA damage and has unique capability being as effector and regulator of p53, forming an autoregulatory feedback loop (25).
Recently, improved computational predictions together with more sensitive RNA sequencing (RNA-seq) technologies have helped to uncover the true extent of the lncRNA transcriptome (26, 27). The latter is important as the expression level of lncRNAs is often quite low compared with mRNAs (28). There are an increasing number of publications detailing the specific roles of individual lncRNAs across a spectrum of p53-regulated signalling pathways (8, 9, 29), most notably the dysregulation of lncRNAs in cancer and their contribution to disease pathophysiology (21, 30). Keeping in mind the growing importance of this research, we designed and implemented a comprehensive database ‘TP53LNC-DB’ providing the accumulated knowledge of human lncRNAs specifically involved or implicated in p53 signalling pathways.
Data source implementation and contents of database
Articles published before 31 May 2018 were retrieved from PubMed using keyword searches [p53 AND long non-coding RNA], [p53 AND lncRNA] and [p53 AND lincRNA]. Abstracts were curated, and information was extracted as a manually annotated record of database fields including linked entries to PMIDs of the relevant articles (Table 1). Abstracts of the retrieved articles were read to confirm if each study was related to the p53 signalling pathway. Thereafter, high-confidence data were collected from these studies to appropriately match data fields required for entry into the database.
Table 1.
Detailed field entry data for TP53LNC database
| Field code | Entry variables | Description of criteria |
|---|---|---|
| P53LncRNA_ID | P53LNCxxxx | System generate unique identifier for TP53LNC database |
| LNCipedia_ID | Unique identifier assigned from the LncRNAdb project with link out service to LNCipedia.org entry | |
| Ensembl_ID | ENSTxxxx | The transcript variant associated with the gene symbol with link out service to Ensemble |
| NONCODE_ID | NONHSATxxxx | The transcript variant associated with the gene symbol with link out service to NONCODE |
| Symbol | The lncRNAs identified by its common HNGC identifier along with available identifying information retrieved from each publication | |
| Verification | Predicted OR Verified (If verified methods specified) | LncRNAs associated with p53-signalling either Predicted (no further experimental evidence available) or Verified (supported by additional experimentation) |
| Method | The experimental methods that were employed to identify the lncRNAs, such as microarray, RNA-seq, Tilling array, etc. | The experimental approach by which a particular or a large dataset(s) LncRNA related to p53 signalling was identified |
| p53_regul. | Up-regulated, Down-regulated, N/A | Up-regulated/ Down-regulated according to positive or negative regulation by p53. N/A if unknown |
| p53_pathway | Pathway(s) OR cellular process(es) such as DNA damage response (DDR), cell cycle, senescence, proliferation, etc. | Particular signalling pathway(s) or processes involved where lncRNAs are Verified |
| Tissue_Cells | Cell line (tissue source/name specified) | The specific cell(s) type OR tissue(s) (cancerous) in which lncRNA(s) are identified |
| PMID | Link out to PMIDs of the relevant article |
First, each lncRNA entered into database was assigned a unique identifying number (P53LncRNA_ID). Entries derived from high-throughput approaches including RNA-seq, deep sequencing and microarrays but not having been verified by secondary approaches were classified as ‘Predicted’ in the ‘Verification’ field. If secondary approaches were used to confirm the lncRNA-p53 association, then these lncRNAs were classified as verified and the methods used were recorded in the ‘Verification’ field. For example, Leveille et al. (31) globally mapped p53-regulated enhancers by treating MCF-7 cells with the p53 activator nutlin3a. After RNA-seq analysis and identification of differentially induced lncRNAs, the nutlin3a-responsive lncRNA LED was confirmed by several experimental approaches including qPCR, ChIP and Northern. Thus LED is listed as a ‘Verified’ lncRNA, while other p53-regulated lncRNAs not investigated are listed as ‘Predicted’. Other examples of ‘Verified’ entries include TRINGS (21) and GUARDIN involved in maintaining genomic stability (30). Both lncRNAs were first identified by microarray analysis using H1299 cells carrying an inducible wild-type p53 expression system. After initial screening, both TRINGS and GUARDIN were confirmed to be p53-responsive lncRNAs using a variety of secondary approaches to reveal their function. The ‘Method’ field captures further information regarding the primary screening approach and other methods used to identify the lncRNAs.
As lncRNAs can be either positively or negatively impacted by p53, their regulatory status is recorded in the ‘p53_regul.’ field in the database as ‘Up regulated’ or ‘Down regulated’. Where the status is undetermined or the lncRNA expression unaffected by p53, e.g. as part of the TP53 pathway but not regulated by it, the entry is marked not applicable (NA) for this category field. Additional information regarding the function of each lncRNA in p53 signalling is recorded in the ‘p53_pathway field’, such as DNA-damage response, proliferation and apoptosis for the entry for the lncRNA GUARDIN. In some instances, the identified lncRNAs act as regulators of TP53; for example, HSUP1 is indicated to destabilize p53, this information also being captured within the ‘p53_pathway field’. Lastly, the corresponding experimental approaches used together with cells and/or tissue data were captured for the ‘p53_pathway’ and ‘Tissue_Cells’ fields, respectively.
In addition to the annotated data fields derived from PubMed, the lncRNA gene names or transcript names obtained through PubMed were used to retrieve relevant transcript or gene IDs from other data sources. The database includes web links to LNCipedia (https://lncipedia.org/), Ensembl (https://asia.ensembl.org/index.html) and NONCODE (http://www.noncode.org/index.php) to provide detailed lncRNA transcript information. For example, the lncRNA HOTAIR has links to the LNCipedia (HOTAIR), Ensemble (ENST00000424518) and NONCODE (NONHSAT028510) databases. Where particular transcript identifiers are not available in the corresponding databases, the entry is left blank. The ‘Symbol’ field contains the common gene symbol from HUGO Gene Nomenclature Committee (HGNC), if available, together with other alias information or identifiers obtained through the cited paper. It should be noted that several lncRNA entries are repeated in the database with different TP3LNC IDs, resulting from the same gene being identified through different studies. For example, searching for the lncRNA HOTAIR returns entries related to five studies (P53LNC0036 P53LNC0066, P53LNC0140, P53LNC1458, P53LNC2317) the information related to different transcripts as well as listing the specific attributes of each report. Leaving the information in this discrete form ensures the information from separate studies is easier to interpret.
Development of database
The application architecture consists of a PHP presentation layer and MySQL persistent storage (Figure 1). Combined HTML/CSS and JavaScript enable interfaces that are easy to interpret and navigate. TP53LNC-DB is supported by main standards-compliant web browsers including Firefox, Google Chrome, Internet Explorer and Safari.
Figure 1.
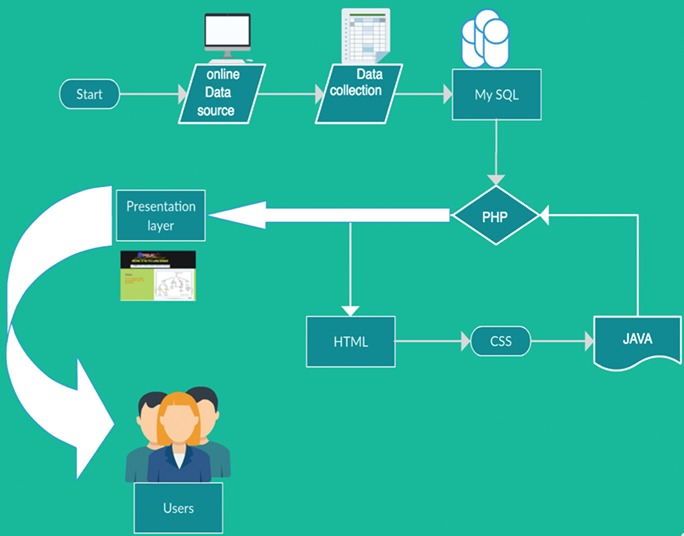
Layout of application architecture. The collected lncRNA in database information was integrated as PHP presentation layer combined with MySQL persistent storage. HTML/CSS and JavaScript interfaces were given for interpretation and navigation.
Data query search and browsing
The front end of TP53LNC-DB is a simple user-friendly web interface (Figure 2A) with access to information provided by text-based search (Figure 2B). Searches are configured to match either full or partial query terms. For example, searching for the keyword ‘DINO’ will retrieve the entry for lncRNA DINO, whereas a partial search term such as ‘DIN’ will retrieve the entries for DINO and GUARDIN. Multi-conditional search options are also configured for key parameters, whether the lncRNA(s) are experimentally verified or predicted, the conditional status of p53 being up-regulated or down-regulated and the implemented experimental approach. Total database entries can also be viewed in the browsing function along with the display of preconfigured search queries aligned with different aspects of p53 biology (Figure 2C; Table 2).
Figure 2.
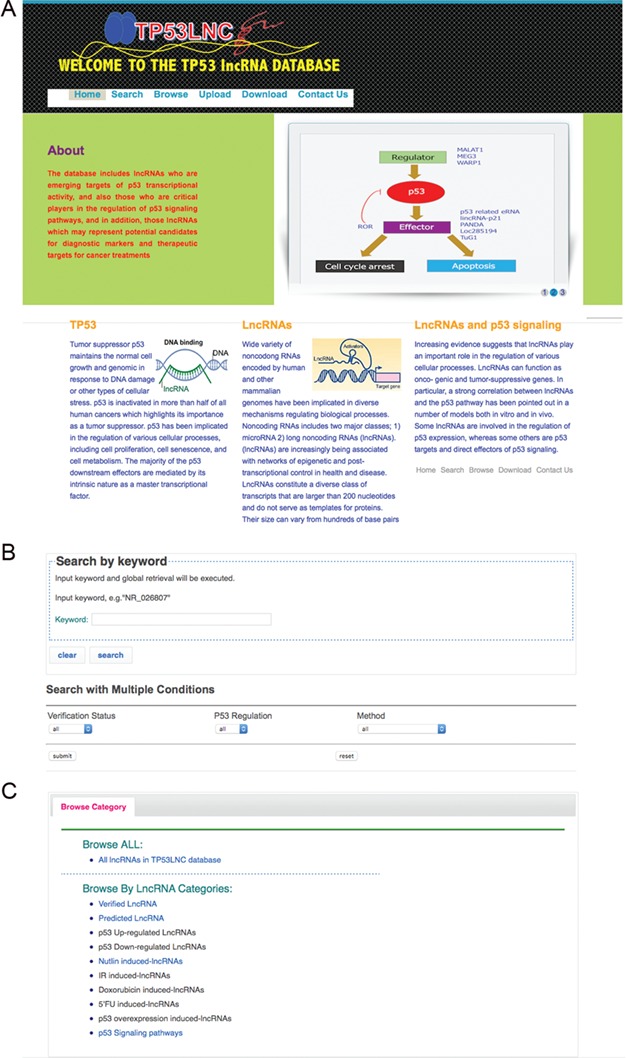
The web interface of TP53LNC-DB. (A) The homepage screen, (B) search screen and (C) preconfigured browsing options.
Table 2.
Summary of preconfigured search terms for browsing TP53LNC-DB entries
| Grouping | Pre-configured database queries | Search term employed | Total entries/ group |
|---|---|---|---|
| Group I Verification status |
Verified lncRNAs | 227 | 4851 |
| Predicted lncRNAs | 4624 | ||
| Group II Regulation status |
p53 Up-regulated lncRNAs | 2680 | 4094 |
| p53 Down-regulated lncRNAs | 1414 | ||
| Group III p53-lncRNA inducing agents and others |
Nutlin-induced lncRNAs | 769 | 4526 |
| IR-induced lncRNA | 785 | ||
| Doxorubicin-induced lncRNAs | 25 | ||
| 5’-FU–induced lncRNAs | 25 | ||
| p53 overexpression–induced lncRNAs | 2804 | ||
| Others | 118 |
Discussion
The knowledgebase around lncRNAs is considerable less than that of protein-coding genes. Notably, however, there are other aspects of lncRNA biology that impede research efforts (26). Annotating the lncRNA transcriptome is inherently difficult for several reasons; their relative low expression causes lncRNA transcripts to be under-sampled in high-throughput RNA-seq, and they lack identifiable features such as open reading frames (ORFs). Moreover, the weak or absent conservation of lncRNAs necessitates a heavy reliance on empirical studies. On the latter basis together with our own research interests in p53, we initiated the TP53LNC-DB project.
Many databases have already been developed to store lncRNA-associated information (8, 32–34); however, an lncRNA database dedicated to a singular gene such as p53 signalling has never been previously implemented. At time of publication, there were 4851 unique lncRNA entries correlated with p53-related signalling pathways in the database, a figure that clearly speaks to the importance of this emerging aspect of p53 biology. However, less than 5% of these entries have been experimentally verified either in vitro or in vivo (Table 2). We anticipate the database will help de-convolve this complexity of information around p53 and lncRNAs and prove a useful resource to the research community.
Acknowledgements
The Translational Research Institute gratefully acknowledges funding support from the Henan Provincial People’s Hospital.
Funding
MRK was supported through an Initial Funding Postdoctoral Research Project grant awarded by Henan Province.
Conflict of interest. None declared.
Database URL: http://www.trihpph.net/TP53LNC/index.php
References
- 1. Derrien T., Johnson R., Bussotti G. et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res., 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ransohoff J.D., Wei Y. and Khavari P.A. (2018) The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol., 19, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carlevaro-Fita J., Rahim A., Guigo R. et al. (2016) Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA, 22, 867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Djebali S., Davis C.A., Merkel A. et al. (2012) Landscape of transcription in human cells. Nature, 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Melissari M.T. and Grote P. (2016) Roles for long non-coding RNAs in physiology and disease. Pflugers Arch., 468, 945–958. [DOI] [PubMed] [Google Scholar]
- 6. Mercer T.R. and Mattick J.S. (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol., 20, 300–307. [DOI] [PubMed] [Google Scholar]
- 7. Batista P.J. and Chang H.Y. (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell, 152, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang A., Xu M. and Mo Y.Y. (2014) Role of the lncRNA-p53 regulatory network in cancer. J. Mol. Cell Biol., 6, 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huarte M., Guttman M., Feldser D. et al. (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell, 142, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caron de Fromentel C. and Soussi T. (1992) TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes Chromosomes Cancer, 4, 1–15. [DOI] [PubMed] [Google Scholar]
- 11. Chao C., Hergenhahn M., Kaeser M.D. et al. (2003) Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem., 278, 41028–41033. [DOI] [PubMed] [Google Scholar]
- 12. Kruiswijk F., Labuschagne C.F. and Vousden K.H. (2015) p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol., 16, 393–405. [DOI] [PubMed] [Google Scholar]
- 13. Levine A.J. and Oren M. (2009) The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer, 9, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kastenhuber E.R. and Lowe S.W. (2017) Putting p53 in context. Cell, 170, 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halaby M.J. and Yang D.Q. (2007) p53 translational control: a new facet of p53 regulation and its implication for tumorigenesis and cancer therapeutics. Gene, 395, 1–7. [DOI] [PubMed] [Google Scholar]
- 16. Kruse J.P. and Gu W. (2009) Modes of p53 regulation. Cell, 137, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dornan D., Wertz I., Shimizu H. et al. (2004) The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature, 429, 86–92. [DOI] [PubMed] [Google Scholar]
- 18. Meek D.W. (2009) Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer, 9, 714–723. [DOI] [PubMed] [Google Scholar]
- 19. Sheng Y., Laister R.C., Lemak A. et al. (2008) Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat. Struct. Mol. Biol., 15, 1334–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grossi E., Sanchez Y. and Huarte M. (2016) Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim. Biophys. Acta, 1859, 200–208. [DOI] [PubMed] [Google Scholar]
- 21. Khan M.R., Xiang S., Song Z. et al. (2017) The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J., 36, 3483–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hung T., Wang Y., Lin M.F. et al. (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet., 43, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmitt A.M., Garcia J.T., Hung T. et al. (2016) An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet., 48, 1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang X., Gejman R., Mahta A. et al. (2010) Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res., 70, 2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang A., Zhou N., Huang J. et al. (2013) The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res., 23, 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uszczynska-Ratajczak B., Lagarde J., Frankish A. et al. (2018) Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet., 19, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Z., Nair A., Chen X. et al. (2018) Author correction: UClncR: ultrafast and comprehensive long non-coding RNA detection from RNA-seq. Sci. Rep., 8, 5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cabili M.N., Trapnell C., Goff L. et al. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev., 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang E.B., Yin D.D., Sun M. et al. (2014) P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis., 5, e1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu W.L., Jin L., Xu A. et al. (2018) GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol., 20, 492–502. [DOI] [PubMed] [Google Scholar]
- 31. Leveille N., Melo C.A., Rooijers K. et al. (2015) Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun., 6, 6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou Z., Shen Y., Khan M.R. et al. (2015) LncReg: a reference resource for lncRNA-associated regulatory networks. Database, 2015, 1 January 2015, bav083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quek X.C., Thomson D.W., Maag J.L. et al. (2015) lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res., 43, D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Wu D., Chen L. et al. (2014) RAID: a comprehensive resource for human RNA-associated (RNA-RNA/RNA-protein) interaction. RNA, 20, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]


