Abstract
Baratela-Scott syndrome (BSS) is a rare, autosomal-recessive disorder characterized by short stature, facial dysmorphisms, developmental delay, and skeletal dysplasia caused by pathogenic variants in XYLT1. We report clinical and molecular investigation of 10 families (12 individuals) with BSS. Standard sequencing methods identified biallelic pathogenic variants in XYLT1 in only two families. Of the remaining cohort, two probands had no variants and six probands had only a single variant, including four with a heterozygous 3.1 Mb 16p13 deletion encompassing XYLT1 and two with a heterozygous truncating variant. Bisulfite sequencing revealed aberrant hypermethylation in exon 1 of XYLT1, always in trans with the sequence variant or deletion when present; both alleles were methylated in those with no identified variant. Expression of the methylated XYLT1 allele was severely reduced in fibroblasts from two probands. Southern blot studies combined with repeat expansion analysis of genome sequence data showed that the hypermethylation is associated with expansion of a GGC repeat in the XYLT1 promoter region that is not present in the reference genome, confirming that BSS is a trinucleotide repeat expansion disorder. The hypermethylated allele accounts for 50% of disease alleles in our cohort and is not present in 130 control subjects. Our study highlights the importance of investigating non-sequence-based alterations, including epigenetic changes, to identify the missing heritability in genetic disorders.
Keywords: XYLT1, methylation, epigenetic, trinucleotide repeat, repeat expansion, skeletal dysplasia, Desbuquois dysplasia, 16p13 deletion, fragile site
Introduction
Baratela-Scott syndrome (BSS [MIM: 300881]) is a rare syndrome characterized by short stature with skeletal dysplasia, facial dysmorphisms, and developmental delay.1 Here we report that BSS is caused by recessive pathogenic variants in XYLT1 (MIM: 608124), which have also been reported in Desbuquois dysplasia type II (DBQD-II [MIM: 615777]).2, 3, 4, 5, 6, 7 Compound heterozygous or homozygous pathogenic variants in XYLT1 were identified in 12 families with one or more individuals with DBQD-II, including 3 frameshift, 3 missense, 2 splice site, and 2 nonsense variants (Table S1). In addition, a whole gene deletion and an 18 bp deletion encompassing the intron 7-exon 8 junction was reported in an infant with severe short limb skeletal dysplasia but without the typical findings of DBQD-II.8
XYLT1 encodes the xylosyltransferase enzyme, XT1, which catalyzes the first step in the biosynthesis of chondroitin sulfate and dermatan sulfate proteoglycans.9 The promoter region of XYLT1 was recently characterized and found to have 238 bp of sequence (XYLT1-238) that is not present in the reference genome (GRCh37/hg19, GRCh38/hg38) but appears to be fixed in the human population (100/100 individuals tested)10 and is evolutionarily conserved in chimpanzee and mouse. The 238 bp region also contains a variable GGC repeat with a range of 9–20 repeats in 100 individuals tested; repeat length within this range did not influence promoter activity in in vitro assays.10
In this work, we identified a pathogenic GGC repeat expansion in the annotated XYLT1 promoter region associated with hypermethylation of exon 1 (Figure 1) in eight of ten families with BSS with single or no known XYLT1 variants. The hypermethylated and expanded alleles show reduced expression and account for half of the disease alleles in our cohort. We also identified a recurrent 3.1 Mb deletion of 16p13 encompassing XYLT1 that accounts for 20% of disease-associated alleles.
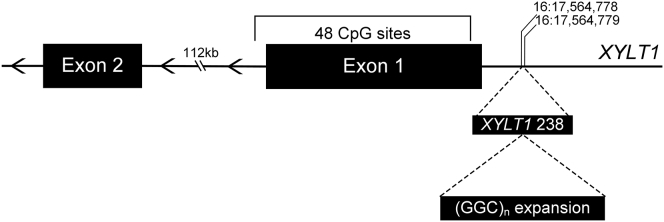
Figure 1.
XYLT1 Exon 1 and Surrounding Region
Schematic of XYLT1 (GRCh37/hg19, GenBank: NM_022166) exon 1 and surrounding region, noting the region upstream previously described as XYLT1-238, which contains the GGC repeat region; genomic coordinates notated. Methylation-specific PCR was performed on the bracketed segment of exon 1, which includes 48 CpG sites.
Material and Methods
Cohort Recruitment and Sample Collection
Affected individuals, family members, and unaffected control subjects were recruited into this study after obtaining written informed consent. This study was approved by the institutional review boards at University of Washington (IRB# 28853) and Nemours (IRB# 212117). DNA was extracted from white blood cells (WBC), saliva, or fibroblasts using standard procedures. Fibroblasts were cultured using standard protocols.
Exome Sequencing
Sequence capture was performed using the Roche NimbleGen SeqCap EZ Human Exome Library v.2.0 (37 Mb target, Basel, Switzerland) and sequenced via paired-end 50 bp reads on Illumina HiSeq2000/2500 sequencers. BAM files were aligned to human reference (GRCh37/hg19) using Burrows-Wheeler Aligner (BWA) and variants were called using UnifiedGenotyper tool from Genome Analyzer Toolkit (GATK) and annotated with VEP v.73.
Bisulfite Conversion of DNA
Bisulfite treatment of genomic DNA was carried out with the EZ DNA Methylation Direct Kit (Zymo, D5020) using 600 ng DNA and final elution of 12 μL. Bisulfite treatment of genomic DNA for exon 1 methylation controls was done using the EZ DNA Methylation Kit (Zymo, D5003) per the manufacturer’s protocol.
RNA Methods
RNA was extracted from cultured fibroblast cells using Trizol (Ambion) and RNA Clean and Concentrator Kit (Zymo). RNA was extracted from blood using the QIAamp Blood Mini Kit following the manufacturer’s protocols (QIAGEN). For RT-PCR, cDNA was prepared using either SuperScript II (Invitrogen, 18064-014) or the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). RNA-seq libraries were constructed using the TruSeq RNA Library Prep Kit v2 (Illumina) and sequenced with 150 nucleotide paired-end reads on a HiSeq2500. Output fastq files were processed using Spliced Transcripts Alignment to a Reference (STAR); subsequently, output BAM files were visualized as Sashimi Plots with Integrated Genomics Viewer (IGV).
Methylation-Specific PCR
Methylation-specific primers were designed to capture exon 1 of XYLT1 (GenBank: NM_022166.3) as completely as possible. Using the GRCh37/hg19 reference sequence (UCSC genome browser), primers were manually designed to maximize the number of CpG sites captured. The 3′ end of the reverse primer was specifically a CpG site to ensure methylation-specific binding. PCR was carried out with HotStarTaq DNA Polymerase (QIAGEN, 203203) using 2 μL of bisulfite-treated DNA and 0.5 μM of both forward and reverse primer in a final reaction volume of 50 μL. Commercially available fully methylated DNA was combined with fully unmethylated DNA (Zymo, D5014) and diluted to 5% methylation to ensure that this method could detect low levels of methylated alleles.
Southern Blot Analysis
5 μg genomic DNA was digested with restriction enzyme KpnI (New England Biolabs), followed by electrophoresis in a 0.8% agarose gel. Capillary transfer of the separated DNA fragments was carried out with a charged nylon membrane (GE Amersham) overnight and crosslinked by exposure to ultraviolet light. Probe was prepared by PCR amplification of a genomic fragment (Table S2) cloned into a plasmid. The probe was labeled with p32-alpha-dCTP (MegaPrime) and hybridized to the membrane at 65°C overnight. Membrane was washed two times for 15 min each time in 2× SSC, 0.1% SDS and once with 0.2× SSC, 1% SDS at 65°C. The probes were exposed to film anywhere from 24 hr to 7 days at -80°C before development.
Whole-Genome Sequencing and STRetch Analysis
Genomic DNA underwent library preparation using Accel-NGS 2S PCR-free DNA library kit (Swift BioSciences, 20024) for 150 nucleotide paired-end sequencing on a HiSeq4000. Output fastqs were adapter trimmed and aligned to a modified reference genome (hg19) that included XYLT1-238 with BWA. Paired-end whole-genome sequencing data were also interrogated for repeat expansions using STRetch software.11 Default analysis parameters and the provided STRdecoys.sorted.bed file were used for the STRetch analysis pipeline STRetch_wgs_pipeline.groovy. We used WGS data from 75 in-house genomes as controls in the analysis.
Whole-Genome Bisulfite Sequencing
DNA bisulfite conversion was done using EZ DNA Methylation Lightning Kit (Zymo). Library construction was carried out with the TruSeq DNA Methylation Kit (Illumina) for 150 nucleotide paired-end sequencing on a HiSeq4000. Output fastqs were trimmed with TrimGalore-0.4.4 and Cutadapt-0.16. A modified reference genome (hg19) containing XYLT1-238 underwent in silico bisulfite conversion, and trimmed fastqs were aligned using Bismark-0.19.0 and bowtie2. Output BAM files were sorted using PicardTools for “bisulfite mode” visualization in IGV. A methylation report of individual base pair conversion was generated using Bismark methylation extractor v.0.19.0 to confirm results seen in IGV.
Results
Cohort
We identified 12 individuals (11 male) in 10 unrelated families with BSS. The detailed phenotypes of 7 individuals from 6 families were previously described, delineating the BSS phenotype.1 All 12 affected individuals had short stature at birth (length range −3 to −11 SD) that persisted throughout development (present height range −5 to −9 SD) and mild facial dysmorphisms including midface hypoplasia, depressed nasal bridge, and epicanthal folds. Significant and characteristic developmental delays are present (Figure 2, Table 1). Though 11/12 affected individuals in our cohort are males, previous reports of individuals with DBQD-II due to XYLT1 pathogenic variants do not show the same skewing (6 males, 7 females, 3 not reported; Table S1), suggesting that larger cohorts are needed to determine whether a true sex bias exists.
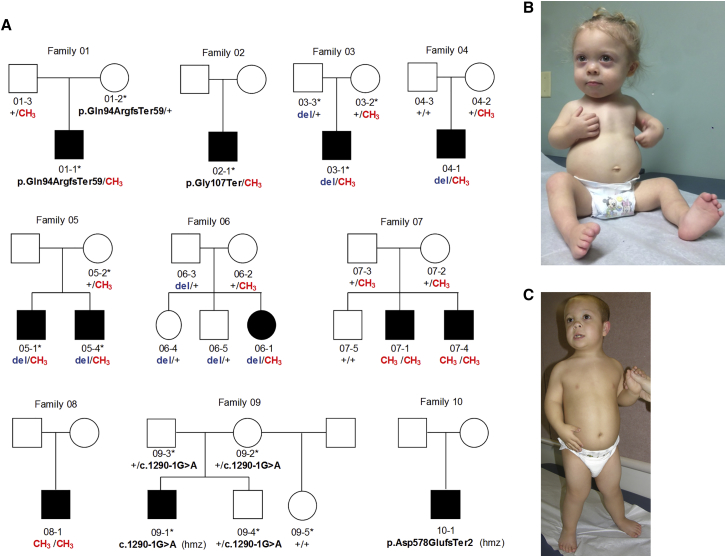
Figure 2.
BSS Cohort
(A) Pedigrees of 10 families with 12 affected individuals included in this study. XYLT1 variants are noted for those individuals tested. CH3, methylated allele; del, 16p13 deletion; ∗, individual underwent exome sequencing; hmz, homozygous.
(B) Proband 06-1 as an infant.
(C) Proband 04-1 at 9 years of age. Dysmorphic features include flat midface, depressed nasal bridge, epicanthal folds.
Table 1.
Anthropometric Data and Craniofacial Features
|
04-1 |
06-1 |
03-1 |
02-1 |
08-1 |
01-1 |
05-1 |
05-4 |
07-1 |
07-4 |
09-1 |
10-1 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Previously Reported Patient No.1 | Patient 2 | Patient 1 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | ||||||
| XYLT1 variant 1 | 16p13 del (de novo) | 16p13 del (pat) | 16p13 del (pat) | c.319G > T | CH3 | CH3 (pat) | 16p13 del | 16p13 del | CH3 (pat) | CH3 (pat) | c.1290−1G>A (pat) | c.1730_1733dup |
| XYLT1 variant 2 | CH3 (mat) | CH3 (mat) | CH3 (mat) | CH3 | CH3 | c.281_306del (mat) | CH3 (mat) | CH3 (mat) | CH3 (mat) | CH3 (mat) | c.1290−1G>A (mat) | c.1730_1733dup |
| Birth weight (kg; SDS) | 2.4 (−2.4) | 1.86 (−3.3) | 3.2 (−1.3) | 3.0 (−1.0) | 2.6 (−2.2) | 2.5 (−2.3) | 3.3 (−1.1) | 2.3 (−2.5) | 2.5 (−1.7) | 2.0 (−2.3) | NA | 2.0 (NA) |
| Birth length (cm; SDS) | 38.1 (−6.1) | 33.5 (−11.1) | 44.1 (−3.5) | 42.0 (−3.0) | 42.0 (−4.4) | 38.7 (−5.9) | 43.0 (−4.0) | 43.0 (−4.0) | 37.0 (−5.0) | 36.0 (−5.1) | 34.2 (−8.0) | 37.0 (NA) |
| Birth OFC (cm; SDS) | 33.0 (−2.0) | 31.0 (−3.9) | 33.0 (−2.2) | 35.0 (−0.4) | 33.7 (−1.6) | 35.0 (−0.7) | NA | NA | 32.7 (−1.5) | 32.0 (−1.7) | NA | NA |
| Age at most recent exam | 11 year | 5 year | 20 year | 13 year 7 mo | 22 year | 16 year | 22 year | 18 year | 4 year 6 mo | 22 mo | 14 year | 3 year 7 mo |
| Current wt (kg; SDS) | 23.4 (−2.9) | 10.2 (−5.9) | 46.8 (−3.1) | 41.1 (−0.9) | 36.7 (−5.6) | 52.3 (−1.0) | 59.0 (−1.2) | 51.0 (−2.0) | 10.8 (−4.9) | 6.8 (−4.9) | 19.0 (−7.2) | 12.2 (−2.3) |
| Current ht (cm; SDS) | 99.0 (−7.0) | 76.5 (−7.9) | 116.0 (−8.2) | 114.6 (−5.5) | 114.0 (−8.5) | 130.0 (−5.0) | 120.0 (−7.7) | 114.0 (−8.0) | 76.9 (−6.4) | 59.7 (−9.0) | 97.0 (−7.2) | 71.6 (−7.5) |
| Current OFC (cm; SDS) | 50.5 (−2.0) | 44.0 (−4.6) | 51.5 (−2.4) | 52.0 (−1.4) | 53.0 (−1.4) | 55.3 (0.1) | 55.7 (0.4) | 57.2 (1.5) | 48.7 (−1.2) | 45.2 (−2.1) | 47.0 (−4.8) | 46.7 (−2.2) |
| Developmental delay | + moderate | + moderate | + (IQ 49) | + | + (IQ 53) | + | + | + | + | + | + | + |
| Craniofacial Features | ||||||||||||
| Depressed nasal bridge | + | + | + | + | + | + | + | + | + | + | + | + |
| Cleft palate/bifid uvula | − | − | + | − | − | + | + | + | − | − | − | − |
| Epicanthal folds | − | − | − | − | + | + | + | + | − | − | + | − |
| Synophrys | − | − | + | − | + | + | − | − | − | − | + | − |
Phenotypic data of affected individuals within our BSS cohort. Identification numbers are provided for those individuals with previously reported phenotypic data.1XYLT1 variants are notated: pat, paternally inherited; mat, maternally inherited; CH3, methylated allele. For proband 10-1, standard deviation scores at birth were not available as gestational age was not known.
Sequence and CNV Analysis Identify XYLT1 Variants in a Minority of Individuals
To determine the genetic etiology of BSS in this cohort, we performed exome sequencing in 14 individuals from 5 families, including 6 affected individuals (5 probands, 1 affected sibling; Figure 2). We identified a homozygous XYLT1 splice site variant in one proband (09-1, c.1290−1G>A) which causes exon 6 skipping (Figure S1), but no variants in XYLT1 in the remaining four probands, although exon 1 was not covered in the exome data, likely due to high GC content (82% GC). CNV analysis of exome data using CoNIFER12 identified a 3.1 Mb heterozygous deletion (approximate coordinates: chr16:15,300,000–18,400,000 [GRCh37/hg19]) that includes all of XYLT1 in two families (03-1 and his father, and 05-1 and his affected sibling), which was confirmed by SNP array in family 03.
Two additional probands (04-1 and 06-1) were identified by clinical chromosome microarray to have a heterozygous 3.1 Mb deletion of 16p13. Targeted capture and next-generation sequence analysis of XYLT1 failed to identify a pathogenic variant on the remaining allele in these two individuals; as with exome sequencing, exon 1 was not well covered. Low-pass whole-genome sequencing was performed in these two individuals to search for structural variants, but none were identified. In three other probands who did not have exome sequencing, Sanger sequencing of XYLT1 identified a homozygous 4 bp duplication (c.1730_1733dup [p.Asp578GlufsTer2], exon 8) in proband 10-1, but no variants in 07-1 or 08-1.
Given that we identified homozygous, likely pathogenic variants in XYLT1 in two affected individuals (09-1, 10-1) and a heterozygous deletion in an additional four case subjects (03-1, 04-1, 05-1, 06-1), we Sanger sequenced exon 1 for the six probands without previous exon 1 coverage. No second deleterious variant was found in the four probands with a heterozygous XYLT1 deletion; however, variants were identified in 02-1 (c.319G>T [p.Gly107Ter]) and 01-1 (c.281_306del [p.Gln94ArgfsTer59]). Both variants appeared homozygous, consistent with a recessive mode of transmission for this disorder; however, further segregation analysis in family 01 revealed that only the mother was heterozygous for the variant, suggesting possible allelic drop-out of the paternal allele. Re-examination of the Sanger sequence data for 02-1 showed a small wild-type peak in both saliva and WBC DNA, suggesting skewed amplification of one allele. The presence of the reference allele in 02-1 was confirmed by allele-specific PCR (Figure S2). Parents of 02-1 were not available for segregation studies. While screening for causal variants in family 05, we suspected similar allele drop-out for the mother. As both affected siblings share a heterozygous 16p13 deletion of XYLT1, which is not inherited from their mother, we expected the remaining allele would be maternal, but found numerous SNPs were not shared with either child. This phenomenon was specific to exon 1 and the 5′ region of the gene, suggesting that specific local DNA modification or conformational changes may interfere with amplification and cause PCR bias toward one allele. Thus, after standard sequencing and CNV analyses, we identified only a single pathogenic allele in six of ten families and no pathogenic alleles in two of ten families.
To investigate the hypothesis that local DNA modification or conformation was interfering with PCR, we digested DNA with BsrF1, a methylation-sensitive restriction endonuclease that cuts at multiple discrete sites in the region of interest. This approach in family 05 revealed a second, methyl-protected allele in the mother, and SNP analysis confirmed its inheritance by the affected siblings (Figure S3). The same approach in probands 02-1 and 01-1 confirmed that the apparently homozygous pathogenic variant in each was heterozygous, and a second methyl-protected allele with wild-type sequence was present. We conclude that BsrF1 digestion cut the unmethylated allele, allowing amplification and detection of the previously “hidden” methylated allele. Together, these results suggested that exon 1 and surrounding sequence are heavily methylated.
Bisulfite Sequence Analysis Confirms Hypermethylation of Exon 1 Alleles without Variants
Bisulfite treatment of DNA converts unmethylated cytosine residues to uracil, while 5-methylcytosine is unaffected. Sequence analysis of bisulfite-treated DNA therefore reveals the pattern of methylation in the sequenced region. We designed methylation-specific PCR (MS-PCR) primers to perform specific amplification of methylated and unmethylated alleles after bisulfite treatment (Table S3). Sequencing of the product, which encompasses the majority of exon 1, revealed methylation of all 48 CpG sites on the chromosome with the presumed missing alleles in our cohort (Figure S4A). Both alleles were fully methylated in individuals 08-1, 07-1, and 07-4, in whom no detectable sequence or copy number variant had been previously identified. For individuals 04-1, 06-1, 05-1, 05-4, and 03-1, the non-deleted allele was fully methylated; 01-1 and 02-1 had methylation of one allele. Segregation analysis using MS-PCR in available family members confirmed that the methylated allele segregated in trans to the variant allele in all case subjects (Figure S4A; Table S4).
Methylated Alleles Are Not Transcribed
We hypothesized that hypermethylation of exon 1 prevents transcription of XYLT1. Sanger sequencing of MS-PCR products in proband 02-1 confirmed that the heterozygous nonsense variant and hypermethylation alleles are in trans. Sequence analysis of cDNA from proband fibroblasts showed that only the unmethylated allele was present in cDNA sequence, indicating that the unmethylated allele was preferentially transcribed (Figure 3A). Using fibroblast cDNA from an individual with one deleted allele and one hypermethylated allele (04-1), we were unable to amplify any product spanning the exon 1-exon 2 splice junction; this result was confirmed by RNA-seq (Figures 3B, 3C, and S5). In addition, preliminary 5′-RACE and RT-PCR experiments suggest the reference annotation for the transcriptional start of XYLT1 may also be incomplete. Sequencing of mRNA from 09-1, 09-2, 02-1 and a control fibroblast line (GM08447) all showed evidence of a transcript that begins 5′ of the reference annotation, within XYLT1-238, and in fact may contain the entire repeat region (Figure S6; Table S5). The transcription start site that we identified is conserved in mouse as well. This suggests that the GGC repeat expansion lies within the 5′ UTR of XYLT1, similar to other trinucleotide repeat disorders with methylation of exon 1 and transcriptional silencing.
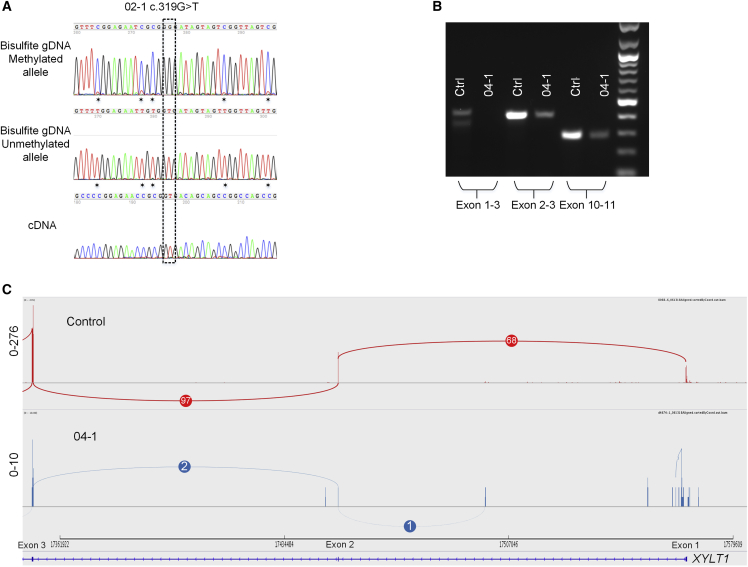
Figure 3.
Methylated Alleles Are Not Transcribed
(A) Sequence analysis of MS-PCR product from bisulfite-treated genomic DNA (top) of individual 02-1 confirms that the XYLT1 point mutation (c.319G>T) is located on the unmethylated allele in genomic DNA. CpG sites are notated (∗). Fibroblast cDNA sequence (bottom) confirms that only the unmethylated allele is transcribed.
(B) RT-PCR of fibroblast cDNA from 04-1 shows that transcription of exon 1-3 is absent and levels of shortened transcripts that begin downstream are reduced compared to the control.
(C) Sashimi Plot of RNA-seq data for 04-1 shows exon 1 is not transcribed and the exon 1-2 junction is absent. Overall transcription levels are severely reduced compared to the control.
Methylation of Exon 1 Is Rare in Control Subjects
To investigate the frequency of XYLT1 exon 1 hypermethylation in a healthy population, we tested 130 unrelated, unaffected individuals using MS-PCR on bisulfite-treated DNA. All control subjects were positive for the unmethylated product, confirming both the successful bisulfite conversion of the DNA and that exon 1 was unmethylated. All control subjects were negative for the methylated product. Studies using a mixture of commercially available fully methylated and unmethylated human genomic DNA showed that we could detect as few as 5% methylated alleles using MS-PCR (Figure S4B).
Southern Blot and WGS Analysis Suggest GGC Repeat Expansion
Using PCR primers upstream of the 5′ UTR region, we confirmed that the XYLT1-238 sequence10 is present in all individuals for whom PCR was successful. In probands with homozygous or hemizygous (with deletion) methylation, we were unable to obtain a PCR product using the same primers. As the XYLT1-238 region contains a GGC repeat ((GGC)n-AGC-(GGC)n-(GGA)n in most individuals), we suspected that individuals with hypermethylation have expanded repeats, preventing amplification of the region.
We performed Southern blots to evaluate the size of the promoter region of XYLT1 in four families. All individuals with a methylated allele had a larger DNA fragment than control subjects, with the increase in fragment size ranging from 300 to 2,500 bp (Figure 4). In two families (04 and 07), the expansion increased in size when transmitted from the unaffected mother. For individuals with hemizygous or homozygous methylated alleles (07-4, 04-1, 06-1), only expanded alleles were seen.
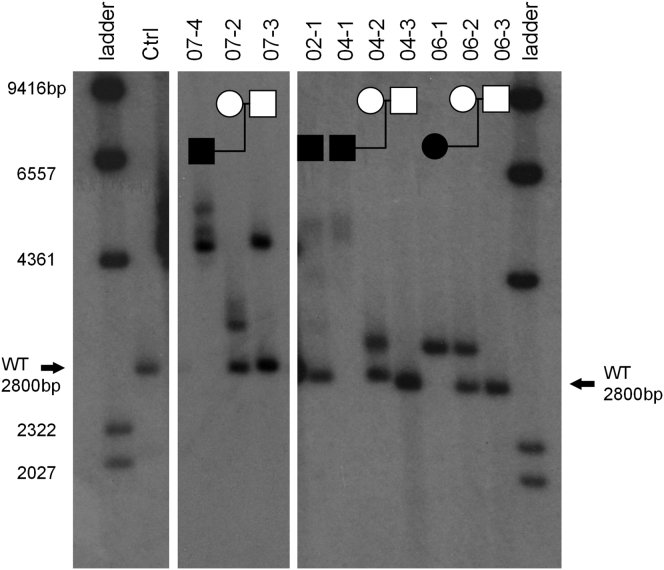
Figure 4.
Southern Blot of Four Affected Individuals and Available Family Members
The expected wild-type fragment (2,800 bp) encompasses exon 1 and surrounding DNA including XYLT1-238. For each individual with one or two methylated alleles, a fragment 300–2,500 bp larger than expected was detected. Individual 07-4 has two methylated alleles; 04-1 and 06-1 each have one methylated and one deleted allele, and 02-1 is heterozygous for methylation. In families 07 and 04, there is further expansion when transmitted from unaffected mother to affected child. Fragments that are >400 bp larger than wild-type exhibit instability in somatic cells as indicated by the non-distinct bands seen; particularly in 02-1 and 04-1. Each panel is from the same Southern blot but with different exposure times (panel 1, 4 day; panel 2, 24 hr; panel 3, 4 day).
To determine whether the increase in fragment size is due to expansion of the GGC repeat, we analyzed whole-genome sequence data for proband 04-1 using STRetch software.11 STRetch is designed to detect short tandem repeat (STR) expansions using paired-end reads by adding STR decoy sequences to the reference genome for mapping. Using an altered reference genome that includes the XYLT1-238 sequence, we found that all reads approaching the proposed expansion site had read pairs mapping to an expanded GGC decoy sequence. In addition, no read pairs span the expansion site, indicating a segment larger than the average insert size of 543 bp (Figure S7). These findings are consistent with the Southern blot data; in addition, the STRetch analysis is strong indication that the expansion comprises primarily GGC repeats.
Genome Bisulfite Sequencing Confirms Methylation of XYLT1 Expanded Repeat Alleles
To gain a broader understanding of the methylation landscape in the XYLT1 region, we performed whole-genome bisulfite sequencing (WGBS) on probands 04-1 and 06-1 who each have a deletion and a methylated allele (Figure S8). For 04-1, methylation stretched 1.7 kb downstream into intron 1 and 1.4 kb upstream of the expansion site (chr16:17,563,050–17,566,200). For 06-1, methylation was present approximately 700 bp both upstream and downstream of the expansion site (chr16:17,564,100–17,565,500). Coverage of the region was not uniform; however, 70% of exon 1 was covered for both probands >30×. Poor mapping prevented analysis of WGBS data across the GGC region. Coverage encompassing exon 1 (chr16:17,564,250–17,564,700) spanned 51 and 52 CpG sites for 04-1 and 06-1, respectively. For 04-1, 84% of sites were fully methylated and 96% of sites were more than 50% methylated. For 06-1, 88% of sites were fully methylated and 94% of sites were greater than 90% methylated. Results confirm the hypermethylation found using Sanger sequencing methods.
Discussion
We identified a trinucleotide repeat expansion upstream of XYLT1 associated with hypermethylation and transcriptional repression of XYLT1 as the most common pathogenic allele (10/20, 50%) in ten families with BSS. In two families, the affected individuals were homozygous for hypermethylated alleles. The hypermethylated allele cannot be detected by standard sequencing methods (i.e., exome, genome, or Sanger sequencing).
More than 40 disorders in humans are caused by expansions of simple repeats, most often trinucleotide repeats. The consequences of the repeat expansion differ depending on the gene and the location of the repeat within the gene structure. For example, the protein-coding CAG repeat in Huntington disease (MIM: 143100) leads to an expanded polyglutamine tract in the huntingtin protein resulting in aberrant properties that are eventually toxic to the cell; the CTG expansion in myotonic dystrophy (MIM: 160900) is located in the 3′ UTR of DMPK (MIM: 605377) and causes disease through RNA toxicity.13
Most similar to our finding, full expansion of a CGG repeat in the 5′ UTR of FMR1 (MIM: 309550), responsible for fragile X syndrome (MIM: 300264), leads to hypermethylation of the promoter and 5′ UTR and transcriptional silencing.14 Several CGG repeat regions have been characterized as folate-sensitive fragile sites, which are ctyogenetically visible “gaps” in the chromosome associated with repeat expansion and methylation. FRA16A is a fragile site on the short arm of chromosome 16 that was cloned and characterized more than two decades ago,15, 16 and manual comparison of the FRA16A published sequence confirmed that the FRA16A sequence corresponds to the XYLT1-238 sequence (which is not present in the reference genome). Our results are consistent with the original characterization of the FRA16A site, though no disease association was made at that time.
We initiated our study to investigate the genetic cause of BSS, the phenotypic features of which were recently delineated.1 During the course of our study, homozygous or heterozygous pathogenic variants in XYLT1 were identified as the cause of a similar disorder, Desbuquois dysplasia type II (DBQD-II),2 and the results of our current study confirm that BSS and what has been described as DBQD-II are allelic. Desbuquois dysplasia is a heterogeneous group of disorders characterized by skeletal dysplasia and dislocation of the large joints and short stature with flat face and prominent eyes. Specific forms of Desbuquois dysplasia are grouped based on the presence (type I and Kim type) or absence (type II) of specific hand anomalies. Unlike DBQD-I (MIM: 251450), caused by pathogenic variants in CANT1 (MIM: 613165), the majority of individuals with BSS have at least mild to moderate intellectual disability, with strong social and verbal skills initially masking their cognitive impairment. A lack of hand and foot abnormalities in BSS is another differentiating factor. Further detailed phenotyping will be necessary to determine whether individuals currently described as DBQD-II have the same degree and types of intellectual impairment as BSS, or whether the two disorders can be distinguished by cognitive abilities as seen in the distinction between Smit-McCourt dysplasia (MIM: 607326) and Dyggve-Melchior-Clausen dysplasia (MIM: 223800), both caused by recessive loss-of-function mutations in DYM.17 Notably, one of the pathogenic variants in four families (five affected individuals) is a 3.1 Mb recurrent deletion of 16p13, which is a known risk factor for a range of neurodevelopmental disorders.18, 19, 20 While the deletion may contribute to the neurodevelopmental features, individuals in our cohort with single nucleotide or repeat expansion alleles also present with developmental delays, suggesting this is a core feature of BSS.
The vast majority of repeat expansion disorders are autosomal dominant, and many cause neurologic or neurodegenerative disease. BSS now represents an autosomal-recessive skeletal dysplasia associated with repeat expansion. Other examples of autosomal-recessive disorders due to repeat expansions include Friedreich’s ataxia (MIM: 229300) (GAA expansion in intron 1 of FXN; see GeneReviews in Web Resources), Unverricht-Lundborg disease (MIM: 254800) (dodecanucleotide expansion in 5′ region of CSTB),22 and Richieri-Costa-Pereira syndrome (MIM: 268305) (CACA expansion in 5′ UTR of EIF4A3).23 In these conditions, the exact mechanism of transcriptional repression remains unclear.
Typically, in repeat expansion disorders, a threshold number of repeats is necessary to confer pathogenicity, though the specific number varies depending on the disorder. The smallest methylated allele in our cohort was approximately 300 bp (∼100 repeats) larger than the wild-type, unmethylated allele (Figure 4, individual 06-1). Faust and colleagues characterized the wild-type repeat region as 54 bp (18 trinucleotides) with the following composition: GGC9-AGC-GGC5-GGA3.10 In 100 healthy blood donors, they identified five additional homozygous alleles that could be accurately sequenced, with the repeat region ranging from 27 to 60 bp (9–20 trinucleotides). When characterizing the FRA16A site, Nancarrow and colleagues found repeat sizes on normal (not exhibiting a fragile site) chromosomes of up to 216 bp (72 repeats).16 Together with our data, this suggests the minimum repeat size associated with hypermethylation is between 216 bp and ∼360 bp. Faust and colleagues found that 45% of control subjects were homozygous; however, given our findings, it is possible that the “homozygous” allele in some cases was in trans with an expanded allele that could not be amplified. Given that BSS is very rare, it is unlikely that expanded and hypermethylated alleles are common in the population; we detected 0/130 control subjects with hypermethylation, but additional studies combining repeat sizing with methylation status are needed to determine the true frequency of expanded alleles, as well as the repeat size at which hypermethylation occurs.
The expanded, hypermethylated allele can be transmitted from an unaffected parent to child. As with many repeat expansions, the region appears to become unstable as the repeat size increases. In two families, we observed expansion of the maternal allele. Our sample size is too small to determine whether there is a parent-of-origin effect on the stability of the allele when transmitted, as has been seen for other triplet repeat expansion disorders.24, 25 However, earlier studies of FRA16A also suggested increased instability of the repeat in maternal transmissions than in paternal transmissions.16 Identification of additional affected individuals and large families in which transmission can be studied would be required. Large expansions also appear to be unstable in somatic tissue (blood and saliva), as shown by indistinct bands on Southern analysis in several individuals with expansions >2,500 bp (Figure 4).
Neither the expansion nor the hypermethylation are readily detected by standard Sanger or exome sequencing. In fact, we were not able to amplify the repeat region in individuals with an expansion and methylation, despite numerous attempts using methods designed for GC-rich regions (not shown). It is not clear whether amplification was prohibited by the repeat size, overall GC content, DNA conformation, methylation, or a combination. The most reliable detection methods in our studies were MS-PCR analysis to identify excess methylation of the region within and 5′ of exon 1 followed by Southern blot to confirm the presence of a larger allele. Using software designed to identify repeat expansions, we were able to confirm GGC expansion in the XYLT1-238 region using short-read genome sequencing; however, an alternative reference was required since the XYLT1-238 region is not present in the reference human genome.
In the clinical setting, dedicated testing will be required. For those individuals in whom a diagnosis of either BSS or DBQD-II is suspected but genetic testing is inconclusive due to the presence of only one or no pathogenic variants in XYLT1, our results open another avenue of testing to successfully diagnose individuals and aid in family planning. Further, segregation analysis for apparently homozygous pathogenic variant calls in exon 1 are necessary to ensure that an expanded and methylated allele has not been overlooked.
Our study illustrates the importance of investigating non-sequence-based alterations, including epigenetic changes, to identify the missing heritability in genetic disorders. The path to discovery in our cohort was circuitous and slow, as initial efforts were heavily focused on traditional sequencing approaches. Targeted sequencing, whole-exome, and even whole-genome sequencing yielded no causal variants for approximately half of our cohort. Despite this, a search for a unifying cause continued, in part due to the similarity of these individuals and the strong belief that all of the individuals in the cohort had the same condition. In the clinical setting, exome sequencing yields a diagnosis in less than 50% of all case subjects26 and in approximately one third of children who present with short stature.27 Our study highlights that alternate avenues of testing are needed for inherited disorders that continue to lack a clear genetic cause and persistently remain elusive despite extensive study. As the knowledge of the genetic landscape of Mendelian disorders continues to improve, our study provides further insight into genetic mechanisms that are currently not well understood and remain under-explored in the search for the genetic causes of disease.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
Research reported by K.S.-C. in this publication was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under grants P30GM114736 and P20GM103446. This work was also supported by Nemours Genetics Cluster funding (A.I. duPont Hospital for Children, Wilmington, DE, USA) and by the NIH IDeA program. Exome sequencing was provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by NHGRI and NHLBI grants UM1 HG006493 and U24 HG008956. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Genome library preparation was conducted by Molly Weaver at the University of Washington Center for Precision Diagnostics. We thank the patients and families for their participation.
Published: December 13, 2018
Footnotes
Supplemental Data include eight figures and five tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.11.005.
Contributor Information
Heather C. Mefford, Email: hmefford@uw.edu.
Katia Sol-Church, Email: ks5uq@virginia.edu.
Web Resources
CDC Growth Charts, https://www.cdc.gov/growthcharts/who_charts.htm
GeneReviews Resource Materials, https://www.ncbi.nlm.nih.gov/books/NBK481802/
GeneReviews, Bidichandani, S.I., and Delatycki, M.B. (1993). Friedreich ataxia. https://www.ncbi.nlm.nih.gov/books/NBK1281/
OMIM, http://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Baratela W.A., Bober M.B., Tiller G.E., Okenfuss E., Ditro C., Duker A., Krakow D., Stabley D.L., Sol-Church K., Mackenzie W. A newly recognized syndrome with characteristic facial features, skeletal dysplasia, and developmental delay. Am. J. Med. Genet. A. 2012;158A:1815–1822. doi: 10.1002/ajmg.a.35445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bui C., Huber C., Tuysuz B., Alanay Y., Bole-Feysot C., Leroy J.G., Mortier G., Nitschke P., Munnich A., Cormier-Daire V. XYLT1 mutations in Desbuquois dysplasia type 2. Am. J. Hum. Genet. 2014;94:405–414. doi: 10.1016/j.ajhg.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo L., Elcioglu N.H., Iida A., Demirkol Y.K., Aras S., Matsumoto N., Nishimura G., Miyake N., Ikegawa S. Novel and recurrent XYLT1 mutations in two Turkish families with Desbuquois dysplasia, type 2. J. Hum. Genet. 2017;62:447–451. doi: 10.1038/jhg.2016.143. [DOI] [PubMed] [Google Scholar]
- 4.Jamsheer A., Olech E.M., Kozłowski K., Niedziela M., Sowińska-Seidler A., Obara-Moszyńska M., Latos-Bieleńska A., Karczewski M., Zemojtel T. Exome sequencing reveals two novel compound heterozygous XYLT1 mutations in a Polish patient with Desbuquois dysplasia type 2 and growth hormone deficiency. J. Hum. Genet. 2016;61:577–583. doi: 10.1038/jhg.2016.30. [DOI] [PubMed] [Google Scholar]
- 5.Silveira C., Leal G.F., Cavalcanti D.P. Desbuquois dysplasia type II in a patient with a homozygous mutation in XYLT1 and new unusual findings. Am. J. Med. Genet. A. 2016;170:3043–3047. doi: 10.1002/ajmg.a.37858. [DOI] [PubMed] [Google Scholar]
- 6.Al-Jezawi N.K., Ali B.R., Al-Gazali L. Endoplasmic reticulum retention of xylosyltransferase 1 (XYLT1) mutants underlying Desbuquois dysplasia type II. Am. J. Med. Genet. A. 2017 doi: 10.1002/ajmg.a.38244. Published online April 30, 3017. [DOI] [PubMed] [Google Scholar]
- 7.Schreml J., Durmaz B., Cogulu O., Keupp K., Beleggia F., Pohl E., Milz E., Coker M., Ucar S.K., Nürnberg G. The missing “link”: an autosomal recessive short stature syndrome caused by a hypofunctional XYLT1 mutation. Hum. Genet. 2014;133:29–39. doi: 10.1007/s00439-013-1351-y. [DOI] [PubMed] [Google Scholar]
- 8.van Koningsbruggen S., Knoester H., Bakx R., Mook O., Knegt L., Cobben J.M. Complete and partial XYLT1 deletion in a patient with neonatal short limb skeletal dysplasia. Am. J. Med. Genet. A. 2016;170A:510–514. doi: 10.1002/ajmg.a.37453. [DOI] [PubMed] [Google Scholar]
- 9.Prydz K., Dalen K.T. Synthesis and sorting of proteoglycans. J. Cell Sci. 2000;113:193–205. doi: 10.1242/jcs.113.2.193. [DOI] [PubMed] [Google Scholar]
- 10.Faust I., Böker K.O., Lichtenberg C., Kuhn J., Knabbe C., Hendig D. First description of the complete human xylosyltransferase-I promoter region. BMC Genet. 2014;15:129. doi: 10.1186/s12863-014-0129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dashnow H., Lek M., Phipson B., Halman A., Sadedin S., Lonsdale A., Davis M., Lamont P., Clayton J.S., Laing N.G. STRetch: detecting and discovering pathogenic short tandem repeat expansions. Genome Biol. 2018;19:121. doi: 10.1186/s13059-018-1505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumm N., Sudmant P.H., Ko A., O’Roak B.J., Malig M., Coe B.P., Quinlan A.R., Nickerson D.A., Eichler E.E., NHLBI Exome Sequencing Project Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Spada A.R., Taylor J.P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen R.S., Gartler S.M., Scott C.R., Chen S.H., Laird C.D. Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum. Mol. Genet. 1992;1:571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- 15.Nancarrow J.K., Holman K., Mangelsdorf M., Hori T., Denton M., Sutherland G.R., Richards R.I. Molecular basis of p(CCG)n repeat instability at the FRA16A fragile site locus. Hum. Mol. Genet. 1995;4:367–372. doi: 10.1093/hmg/4.3.367. [DOI] [PubMed] [Google Scholar]
- 16.Nancarrow J.K., Kremer E., Holman K., Eyre H., Doggett N.A., Le Paslier D., Callen D.F., Sutherland G.R., Richards R.I. Implications of FRA16A structure for the mechanism of chromosomal fragile site genesis. Science. 1994;264:1938–1941. doi: 10.1126/science.8009225. [DOI] [PubMed] [Google Scholar]
- 17.Ehtesham N., Cantor R.M., King L.M., Reinker K., Powell B.R., Shanske A., Unger S., Rimoin D.L., Cohn D.H. Evidence that Smith-McCort dysplasia and Dyggve-Melchior-Clausen dysplasia are allelic disorders that result from mutations in a gene on chromosome 18q12. Am. J. Hum. Genet. 2002;71:947–951. doi: 10.1086/342669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannes F.D., Sharp A.J., Mefford H.C., de Ravel T., Ruivenkamp C.A., Breuning M.H., Fryns J.P., Devriendt K., Van Buggenhout G., Vogels A. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J. Med. Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinzen E.L., Radtke R.A., Urban T.J., Cavalleri G.L., Depondt C., Need A.C., Walley N.M., Nicoletti P., Ge D., Catarino C.B. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am. J. Hum. Genet. 2010;86:707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ullmann R., Turner G., Kirchhoff M., Chen W., Tonge B., Rosenberg C., Field M., Vianna-Morgante A.M., Christie L., Krepischi-Santos A.C. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum. Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 22.Lalioti M.D., Scott H.S., Antonarakis S.E. Altered spacing of promoter elements due to the dodecamer repeat expansion contributes to reduced expression of the cystatin B gene in EPM1. Hum. Mol. Genet. 1999;8:1791–1798. doi: 10.1093/hmg/8.9.1791. [DOI] [PubMed] [Google Scholar]
- 23.Favaro F.P., Alvizi L., Zechi-Ceide R.M., Bertola D., Felix T.M., de Souza J., Raskin S., Twigg S.R., Weiner A.M., Armas P. A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am. J. Hum. Genet. 2014;94:120–128. doi: 10.1016/j.ajhg.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg Y.P., Kremer B., Andrew S.E., Theilmann J., Graham R.K., Squitieri F., Telenius H., Adam S., Sajoo A., Starr E. Molecular analysis of new mutations for Huntington’s disease: intermediate alleles and sex of origin effects. Nat. Genet. 1993;5:174–179. doi: 10.1038/ng1093-174. [DOI] [PubMed] [Google Scholar]
- 25.Brisson D., Tremblay M., Prévost C., Laberge C., Puymirat J., Mathieu J. Sibship stability of genotype and phenotype in myotonic dystrophy. Clin. Genet. 2002;62:220–225. doi: 10.1034/j.1399-0004.2002.620306.x. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauer N.N., Popp B., Schoeller E., Schuhmann S., Heath K.E., Hisado-Oliva A., Klinger P., Kraus C., Trautmann U., Zenker M. Clinical relevance of systematic phenotyping and exome sequencing in patients with short stature. Genet. Med. 2018;20:630–638. doi: 10.1038/gim.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.