Abstract
The use of whole-exome and whole-genome sequencing has been a catalyst for a genotype-first approach to diagnostics. Under this paradigm, we have implemented systematic sequencing of neonates and young children with a suspected genetic disorder. Here, we report on two families with recessive mutations in NCAPG2 and overlapping clinical phenotypes that include severe neurodevelopmental defects, failure to thrive, ocular abnormalities, and defects in urogenital and limb morphogenesis. NCAPG2 encodes a member of the condensin II complex, necessary for the condensation of chromosomes prior to cell division. Consistent with a causal role for NCAPG2, we found abnormal chromosome condensation, augmented anaphase chromatin-bridge formation, and micronuclei in daughter cells of proband skin fibroblasts. To test the functional relevance of the discovered variants, we generated an ncapg2 zebrafish model. Morphants displayed clinically relevant phenotypes, such as renal anomalies, microcephaly, and concomitant increases in apoptosis and altered mitotic progression. These could be rescued by wild-type but not mutant human NCAPG2 mRNA and were recapitulated in CRISPR-Cas9 F0 mutants. Finally, we noted that the individual with a complex urogenital defect also harbored a heterozygous NPHP1 deletion, a common contributor to nephronophthisis. To test whether sensitization at the NPHP1 locus might contribute to a more severe renal phenotype, we co-suppressed nphp1 and ncapg2, which resulted in significantly more dysplastic renal tubules in zebrafish larvae. Together, our data suggest that impaired function of NCAPG2 results in a severe condensinopathy, and they highlight the potential utility of examining candidate pathogenic lesions beyond the primary disease locus.
Keywords: chromosome condensation, microcephaly, NPHP1, renal cyst, zebrafish, genetic interaction, NCAPH2, NCAPD3, micronuclei, cell cycle delay
Introduction
Neurodevelopmental disorders (NDDs) are a clinically and genetically heterogeneous group of pediatric conditions hallmarked by structural and/or functional abnormalities of the central nervous system (CNS). These disorders can manifest neurological phenotypes such as learning, speech, memory, and mobility deficits1 and can also be coincident with syndromic phenotypes affecting multiple organ systems. Although the term NDDs encompasses a broad range that can include either behavioral or neuroanatomical defects with diverse molecular underpinnings, the term can be useful for clinical assessment and management of affected children2 and has guided the genomics-based approach for the discovery of a host of causal loci.3 However, despite a marked acceleration in the rate of identifying the genetic basis of NDDs, >50% of cases assessed on any genetic platform, including whole-genome sequencing (WGS), whole-exome sequencing (WES), and copy number variant (CNV) analysis, still await a molecular diagnosis.4, 5, 6, 7, 8, 9 Considering that functional testing of candidate alleles adds value to WES and WGS, we have established a paradigm in which young individuals with suspected genetic defects are offered a WES and functional analysis as a means of expediting a diagnosis.10
Altered cell division has emerged as a frequent contributor to a subset of NDDs in which primary microcephaly is a predominant feature.11 In most eukaryotes, including humans, cell division is facilitated by two condensin complexes, condensin I and condensin II, each of which is composed of five subunits.12 Both complexes contain two proteins from the family of proteins responsible for the structural maintenance of chromosomes (encoded by SMC2 [MIM: 605576] and SMC4 [MIM: 605575]) and three unique non-SMC subunits. In humans, the non-SMC subunits of condensin I are encoded by NCAPD2 (MIM: 615638), NCAPG (MIM: 606280), and NCAPH (MIM: 602332); the respective counterparts in the condensin II complex are encoded by NCAPD3 (MIM: 609276), NCAPG2 (MIM: 608532), and NCAPH2 (MIM: 611230).13, 14 Condensin I and II perform distinct functions in the cell cycle and differ in their cellular localization and spatio-temporal interaction with chromosomes. Condensin II is localized typically to the nucleus and binds chromatin during early condensation in prophase, whereas condensin I localizes to the cytoplasm and interacts with chromosomes only after the nuclear membrane is dissolved.14 Not surprisingly, all reported mutant models of condensin-complex genes display embryonic lethality and/or neuroanatomical defects (Table S1). Recently, mutations in NCAPD2, NCAPD3, and NCAPH were reported in humans with microcephaly, growth retardation, and mild intellectual disability (MCPH21 [MIM: 617983], MCPH22 [MIM: 617984], and MCPH23 [MIM: 617985], respectively).15
Here, we report the identification of NCAPG2 variants that segregate under a recessive paradigm in two unrelated pediatric individuals with overlapping neurodevelopmental syndromic features. We used trio-based WES to identify rare NCAPG2 missense changes, and we established relevance to clinical phenotype and variant pathogenicity by using primary fibroblast cell lines, as well as zebrafish models. We also noted a marked difference in renal and urogenital phenotypes between the two affected individuals; this difference was coincident with a heterozygous NPHP1 (MIM: 607100) deletion, a known pathogenic allele for recessive nephronophthisis (NPHP [MIM: 256100]). To test for genetic interaction between NCAPG2 and NPHP1, we co-suppressed ncapg2 and nphp1 and phenotyped the pronephri of zebrafish larvae. We found significant structural renal defects, suggesting a role for these two trans alleles in the development of that pathology.
Material and Methods
Human Subjects, Ethics Approval, and Sample Ascertainment
We obtained peripheral blood, saliva, DNA, or skin biopsies from research participants subsequent to informed consent. All research on human subjects was performed according to protocols approved by the Duke University Medical Center institutional review board. We extracted DNA from blood (Gentra PureGene, Qiagen) or saliva (OG500 DNA collection kit, Oragene) according to the manufacturer’s instructions. We established primary skin fibroblast cultures at the Duke Clinical Cytogenetics Laboratory according to standard procedures.
Karyotype Analysis
Chromosome analysis was performed on 2 ml of heparinized peripheral blood that was set in culture and harvested by a standard synchronized method. Cells were fixed with 3:1 methanol/acetic acid, and GTW bands were visualized with Wright’s stain at 550–650 band resolution (n = 20 cells). Karyotyping was done with Applied Spectral Imaging.
WES and Variant Filtration in Family 1
We performed trio-based WES as described.10 For paired-end pre-capture library generation, we fragmented DNA by sonication and ligation to Illumina multiplexing PE adapters. We amplified adapter-ligated DNA by using PCR with barcoded primers. For exome capture, we enriched the library by hybridizing to biotin-labeled VCRome 2.116 in-solution Exome Probes at 47°C for 64–72 hr. The post-capture library DNA was analyzed on an Illumina Hiseq platform, which produced 100 base pair (bp) paired-end reads. The output data were converted to FastQ files in CASAVA 1.8 and mapped in BWA. We achieved a mean coverage of 126× for the target region (Table S2). We used Atlas-SNP and Atlas-indel to perform variant calling.17 We filtered data so that we would retain functional variants (predicted to alter mRNA splicing or amino acid sequences) that had a minor-allele frequency (MAF) of ≤1% in public SNP databases (dbSNP, 1000 Genomes); the National Heart, Lung and Blood Institute (NHLBI) Exome Sequencing project (n = 6,500 exomes); and the Atherosclerosis Risk in Communities cohort (ARIC; n = 2,300 exomes). We confirmed variants visually by using the Integrated Genomics Viewer (IGV, Broad Institute). We filtered IGV-confirmed variants further by comparing them to data from the Exome Aggregation Consortium (ExAC) and genome Aggregation Database (gnomAD) to ensure a MAF of <1%. For the remaining list of variants (Table S3), bidirectional Sanger sequencing was conducted on an ABI 3730 with BigDye Terminator chemistry (Applied Biosystems).
WES and Variant Filtration in Family 2
Trio-based WES was performed in a clinical setting (XomeDx; GeneDx). We used the Agilent SureSelectXT2 All Exon V4 Kit (Agilent Technologies) for library generation. Targeted regions were sequenced on an Illumina HiSeq 2000, resulting in 100 bp paired-end reads. The DNA sequence was mapped to the reference genome (UCSC hg19). We obtained a mean coverage of 83× and mapped 97.8% of targets at a coverage of 10×. Xome Analyzer evaluated sequence changes in the family 2 proband compared to the parents. WES data were re-analyzed in Enlis Genome Research software (Enlis, LLC) with a goal of retaining functional variants with a MAF <1%. Candidate variants were confirmed visually with IGV and experimentally validated with bidirectional Sanger sequencing in DNA samples from all available family members (Table S3).
NCAPG2 Immunoblotting
We established transformed lymphocyte cell lines (LCLs) from the proband of family 1 and a matched control according to standard procedures.18 Protein was harvested with RIPA lysis buffer (50 mM HEPES [pH 7.6], 1% Triton X-100, 0.1% SDS, 50 mM NaCl, 0.5% sodium deoxycholate, 1 mM NaF, and 0.02 mM DTT) and a protease inhibitor cocktail (Sigma Aldrich). We quantified protein with a BCA assay (ThermoFisher Scientific), and 50 μg/lane was separated by SDS-PAGE (4%–15%, Bio-Rad), followed by transfer to a polyvinylidene difluoride (PVDF) membrane. We incubated membranes overnight at 4°C in antibody solution (PBS; 3% BSA; rabbit anti-CAP-G2 antibody [Bethyl Laboratories, A300-605A-T; 1:1000 dilution]); glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control (Santa Cruz Biotechnology, SC-47727; 1:2000 dilution). Secondary detection was performed with antibody solution (PBS; 3% BSA, anti-rabbit IgG-HRP conjugate [A0318; 1:5000 dilution]). Immunoblots were developed via SuperSignal West Pico PLUS chemiluminescent substrate (ThermoFisher Scientific) according to the manufacturer’s instructions, and a ChemiDoc Imaging System (Bio-Rad) was used for chemiluminescence detection.
Cellular Assays of Primary Skin Fibroblasts
We cultured primary dermal fibroblasts in DMEM (Sigma Aldrich), 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin. We synchronized cells by reducing serum content to 0.1% for 24 hr, then by stimulating the cells for 24 hr with 10% FBS.19 We performed the analyses noted below on cells passaged three times (matched for proband and control cells).
Immunofluorescent Staining and Micronuclei Quantification
Fibroblasts were grown on coverslips and fixed in 4% paraformaldehyde (PFA) in PHEM (25 mM HEPES-NaOH at pH 6.8, 100 mM EGTA, 60 mM PIPES, 2 mM MgCl2) for 10 min. After three washes of PBS (>15 min/wash), cells were permeabilized in 0.3% Triton X-100 in PHEM for 10 min. Cells were blocked in 5% FBS in PBS for 1 hr at room temperature, followed by o/n incubation in anti-NCAPH2 antibody (Bethyl Laboratories; A302-276A-T, 1 μg/ ml) at 4°C. We washed cells 3–5 times in PBS (>15 min/wash), followed by secondary detection (anti-Rabbit IgG, Alexa Fluor 594; Invitrogen, 1:500 dilution). The coverslips were mounted in VECTASHIELD with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Labs) and visualized on a Zeiss Axio Imager Z2 microscope; images were acquired with an Apotome.2 system and Zen Pro (Zeiss). We quantified micronuclei and calculated statistical differences by using a χ2 test (n = 3513–6946 nuclei per condition), repeated with the investigator masked to the experiment.
In Situ Apoptosis Detection (TUNEL)
We used the Apotag Red In Situ apoptosis detection kit (Millipore). Fibroblasts were fixed in 4% paraformaldehyde in PHEM (25 mM HEPES-NaOH at pH 6.8, 100 mM EGTA, 60 mM PIPES, and 2 mM MgCl2) and post-fixed in prechilled ethanol:acetic acid (2:1) at −20°C. The samples were incubated in equilibration buffer, then incubated in TdT Enzyme at 370 C, and then washed in stop/wash buffer. Next, the samples on coverslips were treated with anti-digoxigenin conjugate (rhodamine) and the coverslips were mounted in mounting media with DAPI. Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)-positive nuclei were visualized on a Zeiss Axio Imager Z2 microscope, and images were acquired with an Apotome.2 system and Zen Pro software (Zeiss). Statistical comparisons were performed via a χ2 test (n = 2,504–4,794 nuclei per condition), repeated with the investigator masked to the experimental condition.
Intrinsic Chromosome Structure Assay
We performed the intrinsic chromosome structure (ICS) assay as described.15, 20, 21 We coated coverslips with poly-L-lysine and seeded cells in 6-well plates, then treated them for 12 hr with 75 ng/ml nocodazole and incubated them in 75 mM KCl buffer for 5 min; this was followed by a TEEN-RSB (TEEN buffer [0.5 mM triethanolamine-HCl [pH 8.5]; 0.1 mM Na EDTA; and 12.5 mM NaCl] for 30 min followed by RSB [10 mM Tris-HCl (pH 7.4); 210 mM NaCl; and 5 mM MgCl2] for 10 min). We fixed cells for 30 min with methanol and acetic acid (3:1 ratio). The coverslips were mounted in DAPI. Nuclei were visualized on a Zeiss Axio Imager Z2 Microscope (100×/1.4 Plan-Apochromat oil differential interference contrast lens [420780-02]; numerical aperture, 1.40), and images were acquired with Apotome.2 (Zeiss) and Zen Pro software (Zeiss). We scored n = 53–115 nuclei/condition and repeated the scoring with the investigator masked to experimental condition.
Flow Cytometry
We quantified DNA content with a propidium iodide (PI) flow-cytometry kit (Abcam) according to the manufacturer’s instructions. In brief, primary fibroblasts were fixed o/n at 4°C in prechilled 66% ethanol at ∼70% confluency. Cells were rehydrated in PBS and then incubated at 37°C in PI solution in the dark (PI + RNase staining solution). The solution containing the cell aggregates was passed through a 30 μm cell strainer prior to analysis. We acquired fluorescence-activated cell sorting (FACS) data with a FACSCalibur (Becton Dickinson) instrument and FACSDiva software (Becton Dickinson).
Molecular Cloning of NCAPG2 Plasmids and In Vitro Transcription
We obtained a full-length wild-type (WT) open-reading frame NCAPG2 (GenBank: NM_017760.6) construct (GeneArt, ThermoFisher) and cloned it into the pCS2+ vector via LRII clonase-mediated recombination (ThermoFisher). We generated mutant constructs by site-directed mutagenesis as described;22 amplification reactions were performed with 2× Phusion PCR Master Mix (ThermoFisher). The sequences of WT and mutant constructs were confirmed by standard methodology on an Applied Biosystems 3730 DNA sequencer. For rescue experiments, we generated WT and mutant mRNAs with a mMessage mMachine SP6 transcription kit (ThermoFisher) and used linearized pCS2+ constructs as a template.
Reagents for Transient Suppression and CRISPR-Cas9 Genome Editing in Zebrafish
We identified a single ncapg2 ortholog (Ensembl: ENSDART00000148390.3, GRCz10; 45% similarity versus human). We designed and injected a splice blocking (sb) morpholino (MO) that targeted the exon 8-intron 8 boundary of ncapg2. At 2 days post-fertilization (dpf), embryos were harvested in Trizol (Invitrogen), and RNA was extracted. cDNA was synthesized with the QuantiTect Reverse Transcription Kit (Qiagen) and used as a template for RT-PCR and subsequent sequencing. CRISPR single-guide RNA (sgRNA) targets were identified with ChopChop2.23, 24 We synthesized two sgRNAs (sgRNA1 and sgRNA2) with the GeneArt Precision gRNA Synthesis Kit (ThermoFisher).25 For each injection, 100 pg of sgRNA and 200 pg of Cas9 (PNA Bio, CP01) were injected into the cell of 1-cell stage embryos. We determined sgRNA targeting efficiency by extracting DNA from individual F0 embryos at 2 dpf by proteinase K digestion (Life Technologies, AM2548). The targeted site of each sgRNA was amplified by PCR, denatured, and reannealed slowly. Heteroduplexes were detected by polyacrylamide gel electrophoresis on a 15% gel26 (n = 6 embryos tested/condition), followed by TOPO-TA cloning of PCR amplicons and sequencing to estimate mosaicism (n = 3 embryos/sgRNA, 24 clones each).
Zebrafish Embryo Injections
All zebrafish experiments were approved by the Duke Institutional Animal Care and Use Committee (IACUC). To determine MO efficiency and titration, we injected 1 nL of MO into the yolk of 1- to 4-cell-stage embryos (1, 3, and 5 ng/embryo; 42–50 embryos/injection). For complementation experiments, we injected 1 nL of cocktail (3 ng MO and/or 150 pg of mRNA) into the yolk of 1- to 4-cell stage embryos. For CRISPR-Cas9 experiments, we injected 1 nL of CRISPR cocktail (100 pg sgRNA and 200 pg Cas9 protein) into the cell at the 1-cell stage. For ncapg2-nphp1 genetic interaction experiments, we injected 1 nL of injection solution containing 1 ng ncapg2 and/or 0.5 ng nphp1 sb-MO.27 The embryos were maintained in fresh embryo media (0.3 g/L NaCl, 75 mg/L CaSO4, 37.5 mg/L NaHCO3, and 0.003% methylene blue) at 28°C. The larvae were phenotyped for apoptosis and cell proliferation at 2 dpf, head size and brain structures at 3 dpf, and renal structures at 4 dpf. All injections and phenotyping studies were performed with the investigator masked to experimental conditions.
Zebrafish Phenotyping
Head Size Phenotyping
We anesthetized larvae with tricaine and acquired bright-field dorsal images from a Nikon SMZ745 microscope and Nikon NIS Elements Software. We assessed head size with ImageJ (NIH) by measuring the anterior-to-posterior length of the head from the mouth to the pectoral-fin attachment.
Whole-Mount TUNEL Assay
We used TUNEL to detect apoptosis in zebrafish whole mounts.28 In brief, embryos at 2 dpf were dechorionated and fixed in 4% PFA at 4°C overnight, then transferred to 100% methanol at −20°C for 2 hr. After rehydration in PBS, embryos were permeabilized with proteinase K (10 μg/mL) and post-fixed with 4% PFA; they were next incubated in equilibration buffer and then with TdT enzyme, followed by anti-digoxigenin provided in the ApopTag Red In Situ Apoptosis Detection Kit (Millipore).
Mitotic Progression Assay
We dechorionated embryos at 2 dpf and fixed them in Dent's solution overnight at 4°C; they were then rehydrated in PBS with a stepwise reduced concentration of methanol. Embryos were then bleached as described,29 post-fixed with 4% PFA, permeabilized with proteinase-K, washed twice in IF buffer (1% BSA, 0.1% Tween-20 in 1× PBS), and incubated overnight with anti-p-histone H3 (PH3; 1:500, Santa Cruz Biotechnology, sc-8656-R). After two washes in IF buffer, embryos were placed in secondary antibody solution containing Alexa Fluor 488 goat anti-rabbit IgG (1:500; ThermoFisher) in blocking solution for 1 hr at room temperature.
CNS Phenotyping
Larvae were anesthetized and fixed in Dent’s solution at 3 dpf. Standard whole-mount immunostaining was then performed30 by staining with anti-α-acetylated tubulin (T7451, mouse, Sigma-Aldrich, dilution 1:1000) and Alexa Fluor 488 goat anti-mouse IgG (ThermoFisher, dilution 1:500).
Renal Phenotyping
Larvae at 4 dpf were anesthetized with tricaine and fixed overnight in Dent’s solution, then stained with anti-Na+-K+ ATPase α-1 subunit (α6F; DSHB, 1:20 dilution) and Alexa Fluor rabbit anti-mouse IgG (ThermoFisher, 1:500 dilution) as a secondary antibody according to established methods.31
Fluorescence Microscopy of Zebrafish Larvae
We acquired z-stacked images of the fluorescent signal with a Nikon AZ100 Microscope equipped with a Nikon digital-sight black-and-white camera and Nikon NIS Elements software (TUNEL, PH3, α-acetylated tubulin—dorsal; Na+-K+ ATPase α-1—lateral). We quantified TUNEL and PH3 staining by counting positive cells in a defined region of the head in ImageJ software (NIH). We measured head size in ImageJ (NIH) software as described.30 We also measured the diameter of the renal tubule at a consistent position immediately posterior to the proximal convolution in ImageJ software (NIH) as described.32
Statistical Analysis
For the TUNEL, ICS, and micronuclei assays, we used a χ2 test to perform pairwise statistical comparisons across experimental conditions. For quantitative zebrafish phenotyping assays, we performed pairwise comparisons by using a Student’s t-test; for qualitative phenotyping assays, we performed pairwise comparisons by using a Fisher’s exact test (GraphPad Prism).
Transcriptomic Analysis of ncapg2 F0 Mutants
We extracted total RNA from the zebrafish embryo heads from five independent embryo pools per condition (gRNA alone; gRNA + Cas9) at 2 dpf with Trizol (ThermoFisher); quality and concentration were assessed on a 2100 Bioanalyzer (Agilent) and Qubit 2.0 (ThermoFisher), respectively. Only extracts with RNA integrity number (RIN) > 7 were processed for sequencing. RNAseq libraries were prepared with the Stranded mRNA-Seq Kit (KAPA) according to the manufacturer’s protocol. In brief, mRNA transcripts were captured with magnetic oligo-dT beads, fragmented with heat and magnesium, and reverse-transcribed with random primers. Illumina sequencing adapters were ligated to dscDNA fragments and amplified to produce RNA-seq libraries. Libraries were dual indexed; fragment length distribution and QC were assessed on a 2100 Bioanalyzer with the High-Sensitivity DNA Kit (Agilent Technologies). All libraries were pooled in an equimolar ratio (5 libraries/lane) and sequenced on an Illumina HiSeq 4000 flow cell to yield ∼66 million 50 bp single-end sequences per sample. Data were demultiplexed and FastQ files generated via Bcl2Fastq (Illumina). RNAseq data were processed with the TrimGalore toolkit, which employs Cutadapt33 to trim low-quality bases and sequencing adapters from the 3′ ends of the reads. Only reads >20 nt after trimming were kept. Reads were mapped to the Zv10r87 version of the zebrafish genome and transcriptome34 with the STAR RNA-seq alignment tool.35 Gene counts were compiled with the HTSeq tool. Only genes that had >10 reads were used in subsequent analysis. Normalization and differential expression were carried out with the DESeq236 Bioconductor37 package with the R statistical programming environment. We calculated the false-discovery rate to control for multiple hypothesis testing and performed gene-set enrichment analysis38 to identify gene ontology terms associated with altered gene expression.
Results
Clinical Ascertainment of Unrelated Children with a Complex Congenital Syndrome
The proband of family 1 presented with a host of congenital anomalies (Figure 1A, Table 1, Supplemental Material and Methods). She was born at 42 weeks via normal spontaneous vaginal delivery to European American parents: a 37-year-old G3P3 mother and a 35-year old father with no known contributory family history or consanguinity. A prenatal ultrasound at 41 weeks revealed multiple fetal anomalies, including frontal bossing, a two-vessel cord, renal findings interpreted as a possible cyst or hydronephrosis, and dilated cerebral ventricles. Some of her growth parameters were notable but not extreme; she had a birth weight, length, and head circumference of 3,463 g (55th percentile), 48 cm (30th percentile), and 33.5 cm (22nd percentile), respectively. A physical examination at birth showed severe hypotonia, bilateral postaxial polydactyly on the feet, a sacral dimple, a weak cry, a weak suck, and a poorly coordinated swallow. During the first month of life, she developed febrile pyelonephritis and was diagnosed with bilateral grade IV vesicoureteral reflux.
Figure 1.
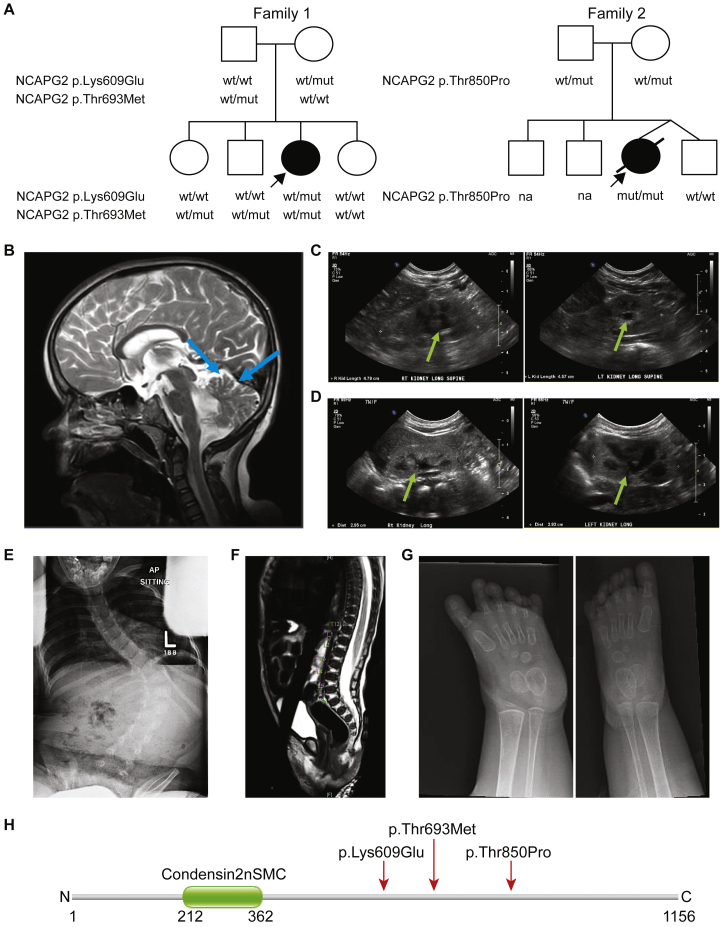
Two Unrelated Pedigrees with Complex Syndromic Features Harbor Recessive NCAPG2 Mutations
(A) Pedigrees of two families harboring NCAPG2 missense mutations. Family 1 has one affected female with compound-heterozygous missense NCAPG2 mutations, c.1825A>G (p.Lys609Glu) and c.2078C>T (p.Thr693Met); three healthy siblings are either wild-type (WT) or heterozygous for the mutations (mut). An affected female from an unrelated pedigree (family 2) inherited a homozygous missense NCAPG2 mutation, c.2548A>C (p.Thr850Pro), from carrier parents. The healthy dizygotic twin sibling of the proband is homozygous for the WT allele. Affected individuals (shaded symbols) are denoted with an arrow in each pedigree; NA = sample not available.
(B) Brain MRI without contrast (family 1 proband, 9 years of age) indicates cerebellar vermian hypoplasia (blue arrows).
(C) A renal ultrasound (US; family 1 proband, 1 year of age) shows bilateral grade I hydronephrosis (green arrows); right kidney, 4.8 cm; left kidney, 4.6 cm.
(D) A renal US (family 2 proband, 2 months of age) shows bilateral grade II hydronephrosis (green arrows); right kidney, 3.0 cm; left kidney, 2.9 cm.
(E) A radiograph (family 1 proband, 5 years of age) indicates thoracolumbar S-shaped scoliosis. L = left.
(F) A radiograph (family 2 proband, 1 month of age) shows a low-lying conus medullaris terminating at the bottom of L3; there is no evidence of a thickened filum terminale or spinal dysraphism.
(G) Radiographs (family 1 proband, 19 months of age) show bilateral polydactyly of the feet. There are 6 digits in each foot and each has a proximal, middle, and distal phalanx.
(H) Schematic of the NCAPG2 protein (GenBank: NP_060230.5) showing domain structure and localization of mutations identified in affected individuals (indicated by red arrow). Numbers indicate amino acid position; N = amino-terminus; C = carboxyl-terminus.
Table 1.
Clinical Features of the Probands of Families 1 and 2
| Family 1 (DM074) | Family 2 (DM516) |
|---|---|
| Age/Sex | |
| 11 years/female | 4 months (deceased)/female |
| Ethnicity | |
| European American | Mexican |
| NCAPG2 Mutation (GenBank:NM_017760.6) | |
| c.1825A>G (p.Lys609Glu) heterozygous-maternal | c.2548A>C (p.Thr850Pro) homozygous-paternal/maternal |
| c.2078C>T (p.Thr693Met) heterozygous-paternal | |
| Head and neck phenotypes | |
| microcephaly (HP: 0000252) | microcephaly (HP: 0000252) |
| frontal bossing (HP: 0002007) | micrognathia (HP: 0000347) |
| delayed closure of anterior fontanelle (HP: 0001476) | arched eyebrows (HP: 0002553) |
| short nose (HP: 0003196) | |
| triangular mouth (HP: 0000207) | |
| Central Nervous System (Structure and Function) | |
| cerebellar vermis hypoplasia (HP: 0001320) | hypertonia (HP: 0001276) |
| colpocephaly (HP:0030048) | low-lying conus medullaris (HP: 0002143) |
| history of dilated cerebral ventricles (HP: 0002119) | |
| history of tethered cord (HP: 0002144) | |
| intellectual disability, moderate (HP: 0002342) | |
| non-verbal (HP: 0001344) | |
| developmental delay (HP: 0001263) | |
| hypotonia (HP: 0001290) | |
| Urogenital Abnormalities | |
| history of hydronephrosis (HP: 0000126) | hydronephrosis (HP: 0000126) |
| small right kidney (HP: 0012583) | small kidneys (HP: 0000089) |
| renal cysts (HP: 0000107) | |
| history of bilateral vesicoureteral reflux (HP: 0000076 | |
| ureteral duplication (HP: 0000073) | |
| absent clitoris (HP: 0040255) | |
| Ocular Abnormalities | |
| vision loss (HP: 0000572) | vision loss (HP: 0000572) |
| history of strabismus (HP: 0000486) | Peters anomaly (HP: 0000659) |
| history of nystagmus (HP: 0000639) | glaucoma (HP: 0000501) |
| pigmentary retinopathy (HP: 0000580) | buphthalmos (HP: 0000557) |
| history of epiblepharon (HP: 0011225) | |
| corneal scarring (HP: 0000559) | |
| history of eye-lash inversion (HP: 0001128) | |
| Skeletal and Digit Abnormalities | |
| bilateral post axial polydactyly on feet (HP: 0100259) | clinodactyly, fifth finger, right hand (HP: 0030084) |
| history of scoliosis (HP: 0002650) | absent fourth and fifth digits, left foot; absent fifth digit, right foot (HP: 0006209) |
| mild pectus excavatum (HP: 0000767) | contractures of left arm and leg (HP: 0001371) |
| Growth Defects | |
| short stature (HP: 0004322) | Intrauterine growth restriction (HP: 0001511) |
| small for gestational age (HP: 0001518) | |
| failure to thrive (HP: 0001508) | |
| Other Phenotypes | |
| sacral dimple (HP: 0000960) | sacral dimple (HP: 0000960) |
| sensorineural hearing impairment (HP: 0000407) | patent ductus arteriosus after premature birth (HP: 0011649) |
| sleep apnea (HP: 0010535) | patent foramen ovale (HP: 0001655) |
| anus malposition (HP: 0004397) | tricuspid regurgitation (HP: 0005180) |
| eczema (HP: 0000964) | heart murmur (HP: 0030148) |
| bilateral superior vena cava with no bridging vein (HP: 0011668), | |
| anemia (HP: 0001903) | |
| lymphopenia (HP: 0001888) | |
| neutropenia (HP: 0001875) | |
| oral-pharyngeal dysphagia (HP: 0200136) | |
Human Phenotype Ontology (HPO) terms are indicated in parentheses; HP = human phenotype
Her syndromic presentation at birth, coupled with failure to reach multiple developmental milestones during childhood, led to extensive clinical evaluation (Figures 1B, 1C, 1E, and 1G). Brain MRI conducted at 6 months of age raised a concern for Joubert syndrome (JBTS [MIM: 213300]) on the basis of a possible molar-tooth sign, but repeat MRI at 22 months of age did not support these initial conclusions. However, subsequent brain imaging revealed mild volume loss, colpocephaly, and cerebellar vermian hypoplasia (Figure 1B). At her latest clinical assessment (11 years of age), she had moderate intellectual disability, was non-verbal, and used limited sign language. Additionally, the proband has been monitored for renal structure and function since birth; a renal ultrasound at 12 months of age revealed bilateral grade I hydronephrosis (Figure 1C), and her follow-up imaging at 8 years of age continued to show hydronephrosis and bilateral vesicoureteral reflux. However, her last renal ultrasound at 10 years of age was normal except for a small right kidney (5.9 cm; average normal size for age is 8.5 cm). Furthermore, she has multiple ocular anomalies, including strabismus, nystagmus, epiblepharon, and pigmentary retinopathy. She had surgical repair of left thoracolumbar scoliosis at 7 years of age (Figure 1E). She also has sensorineural hearing impairment, sleep apnea, and a short stature. The proband has three siblings who do not display any aspects of this complex phenotype (Figure 1A).
We conducted multiple clinical genetic tests, but we were unable to identify a likely cause. Analysis of cells from whole peripheral blood showed a 46,XX karyotype with normal band length and morphology (Figure S1). Chromosomal microarray (CMA) revealed a paternally inherited interstitial deletion of ∼153.7 kb on 2q13 (minimally deleted segment, chr2: 110833630–110987359 [hg19]; CytoScan HD, Affymetrix), which encompasses NPHP1, a frequent cause of juvenile nephronopthisis, Senior-Loken syndrome (SLS [MIM: 266900]), and Joubert syndrome,39, 40, 41, 42 but high-resolution CNV analysis by PCR and mutational screening of the coding exons and splice junctions at the NPHP1 locus were negative for bi-allelic deletions or point mutations. Furthermore, clinical bidirectional sequencing of three additional candidate genes that cause Joubert syndrome43, 44, 45 (AH11 [MIM: 608894], CEP290 [MIM: 610142], and TMEM67 [MIM: 609884]), Rett syndrome (RTT [MIM: 312750]: MECP2 [MIM: 300005]),46 or Robinow syndrome ([MIM: 268310] ROR2 [MIM: 602337])47 were also negative (Supplemental Material and Methods). We could not exclude the possibility that these approaches might have missed deep intronic variants in any of these phenotype-relevant candidate genes.
Independently, we examined the proband of family 2, who was born prematurely at 34 weeks to Mexican parents: a 39-year-old G3P4 mother and a 33-year-old father (Figure 1A, Table 1, Supplemental Material and Methods). The pregnancy was complicated by advanced maternal age (AMA), twin gestation, and severe intrauterine growth restriction (IUGR) of the proband, or twin “B.” The delivery was performed via C-section due to the severe IUGR of twin “B” and the breech presentation of twin “A.” The proband’s birth weight, length, and head circumference were 840 g (<1st percentile), 34 cm (<1st percentile), and 25 cm (<1st percentile), respectively. We noted multiple congenital anomalies at birth; these included microcephaly, facial dysmorphisms, digit abnormalities (postaxial absent toes and clinodactyly of the 5th finger), ocular phenotypes (Peters anomaly, bilateral glaucoma, and buphthalmos of the left eye), contractures, neonatal hypertonia, and a sacral dimple accompanied by a low-lying conus medullaris but not a tethered cord (Figure 1F). Her neonatal course was complicated by prematurity, anemia, neutropenia, lymphopenia, failure to thrive, and cardiovascular defects.
Imaging studies of internal organs added further information about the extent of the family 2 proband’s syndrome. The newborn underwent echocardiography and demonstrated a large patent ductus arteriosus, a large patent foramen ovale, and a mild tricuspid regurgitation. Follow-up imaging revealed a bilateral superior vena cava with an absent bridging vein but did not show a patent ductus arteriosus. Although she was microcephalic, brain MRI at one month of age showed a morphologically normal appearance of the brain. A renal ultrasound at two days of life revealed small but otherwise normal kidneys. At 6 weeks of age, the proband developed bilateral grade 2 hydronephrosis (Figure 1D), which resolved after 2 months. Barium swallow imaging revealed oropharyngeal dysphasia, and at the age of 3 months, a gastric tube was placed. The proband was discharged as stable after 4 months. However, she passed away four days later due to unknown causes. She is survived by three siblings, including her twin brother, who has heart murmur and reactive airway disease. The other two siblings are reported to have no congenital malformations.
Standard clinical testing of the family 2 proband was not diagnostic. Fluorescence in situ hybridization (FISH) was normal. CMA revealed several independent regions showing absence of heterozygosity (AOH), encompassing ∼ 3% of the genome. Finally, sequencing and deletion testing of the mitochondrial genome was also negative.
Whole-Exome Sequencing Identifies Rare Recessive Mutations in NCAPG2
Given that diagnostic genetic testing yielded no likely genetic-driver loci for the family 1 proband and that the family fulfilled our enrollment criteria, we offered participation in the Task Force for Neonatal Genomics (TFNG). Under this paradigm, we employed trio-based WES. We generated sequencing reads on an Illumina platform to a mean coverage of 126× with ∼94% of bases covered ≥20× for the proband and both parents (Table S2). We then filtered the WES data for rare (MAF <1%), nonsynonymous coding or splice-site-disruptive variants segregating under autosomal-recessive or -dominant de novo inheritance patterns. Variants fulfilling these criteria were present in three genes (GLRA3 [MIM: 600421], TTN [MIM: 188840], and NCAPG2); Sanger sequencing confirmed segregation with disease in the pedigree (Figure 1A, Table S3).
We identified a de novo variant in glycine receptor, alpha 3 subunit (GLRA3; GenBank: NM_006529.3: c.76G>A [p.Val26Ile]). This variant is present in heterozygosity in four individuals in gnomAD (Table S3). Additionally, the Val26 codon is polymorphic: gnomAD reports rare changes to each of Phe, Ala, and Leu. GLRA3 encodes a subunit of a glycine receptor,48 ablation of which in mice results in either reduced pain sensitization or compromised auditory nerve activity, but no gross morphological abnormalities.49, 50 We likewise evaluated the missense titin (TTN; GenBank: NM_133378.4) variants c.9073C>G (p.Leu3025Val) and c.44105G>T (p.Ser14702Ile) and decided that they are unlikely to drive pathology; both have been found in healthy controls (Table S3), and although TTN plays established roles in cardiomyopathy phenotypes,51, 52, 53, 54, 55 the features of cardiomyopathy are non-overlapping with the proband’s phenotype.
The third candidate consisted of rare, compound-heterozygous, nonsynonymous changes in non-SMC condensin II complex subunit G2 (NCAPG2; GenBank: NM_017760.6): c.1825A>G (p.Lys609Glu) and c.2078C>T (p.Thr693Met). The first of these is absent from public databases, and the latter is present in seven heterozygous individuals in gnomAD (Table S3). NCAPG2 is a member of the condensin II complex, which plays essential roles in mitotic chromosome assembly and segregation.56 Ncapg2−/− mouse embryos exhibit impaired inner-cell-mass expansion and increased apoptosis, and they die shortly after implantation (Table S1).57 NCAPG2 mutations have not been implicated in human phenotypes in isolation. However, heterozygous multi-gene deletions encompassing NCAPG2 on chr7 and MCPH1 on chr8 have been reported in a case of severe microcephaly and intellectual disability.58
We used GeneMatcher59 to identify family 2. Independent of, and concurrent with, our TFNG work, trio-based WES was performed in a clinical laboratory. The only variant reported was a homozygous NCAPG2 change (GenBank: NM_017760.6: c.2548A>C [p.Thr850Pro]) that was absent from public databases (Table S3). Enrollment of family 2 in our research protocol and reanalysis of the WES data identified additional compound-heterozygous mutations in two genes; carboxypeptidase Z (CPZ [MIM: 603105]: GenBank: NM_003652.3; c.1165A>G [p.Lys389Glu] and c.1694G>A [p.Arg565His]) and family with sequence similarity 184 member A (FAM184A: GenBank: NM_024581.5, c.3319A>G [p.Lys1107Glu] and c.80C>A [p.Ala27Glu]). All four variants are present in public databases, but three of them are reported in homozygosity in healthy control individuals in gnomAD (Table S3), diminishing their candidacy. In sum, we considered NCAPG2 variants to be the most likely molecular drivers of disease because: (1) rare variants segregated with disease in both pedigrees (Figure 1A); (2) both cases displayed overlapping clinical features (Table 1); and (3) mutations in condensin-complex genes have been implicated in microcephaly in humans15 and other model systems (Table S1).
Primary Cells Harboring NCAPG2 Mutations Display Aberrant Chromosomal Condensation, Cell-Cycle Defects, and Increased Apoptosis
Although cell lines were not available from the proband of family 2, we established LCLs from the family 1 proband and detected NCAPG2 in whole-cell protein lysates by immunoblotting; we observed no differences in protein abundance or size between the family 1 proband and control, indicating that protein stability was most likely unaltered (Figure S2). We then asked whether the integrity of the condensin II complex was destabilized and whether this destabilization had potentially resulted in mislocalization of direct binding partners. Subunit-subunit interaction studies have shown previously that NCAPG2 interacts directly with the C-terminal region of NCAPH2.60 We therefore performed NCAPH2 immunostaining of primary skin fibroblasts from the family 1 proband and a matched healthy control. However, we observed the expected nuclear localization of this non-SMC condensin II subunit (Figure S3).
Previous studies have shown that mutations in condensin II components can impair chromosome structural integrity during cell division and thereby lead to increased apoptosis.15 We performed TUNEL staining to monitor cell death in primary fibroblasts from the family 1 proband, and we observed a 6-fold increase in cell death in cultures from the index individual compared to the control (p < 0.0001; n = 2,504–4,794 nuclei per condition, replicated; Figures 2A and 2B). To determine whether these enhanced apoptotic events could be attributed to chromosomal defects seen in other condensin II mutants, we evaluated mitotic chromosome structures via an intrinsic chromosome structure (ICS) assay, which subjects mitotic chromosomes to repeated unfolding and refolding, followed by visualization with DAPI. Chromosomes from the mother of family 1 and a healthy control could recover to the expected condensed morphology. In contrast, 30% of nuclei from the proband displayed either an intermediate phenotype (lost lateral compaction) or severe appearance (totally disorganized; p < 0.0001; n = 53–115 per batch, replicated; Figures 2C and 2D).
Figure 2.
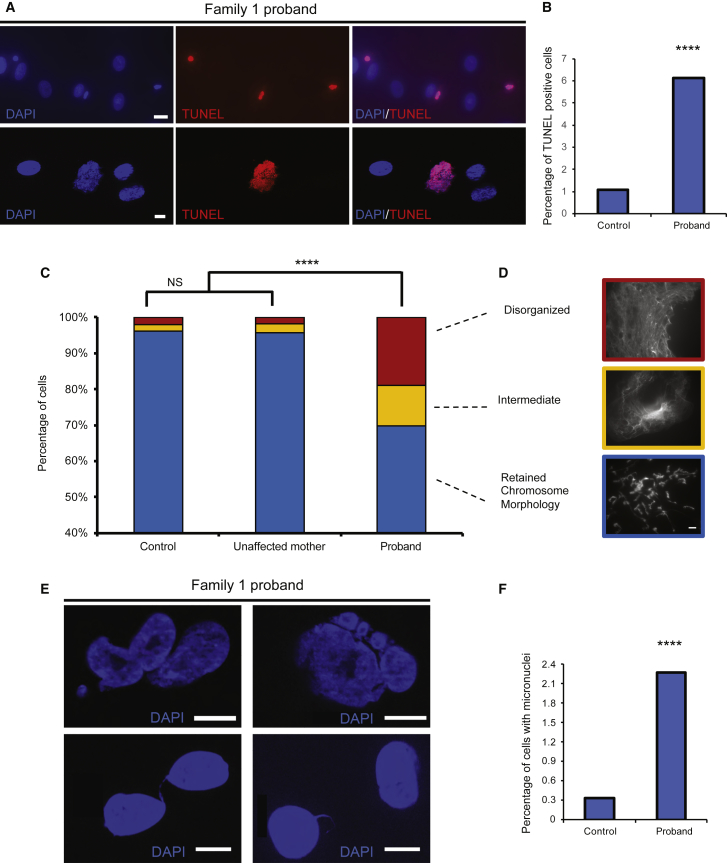
Primary Fibroblasts from the Family 1 Proband Display Augmented Apoptosis, Possibly Due to Mitotic Chromosome Condensation Defects
(A) Representative epifluorescent images of TUNEL positive nuclei; DAPI = blue; TUNEL positive nuclei = red. Top: scale bar, 20 μm (40x objective). Bottom: scale bar, 10 μm (63x objective).
(B) Qualitative scoring of TUNEL positive nuclei in control and proband-derived fibroblasts; n = 2,504–4,794 nuclei per condition, repeated.
(C) Qualitative scoring of primary skin fibroblast cells subjected to the ICS assay. DAPI-stained nuclei originating from an ethnically matched unrelated healthy control, the unaffected mother, and the proband of family 1 were scored according to objective criteria. Blue = retained chromosome morphology, yellow = lost lateral compaction, and red = were totally disorganized; n = 53–115 nuclei per condition, repeated. Statistical comparisons were performed with a χ2 test; NS = not significant
(D) Epifluorescent images of mitotic chromosomes representative of different qualitative categories; the scale bar represents 10 μm.
(E) Representative epifluorescent images of DAPI stained nuclei with displaying various abnormalities subsequent to aberrant chromosome condensation; these include micronuclei (top), lagging chromatin and chromatin bridges (bottom); scale bar, 10 μm.
(F) Qualitative scoring of micronuclei in control and proband derived fibroblasts stained with DAPI; n=3513-6946 nuclei per condition, repeated. Statistical comparisons were performed using a χ2 test; ∗∗∗∗, p < 0.0001.
Previous studies have also shown an abundance of micronuclei in condensin II mutant cells as a result of chromosome instability, and concomitant cell-cycle delay or arrest and DNA damage.15, 61 Systematic blind scoring of fixed and DAPI-stained primary fibroblasts from the index individual and a matched control showed a 7-fold increase of cells with micronuclei (p < 0.0001; n = 3,513–6,946 nuclei per condition, replicated; Figures 2E and 2F). We also observed macroscopic defects, including lagging chromatin fragments and chromatin bridges, in cells from the index individual, and we did not observe these in the control (2/15 versus 0/15 high-exposure DAPI fields for the index individual versus the control; Figure 2E). These cytopathologies were consistent with cell-cycle studies of the same primary fibroblast cell lines that were stained with propidium iodide and subjected to FACS to reveal increased G2/M arrest in cells from the family 1 proband (Table S4). Together, these observations support the hypothesis that deleterious missense mutations in NCAPG2 can lead to a structural abnormality that occurs in mitotic chromosomes and most likely leads to segregation defects, genome instability, and increased cell death.
Suppression and CRISPR-Cas9-Based Targeting of ncapg2 Results in Microcephaly in Zebrafish
We next sought in vivo functional evidence to test the candidacy of NCAPG2 in disease. First, we focused on microcephaly, a feature shared by the probands from families 1 and 2 and a phenotype shown to be experimentally tractable in zebrafish.62, 63, 64, 65 We identified the sole NCAPG2 ortholog in the zebrafish genome (46% identity, 64% similarity; Figure S4A). Consistent with human expression data (GTEx; Human Protein Atlas), ncapg2 is expressed nearly ubiquitously, and predominant expression is in the developing brain and spinal cord of larval zebrafish.66 We designed an sb MO that targeted the exon 8-intron 8 junction, and we established by RT-PCR that it induces exon 8 skipping, resulting in a frameshift and premature termination codon (Figure S4A and S4B and Table S5). As a proxy for microcephaly, we assessed the head size of zebrafish larvae at 3 dpf by measuring the anterior-to-posterior length of the head in dorsal bright-field images. We found a dose-dependent, significant reduction in head size for ncapg2 morphants in comparison to controls (p < 0.0001; Figures 3A, 3B, and S4C); this defect could be ameliorated with the co-injection of WT human NCAPG2 mRNA (p < 0.0001 versus MO, n = 45–71 larvae/batch, repeated; Figures 3A, 3B, and S5); expression of WT human NCAPG2 mRNA did not result in neuroanatomical or gross morphological defects.
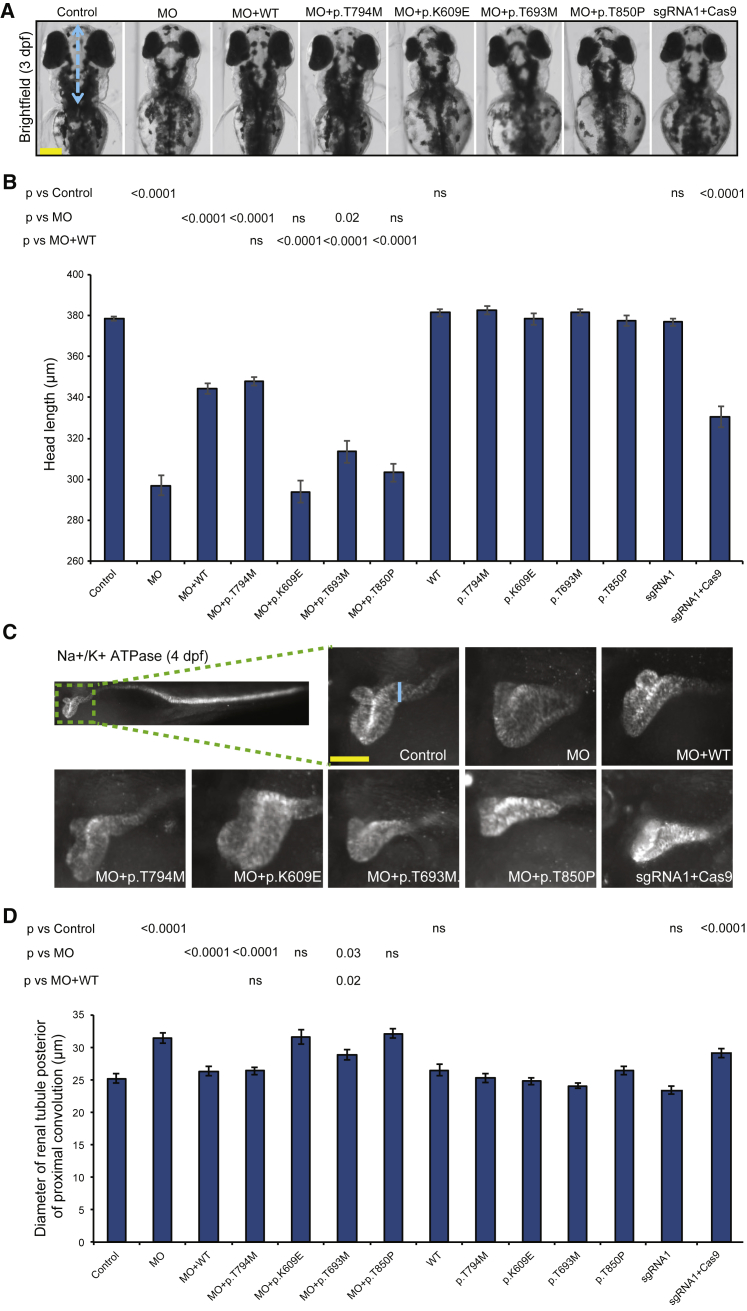
Figure 3.
Morpholino-Induced Suppression and CRISPR-Cas9 Mutation of ncapg2 Results in Neuroanatomical and Renal Phenotypes Relevant to Human Pathology
(A) Representative bright-field dorsal images of zebrafish larvae at 3 days post-fertilization (dpf) show a reduction in head size (length from the pectoral fin attachment to the mouth, blue dotted line) in morphants (MO) and CRISPR-Cas9 F0 mutants (sgRNA1 + Cas9). The scale bar (yellow line) represents 100 μm.
(B) Quantification of head-size measurements acquired for larvae at 3 dpf demonstrates pathogenicity of variants identified in affected individuals. NCAPG2 mRNA harboring a common variant was included so that specificity could be demonstrated (dbSNP: rs10248318: c.2381C>T [p.Thr794Met]; allele frequency 0.2% and present in five homozygous individuals in gnomAD). (n = 32–71 larvae/condition, repeated).
(C) Representative lateral images of zebrafish larvae stained with Na+-K+ ATPase α-1 subunit antibody at 4 dpf. Larvae display an increase in the diameter of the renal tubule posterior to the proximal convolution (blue line) in morphants and CRISPR-Cas9 F0 mutants. The scale bar (yellow line) (represents 50 μm.
(D) Quantification of renal-tubule measurements is consistent with assay results from head-size measurements (n = 26–50 larvae/condition, repeated). p values calculated with a Student’s t-test are shown on top of the bar charts; ns = not significant; error bars represent standard error of the mean (SEM).
To test the specificity of transient ncapg2-suppression phenotypes in zebrafish, and to complement our observations with an independent method, we targeted the ncapg2 locus by using CRISPR-Cas9 genome editing. We designed two sgRNAs (sgRNA1 and sgRNA2, targeted to exons 5 and 3, respectively [Figure S6A and Table S5]) and co-injected each along with Cas9 into 1-cell zebrafish embryos. Through heteroduplex analysis, cloning, and sequencing we found an average of 70% (sgRNA1) and 75% (sgRNA2) mosaicism in F0 mutants (Figures S6B–S6E). As was the case in morphants, there was a significant reduction in the anterior structures of ncapg2 CRISPR F0 mutants compared to either control larvae or larval batches injected with equivalent doses of sgRNA alone (p < 0.0001, n = 58–80 larvae/batch, repeated; Figures 3A, 3B, and S6F).
To evaluate further the neuroanatomical structures of ncapg2 morphants and F0 mutants, we immunostained zebrafish larvae at 3 dpf with anti-α-acetylated tubulin. Consistent with the head-size measurements, we saw a significant reduction in size of the optic tecta (p < 0.0001, n = 82–92 larvae; Figures S5 and S7A–S7C) and cerebellar hypoplasia in ncapg2 morphants (p < 0.0001, n = 41–92 larvae, repeated; Figure S7A), both of which were rescued by co-injection of MO with WT NCAPG2 mRNA (p < 0.0001, n = 92 larvae, repeated; Figures S7A, S7B, and S7D). Importantly, we reproduced both phenotypes in ncapg2 CRISPR F0 mutants (p < 0.0001 versus control for optic tecta, Figures S7A and S7C; and p = 0.007 versus control for cerebellar hypoplasia, n = 27–80 larvae/batch, Figures S7A and S7D).
Next, we employed in vivo complementation22 to test the pathogenicity of missense NCAPG2 variants. We co-injected MOs with mRNA encoding c.1825A>G (p.Lys609Glu), c.2078C>T (p.Thr693Met), and c.2548A>C (p.Thr850Pro) or another eight missense variants present in at least one homozygous individual in ExAC (Figure S8A) and compared rescue efficiency to that of WT NCAPG2 mRNA by using the head-size readout. Variants associated with affected individuals did not rescue the head-size abnormality and conferred either a severe effect (not significantly different from MO; MO + p.Lys609Glu and MO + p.Thr850Pro, n = 45–60 larvae/batch, repeated) or an intermediate effect (p = 0.02 and < 0.0001, MO + p.Thr693Met versus MO and MO + WT, respectively, n = 45–51 larvae/batch, repeated; Figures 3A and 3B). In contrast, all eight NCAPG2 mRNAs harboring homozygous, nonsynonymous changes mined from ExAC rescued the head size similarly to WT NCAPG2 (p < 0.0001 versus MO, n = 31–51 larvae/batch, repeated), supporting the specificity of our assays. Injection of mutant or negative control NCAPG2 mRNA alone resulted in phenotypes that were indistinguishable from those of controls.
ncapg2 Disruption Results in Concomitant Cell Death and Aberrant Mitotic Progression In Vivo
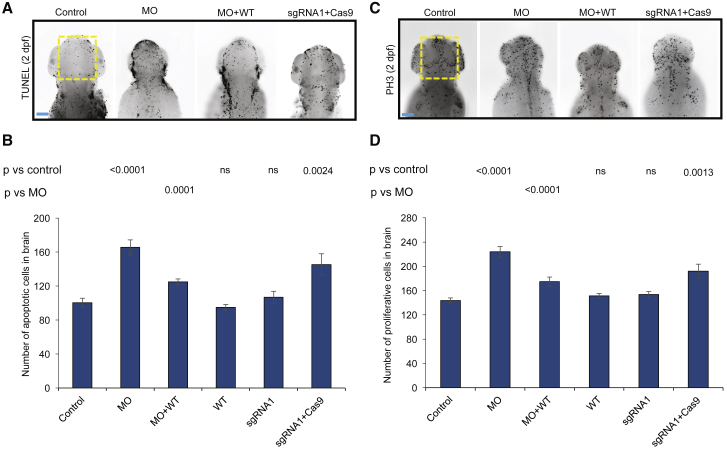
Work in model organisms has shown apoptosis or dysregulated cell proliferation to be pathognomonic of neuroanatomical defects.10, 62, 63 Additionally, mice lacking Ncapg2 show embryonic lethality between embryonic day (E) 8.5 and E10.5 as a result of increased apoptosis in blastocysts (Table S1).57 To investigate the underlying cellular basis of head-size reduction in ncapg2 zebrafish models, we performed TUNEL and PH3 staining. We found a significant increase in cell death in ncapg2 morphants (p < 0.0001 versus control, n = 25–29 larvae/batch, repeated; Figures 4A, 4B, and S5); this increase was rescued significantly by co-injection of MO with WT NCAPG2 mRNA (p < 0.0001, n = 21–29 larvae/batch; Figures 4A, 4B, andS5). TUNEL staining of ncapg2 CRISPR F0 mutants was consistent with that of morphants (p = 0.002 versus control, n = 30–35 larvae/batch, repeated; Figures 4A, 4B, and S5). Furthermore, ncapg2 F0 mutants or morphants showed a significant increase in cell proliferation in a defined region of the head (p < 0.001 versus control, n = 29–35 larvae/batch, repeated; Figures 4C, 4D, and S5). To reconcile the augmented PH3 staining with coincident increases in apoptosis, we speculate that ncapg2 suppression might result in mitotic arrest that could activate the apoptosis machinery and result in increased cell death, consistent with our observations from primary cells (Table S4).
Figure 4.
Suppression and Mutation of ncapg2 Results in Apoptosis and Altered Mitotic Progression in Zebrafish Larvae
(A) Representative dorsal images of zebrafish larvae marked by terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) display increased apoptosis in the forebrain-midbrain region (yellow dotted rectangle) of morphants (MO) and ncapg2 F0 mutants (sgRNA1 + Cas9) at 2 dpf. WT = wild-type; the scale bar (blue line) represents 100 μm
(B) Quantification of TUNEL-stained cells shows significantly increased apoptosis in morphants (MO) and F0 mutants (sgRNA1 + Cas9) compared to controls. Co-injection of MO with WT NCAPG2 mRNA (MO + WT) significantly rescues apoptotic-cell counts. n = 21–35 larvae/condition, repeated.
(C) Representative dorsal images of zebrafish larvae stained at 2 dpf with phospho-histone H3 (PH3) antibody display aberrant cell-cycle progression as indicated by the G2/M cell-cycle transition marker in the forebrain-midbrain region (yellow dotted rectangle) of morphants (MO) and ncapg2 F0 mutants (sgRNA1 + Cas9). The scale bar (blue line) represents 100μm.
(D) Quantification of PH3-stained cells in anterior structures indicates a significantly altered cell-cycle progression in ncapg2 zebrafish models. Co-injection of MO with WT NCAPG2 mRNA (MO + WT) significantly rescues PH3-cell count defects. n = 29–35 larvae/condition, repeated. p values calculated with a Student’s t-test are given on top of the bar charts; ns = not significant; error bars represent standard error of the mean (SEM).
Transcriptome Profiling of ncapg2 F0 Mutants Supports a Role in Embryonic Development and Tissue Morphogenesis
Next, to investigate the global impact of NCAPG2 loss in an in vivo developmental context, we performed transcriptome profiling of zebrafish embryo heads isolated at 2 dpf (heads were injected with gRNA1 alone or gRNA1 + Cas9; 5 biological replicates). Global transcriptome perturbation was modest: of 23,011 unique transcripts detected, ncapg2 was the only coding transcript that met false discovery rate (FDR) thresholds of p < 0.05. However, gene-set enrichment analysis (GSEA) revealed multiple functional gene categories that were altered significantly in F0 mutants (n = 330 enhanced, n = 28 repressed; nominal p < 0.01). We were struck by the observation that the ten most significantly altered functional groups enhanced in F0 mutants (familywise error rate [FWER] p < 0.01) can be mapped back to clinical phenotypes of affected individuals; these are relevant to tissue morphogenesis (GO: 0005201, GO: 0005581, GO: 0001738), eye development (GO: 0048592, GO: 0048593, GO: 0002088, GO: 0048048), hindbrain development (GO: 0030902), and skeletal system development (GO: 0001501; Table S6). By contrast, we did not detect any gene sets that were repressed significantly and met false discovery rate (FDR) or FWER significance thresholds in ncapg2 F0 mutants versus gRNA1-injected batches.
Together, these data validate the relevance of our in vivo ncapg2 models and offer some insights into how condensin II alteration could give rise to syndromic pathophysiology.
Suppression of ncapg2 Results in Hypoplastic Renal Tubules in Zebrafish
Affected individuals in families 1 and 2 displayed renal defects, including hydronephrosis, or a reduction in renal size (Figures 1C and 1D and Table 1). As a second assay to support the relevance of NCAPG2 to this rare congenital syndrome, we employed a renal phenotyping strategy that can assess congenital anomalies of the kidney and urinary tract.25, 31, 67, 68 We ablated ncapg2 function in zebrafish by using either MO-induced suppression or CRISPR-Cas9, immunostained renal tubules at 4 dpf with anti-Na+-K+ ATPase α-1 subunit, and assessed the proximal convoluted tubule. We detected several structural defects in morphants or F0 mutants; such defects included renal aplasia, hypoplasia, and enlargement of the proximal renal convolution tubule (Figure 3C). To quantify these observations, we measured the diameter of the renal tubule posterior to the proximal convolution.32 In ncapg2 morphants, there was a significant increase in renal-tubule diameter compared to that of controls (p < 0.0001, n = 43–50 larvae/batch, repeated); this increase was rescued by co-injection of WT or negative control mRNA (p < 0.0001 versus MO, n = 40–50 larvae/batch, repeated; Figures 3C, 3D, and S5). Injection of sgRNA1 or sgRNA2 with Cas9 recapitulated these observations; we saw a significant increase in the diameter of the renal tubule posterior to the proximal convolution in ncapg2 CRISPR F0 mutants compared to either control larvae or larval batches injected with sgRNA alone (p < 0.0001, n = 30–45 larvae/batch, repeated; Figures 3C, 3D, S5, and S6G). Next, we repeated the in vivo complementation assay by using the renal phenotyping readout. Similar to our neurodevelopmental assays, p.Lys609Glu and p.Thr850Pro behaved as severe pathogenic alleles, and p.Thr693Met is most likely a hypomorph (n = 36–50 larvae/batch, repeated; Figures 3C, 3D, and S5). The control (c.2381C>T [p. Thr794Met], MAF of 2% in African Americans, five homozygotes present in gnomAD]) rescued renal dysplasia similar to NCAPG2 WT, but expression of mutant or negative control NCAPG2 did not show renal defects.
ncapg2 Dysfunction with Concomitant Reduction of nphp1 Exacerbates Renal Defects
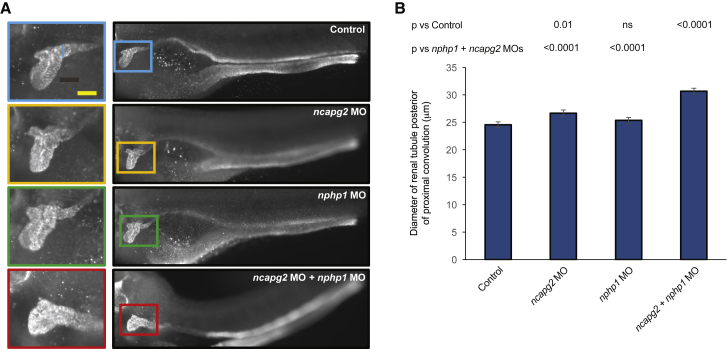
Comparison of the clinical presentation of the two affected children shows substantial overlap in neurological, ocular, digit, and growth phenotypes. However, we noted more complex urogenital features in the proband from family 1 (Table 1 and Supplemental Material and Methods). We hypothesized that this could be due to either (1) stochastic effects or (2) genetic interaction between NCAPG2 and other loci critical for renal development. We noted the presence of a known pathogenic allele in NPHP1 and wondered whether haploinsufficiency at this locus, which by itself is not sufficient to cause disease, might be a contributory factor. Transient gene-suppression models in zebrafish offer a tractable method of assessing genetic interaction in vivo; we have shown that such phenomena can be captured in a renal context, and for nphp1 in particular.27, 32, 69 Therefore, to test an NCAPG2-NPHP1 genetic-interaction hypothesis, we injected a range of MO doses for each gene into embryos and determined a dose at which the diameter of the renal tubule posterior to the convolution was nominally distinguishable from that of controls (p = ≥ 0.01; Figures 5A, 5B, and S5). Next, we co-injected these two reagents and compared the renal phenotypes of larval batches to either controls or single ncapg2 or nphp1 morphant batches. For the double MO versus each single MO comparison, we found a significant exacerbation of the diameter of the renal tubule (p < 0.0001; n = 35–52 per batch, repeated; Figures 5A, 5B, and S5). These observations suggest that a genetic interaction between NPHP1 and NCAPG2 might contribute to the discordant renal phenotypes.
Figure 5.
Genetic Interaction Studies Indicate that Co-suppression of ncapg2 and nphp1 Results in Exacerbation of the Renal Phenotype
(A) Representative images of zebrafish larvae immunostained with Na+-K+ ATPase α-1 subunit antibody at 4 dpf. Right, lateral images of zebrafish pronephric tubules; left, the proximal convolution tubule is magnified. The blue line on the top left image indicates the diameter. The scale bar (yellow line) represents 50 μm.
(B) Quantification of the diameter of the renal tubule positioned just posterior to the proximal convolution (μm); n = 35–52 larvae/batch, repeated; p values were calculated with a Student’s t-test; ns = not significant; error bars represent standard error of the mean (SEM).
Discussion
Here, we report on two unrelated children with a complex congenital syndrome and pathogenic missense NCAPG2 variants that segregate in an autosomal-recessive paradigm. We deployed an array of investigations that culminated in (1) the identification, through GeneMatcher, of a second family with mutations in the same locus and overlapping clinical features, (2) in vitro analysis of proband fibroblasts wherein multiple predicted condensin-II-defective phenotypes21, 47 could be observed, (3) transcriptomic data from ncapg2 zebrafish mutants that intimated perturbation of genes involved in tissue morphogenesis, and (4) in vivo complementation studies in transient suppressant and genetic mosaic ncapg2 zebrafish models, where all discovered variants behaved as pathogenic alleles. Together, these lines of evidence suggest that impaired function at the NCAPG2 locus gives rise to a severe pathology in humans.
Mutations in condensin-complex genes are rare, an observation potentially explained by the critical function of these proteins in chromosome condensation and cell division (Table S1). The only other condensin II component reported, NCAPD3,15 produced a phenotype that was largely restricted to neurological development and growth. By contrast, the clinical features of the individuals with NCAPG2 mutations extend beyond the central nervous system and growth to include ocular, urogenital, skeletal, and digit anomalies (Table 1). Furthermore, mutations in condensin I components NCAPH and NCAPD2 result in phenotypes also confined to growth impairment and neuroanatomical and neurodevelopmental defects. We note that the combination of ICS assay results, augmented micronuclei, persistent chromatin bridges after cytokinesis, and increased cell death in proband fibroblasts are indicators of genome instability in a modest but significant number of cells. However, we do not have direct evidence that the condensin II complex is disrupted in the affected individual in our study, as shown for other condensin II mutants and null cells.20, 70 Characterizing such cellular phenotypes with live-cell imaging and more thoroughly analyzing cells at discrete mitotic stages will require further work. Additionally, although karyotype analysis showed normal band length and morphology, we cannot exclude the possibility of a subtle alteration in chromosome width and length; establishment of such defects might likewise require additional studies.
We also noted the clinical differences between the two affected individuals with NCAPG2 mutations. The family 1 proband, although affected with a syndromic condition, is currently in her second decade of life. By contrast, the family 2 proband presented with more extensive congenital anomalies and was deceased during infancy. One possible explanation is allelism at NCAPG2: family 1 harbors both a mild allele (p.Thr693Met; significantly worse than WT rescue, but improved compared to MO alone in the zebrafish assay) and a severe allele (p.Lys609Glu; not significantly different from MO alone) that together might produce a milder overall clinical presentation in comparison to family 2, which harbors a homozygous severe allele (p.Thr850Pro). However, we must also consider other factors, including alleles in trans. We found it curious that the individual with noted renal pathology also harbored a bona fide heterozygous nephronophthisis allele. We fully concede that this might be a coincidence. However, testing for genetic interaction between ncapg2 and nphp1 in vivo revealed this to be true for renal development, arguing that the observed genetic result might be relevant. Clearly, more examples of such genetic combinations must be observed and reported before we have sufficient evidence to reach robust conclusions, not least because we cannot (yet) measure the prior probability of any two genes or alleles interacting in vivo by chance alone. However, given the limited progress towards understanding differences between individuals who harbor the same primary mutation, and the absence of statistical power to dissect such phenomena at the backdrop of extensive rare genetic variation in human genomes, opportunities such as this should be noted. Indeed, we are encouraged by this and other recent findings71, 72, 73 to begin considering all known pathogenic alleles in the genome of affected individuals as a useful starting point for understanding phenotypic variability.
Consortia
The members of the Task Force for Neonatal Genomics Consortium are as follows: Alexander Allori, Misha Angrist, Patricia Ashley, Margarita Bidegain, Brita Boyd, Eileen Chambers, Heidi Cope, C. Michael Cotten, Theresa Curington, Erica E. Davis, Sarah Ellestad, Kimberley Fisher, Amanda French, William Gallentine, Ronald Goldberg, Kevin Hill, Sujay Kansagra, Nicholas Katsanis, Sara Katsanis, Joanne Kurtzberg, Jeffrey Marcus, Marie McDonald, Mohammed Mikati, Stephen Miller, Amy Murtha, Yezmin Perilla, Carolyn Pizoli, Todd Purves, Sherry Ross, Azita Sadeghpour, Edward Smith, and John Wiener.
Declaration of Interests
N.K. is a paid consultant for and holds significant stock in Rescindo Therapeutics. The other authors declare no competing interests.
Acknowledgments
We are grateful to these two families for their participation in our research. We also thank Mr. Z. Kupchinsky at the Duke Center for Human Disease Modeling for zebrafish husbandry; Ms. E. Phillips (Duke Clinical Cytogenetics Laboratory) for assisting with primary skin fibroblast cell culture; Drs. Y. Gao (Duke Light Microscopy Core), M. Cook (DCI Flow Cytometry Core), N. Devos (Duke Sequencing and Genomic Technologies Core), and D. Corcoran (Duke Genomic Analysis and Bioinformatics Core) for technical assistance; and Ms. M. Cho from GeneDx for facilitating investigator contact through GeneMatcher. This work was supported by grants from the US National Institutes of Health (DK096415, DK075972, and HD042601 to N.K.; DK072301 and DK096493 to N.K. and E.E.D.; and MH106826 to E.E.D.). K.K. was funded by an International Research Support Initiative Program fellowship from the Higher Education Commission of Pakistan. A full list of the Task Force for Neonatal Genomics consortium members and their affiliations is available in the Supplemental Material and Methods.
Published: December 27, 2018
Footnotes
Supplemental Data include Supplemental Material and Methods, eight figures, and six tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.11.017.
Contributor Information
Erica E. Davis, Email: erica.davis@duke.edu.
Nicholas Katsanis, Email: nicholas.katsanis@duke.edu.
Task Force for Neonatal Genomics:
Alexander Allori, Misha Angrist, Patricia Ashley, Margarita Bidegain, Brita Boyd, Eileen Chambers, Heidi Cope, C. Michael Cotten, Theresa Curington, Erica E. Davis, Sarah Ellestad, Kimberley Fisher, Amanda French, William Gallentine, Ronald Goldberg, Kevin Hill, Sujay Kansagra, Nicholas Katsanis, Sara Katsanis, Joanne Kurtzberg, Jeffrey Marcus, Marie McDonald, Mohammed Mikati, Stephen Miller, Amy Murtha, Yezmin Perilla, Carolyn Pizoli, Todd Purves, Sherry Ross, Azita Sadeghpour, Edward Smith, and John Wiener
Accession Numbers
Our zebrafish transcriptomic data were deposited in the NCBI Gene Expression Omnibus under accession number GEO: GSE122932.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
ChopChop Software, http://chopchop.cbu.uib.no/
Ensembl, https://www.ensembl.org
ExAC Browser, http://exac.broadinstitute.org/
Exome Variant Server, http://evs.gs.washington.edu/EVS/
GeneMatcher, https://genematcher.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
Human Phenotype Ontology (HPO), https://hpo.jax.org/app/
Human Protein Atlas,https://www.proteinatlas.org/
ImageJ, https://imagej.nih.gov
NCBI BLAST, https://blast.ncbi.nlm.nih.gov
OMIM, http://www.omim.org/
R Statistical Programming Environment, http://www.r-project.org
TrimGalore Toolkit, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Hu W.F., Chahrour M.H., Walsh C.A. The diverse genetic landscape of neurodevelopmental disorders. Annu. Rev. Genomics Hum. Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thapar A., Cooper M., Rutter M. Neurodevelopmental disorders. Lancet Psychiatry. 2017;4:339–346. doi: 10.1016/S2215-0366(16)30376-5. [DOI] [PubMed] [Google Scholar]
- 3.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krumm N., O’Roak B.J., Shendure J., Eichler E.E. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S., Autism Sequencing Consortium Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evers C., Staufner C., Granzow M., Paramasivam N., Hinderhofer K., Kaufmann L., Fischer C., Thiel C., Opladen T., Kotzaeridou U. Impact of clinical exomes in neurodevelopmental and neurometabolic disorders. Mol. Genet. Metab. 2017;121:297–307. doi: 10.1016/j.ymgme.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Bowling K.M., Thompson M.L., Amaral M.D., Finnila C.R., Hiatt S.M., Engel K.L., Cochran J.N., Brothers K.B., East K.M., Gray D.E. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017;9:43. doi: 10.1186/s13073-017-0433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thevenon J., Duffourd Y., Masurel-Paulet A., Lefebvre M., Feillet F., El Chehadeh-Djebbar S., St-Onge J., Steinmetz A., Huet F., Chouchane M. Diagnostic odyssey in severe neurodevelopmental disorders: Toward clinical whole-exome sequencing as a first-line diagnostic test. Clin. Genet. 2016;89:700–707. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 9.Soden S.E., Saunders C.J., Willig L.K., Farrow E.G., Smith L.D., Petrikin J.E., LePichon J.B., Miller N.A., Thiffault I., Dinwiddie D.L. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan D.M., Frangakis S.G., Golzio C., Cassa C.A., Kurtzberg J., Davis E.E., Sunyaev S.R., Katsanis N., Task Force for Neonatal Genomics Identification of cis-suppression of human disease mutations by comparative genomics. Nature. 2015;524:225–229. doi: 10.1038/nature14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faheem M., Naseer M.I., Rasool M., Chaudhary A.G., Kumosani T.A., Ilyas A.M., Pushparaj P., Ahmed F., Algahtani H.A., Al-Qahtani M.H., Saleh Jamal H. Molecular genetics of human primary microcephaly: An overview. BMC Med. Genomics. 2015;8(Suppl 1):S4. doi: 10.1186/1755-8794-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano T. Condensins: Universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano T., Mitchison T.J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 14.Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- 15.Martin C.A., Murray J.E., Carroll P., Leitch A., Mackenzie K.J., Halachev M., Fetit A.E., Keith C., Bicknell L.S., Fluteau A., Deciphering Developmental Disorders Study Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 2016;30:2158–2172. doi: 10.1101/gad.286351.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis E.E., Savage J.H., Willer J.R., Jiang Y.H., Angrist M., Androutsopoulos A., Katsanis N. Whole exome sequencing and functional studies identify an intronic mutation in TRAPPC2 that causes SEDT. Clin. Genet. 2014;85:359–364. doi: 10.1111/cge.12189. [DOI] [PubMed] [Google Scholar]
- 19.Chen M., Huang J., Yang X., Liu B., Zhang W., Huang L., Deng F., Ma J., Bai Y., Lu R. Serum starvation induced cell cycle synchronization facilitates human somatic cells reprogramming. PLoS ONE. 2012;7:e28203. doi: 10.1371/journal.pone.0028203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green L.C., Kalitsis P., Chang T.M., Cipetic M., Kim J.H., Marshall O., Turnbull L., Whitchurch C.B., Vagnarelli P., Samejima K. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J. Cell Sci. 2012;125:1591–1604. doi: 10.1242/jcs.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson D.F., Vagnarelli P., Gassmann R., Earnshaw W.C. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 2003;5:323–336. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- 22.Niederriter A.R., Davis E.E., Golzio C., Oh E.C., Tsai I.C., Katsanis N. In vivo modeling of the morbid human genome using Danio rerio. J. Vis. Exp. 2013;78:e50338. doi: 10.3791/50338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44(W1):W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Küry S., Besnard T., Ebstein F., Khan T.N., Gambin T., Douglas J., Bacino C.A., Craigen W.J., Sanders S.J., Lehmann A. De novo disruption of the proteasome regulatory subunit PSMD12 causes a syndromic neurodevelopmental disorder. Am. J. Hum. Genet. 2017;100:352–363. doi: 10.1016/j.ajhg.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X., Xu Y., Yu S., Lu L., Ding M., Cheng J., Song G., Gao X., Yao L., Fan D. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 2014;4:6420. doi: 10.1038/srep06420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindstrand A., Davis E.E., Carvalho C.M., Pehlivan D., Willer J.R., Tsai I.C., Ramanathan S., Zuppan C., Sabo A., Muzny D. Recurrent CNVs and SNVs at the NPHP1 locus contribute pathogenic alleles to Bardet-Biedl syndrome. Am. J. Hum. Genet. 2014;94:745–754. doi: 10.1016/j.ajhg.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golzio C., Willer J., Talkowski M.E., Oh E.C., Taniguchi Y., Jacquemont S., Reymond A., Sun M., Sawa A., Gusella J.F. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thisse C., Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 30.Borck G., Hög F., Dentici M.L., Tan P.L., Sowada N., Medeira A., Gueneau L., Thiele H., Kousi M., Lepri F. BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies. Genome Res. 2015;25:155–166. doi: 10.1101/gr.176925.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanna-Cherchi S., Khan K., Westland R., Krithivasan P., Fievet L., Rasouly H.M., Ionita-Laza I., Capone V.P., Fasel D.A., Kiryluk K. Exome-wide association study identifies GREB1L mutations in congenital kidney malformations. Am. J. Hum. Genet. 2017;101:789–802. doi: 10.1016/j.ajhg.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindstrand A., Frangakis S., Carvalho C.M., Richardson E.B., McFadden K.A., Willer J.R., Pehlivan D., Liu P., Pediaditakis I.L., Sabo A. Copy-number variation contributes to the mutational load of Bardet-Biedl syndrome. Am. J. Hum. Genet. 2016;99:318–336. doi: 10.1016/j.ajhg.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinf. in action. 2011;17:10–12. [Google Scholar]
- 34.Kersey P.J., Staines D.M., Lawson D., Kulesha E., Derwent P., Humphrey J.C., Hughes D.S., Keenan S., Kerhornou A., Koscielny G. Ensembl Genomes: An integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–D97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 39.Parisi M.A., Bennett C.L., Eckert M.L., Dobyns W.B., Gleeson J.G., Shaw D.W.W., McDonald R., Eddy A., Chance P.F., Glass I.A. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am. J. Hum. Genet. 2004;75:82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caridi G., Murer L., Bellantuono R., Sorino P., Caringella D.A., Gusmano R., Ghiggeri G.M. Renal-retinal syndromes: Association of retinal anomalies and recessive nephronophthisis in patients with homozygous deletion of the NPH1 locus. Am. J. Kidney Dis. 1998;32:1059–1062. doi: 10.1016/s0272-6386(98)70083-6. [DOI] [PubMed] [Google Scholar]
- 41.Saunier S., Calado J., Heilig R., Silbermann F., Benessy F., Morin G., Konrad M., Broyer M., Gubler M.C., Weissenbach J., Antignac C. A novel gene that encodes a protein with a putative src homology 3 domain is a candidate gene for familial juvenile nephronophthisis. Hum. Mol. Genet. 1997;6:2317–2323. doi: 10.1093/hmg/6.13.2317. [DOI] [PubMed] [Google Scholar]
- 42.Castori M., Valente E.M., Donati M.A., Salvi S., Fazzi E., Procopio E., Galluccio T., Emma F., Dallapiccola B., Bertini E., Italian MTS Study Group NPHP1 gene deletion is a rare cause of Joubert syndrome related disorders. J. Med. Genet. 2005;42:e9. doi: 10.1136/jmg.2004.027375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 44.Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 45.Baala L., Romano S., Khaddour R., Saunier S., Smith U.M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 2007;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 47.Afzal A.R., Rajab A., Fenske C.D., Oldridge M., Elanko N., Ternes-Pereira E., Tüysüz B., Murday V.A., Patton M.A., Wilkie A.O., Jeffery S. Recessive Robinow syndrome, allelic to dominant brachydactyly type B, is caused by mutation of ROR2. Nat. Genet. 2000;25:419–422. doi: 10.1038/78107. [DOI] [PubMed] [Google Scholar]
- 48.Kingsmore S.F., Suh D., Seldin M.F. Genetic mapping of the glycine receptor alpha 3 subunit on mouse chromosome 8. Mamm Genome. 1994;5:831–832. doi: 10.1007/BF00292030. [DOI] [PubMed] [Google Scholar]
- 49.Harvey R.J., Depner U.B., Wässle H., Ahmadi S., Heindl C., Reinold H., Smart T.G., Harvey K., Schütz B., Abo-Salem O.M. GlyR alpha3: An essential target for spinal PGE2-mediated inflammatory pain sensitization. Science. 2004;304:884–887. doi: 10.1126/science.1094925. [DOI] [PubMed] [Google Scholar]
- 50.Dlugaiczyk J., Hecker D., Neubert C., Buerbank S., Campanelli D., Becker C.M., Betz H., Knipper M., Rüttiger L., Schick B. Loss of glycine receptors containing the α3 subunit compromises auditory nerve activity, but not outer hair cell function. Hear. Res. 2016;337:25–34. doi: 10.1016/j.heares.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Gerull B., Gramlich M., Atherton J., McNabb M., Trombitás K., Sasse-Klaassen S., Seidman J.G., Seidman C., Granzier H., Labeit S. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 52.Satoh M., Takahashi M., Sakamoto T., Hiroe M., Marumo F., Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: Identification of a novel disease gene. Biochem. Biophys. Res. Commun. 1999;262:411–417. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 53.Hackman P., Vihola A., Haravuori H., Marchand S., Sarparanta J., De Seze J., Labeit S., Witt C., Peltonen L., Richard I., Udd B. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am. J. Hum. Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lange S., Xiang F., Yakovenko A., Vihola A., Hackman P., Rostkova E., Kristensen J., Brandmeier B., Franzen G., Hedberg B. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 55.Carmignac V., Salih M.A., Quijano-Roy S., Marchand S., Al Rayess M.M., Mukhtar M.M., Urtizberea J.A., Labeit S., Guicheney P., Leturcq F. C-terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann. Neurol. 2007;61:340–351. doi: 10.1002/ana.21089. [DOI] [PubMed] [Google Scholar]
- 56.Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 57.Smith E.D., Xu Y., Tomson B.N., Leung C.G., Fujiwara Y., Orkin S.H., Crispino J.D. More than blood, a novel gene required for mammalian postimplantation development. Mol. Cell. Biol. 2004;24:1168–1173. doi: 10.1128/MCB.24.3.1168-1173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perche O., Menuet A., Marcos M., Liu L., Pâris A., Utami K.H., Kervran D., Cacheux V., Laudier B., Briault S. Combined deletion of two Condensin II system genes (NCAPG2 and MCPH1) in a case of severe microcephaly and mental deficiency. Eur. J. Med. Genet. 2013;56:635–641. doi: 10.1016/j.ejmg.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 59.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Onn I., Aono N., Hirano M., Hirano T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007;26:1024–1034. doi: 10.1038/sj.emboj.7601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blank M., Lerenthal Y., Mittelman L., Shiloh Y. Condensin I recruitment and uneven chromatin condensation precede mitotic cell death in response to DNA damage. J. Cell Biol. 2006;174:195–206. doi: 10.1083/jcb.200604022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stankiewicz P., Khan T.N., Szafranski P., Slattery L., Streff H., Vetrini F., Bernstein J.A., Brown C.W., Rosenfeld J.A., Rednam S., Deciphering Developmental Disorders Study Haploinsufficiency of the chromatin remodeler BPTF causes syndromic developmental and speech delay, postnatal microcephaly, and dysmorphic features. Am. J. Hum. Genet. 2017;101:503–515. doi: 10.1016/j.ajhg.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brooks S.S., Wall A.L., Golzio C., Reid D.W., Kondyles A., Willer J.R., Botti C., Nicchitta C.V., Katsanis N., Davis E.E. A novel ribosomopathy caused by dysfunction of RPL10 disrupts neurodevelopment and causes X-linked microcephaly in humans. Genetics. 2014;198:723–733. doi: 10.1534/genetics.114.168211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim H.T., Lee M.S., Choi J.H., Jung J.Y., Ahn D.G., Yeo S.Y., Choi D.K., Kim C.H. The microcephaly gene aspm is involved in brain development in zebrafish. Biochem. Biophys. Res. Commun. 2011;409:640–644. doi: 10.1016/j.bbrc.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 65.Nakayama T., Al-Maawali A., El-Quessny M., Rajab A., Khalil S., Stoler J.M., Tan W.H., Nasir R., Schmitz-Abe K., Hill R.S. Mutations in PYCR2, encoding pyrroline-5-carboxylate reductase 2, cause microcephaly and hypomyelination. Am. J. Hum. Genet. 2015;96:709–719. doi: 10.1016/j.ajhg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kudoh T., Tsang M., Hukriede N.A., Chen X., Dedekian M., Clarke C.J., Kiang A., Schultz S., Epstein J.A., Toyama R., Dawid I.B. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- 67.Davis E.E., Frangakis S., Katsanis N. Interpreting human genetic variation with in vivo zebrafish assays. Biochim. Biophys. Acta. 2014;1842:1960–1970. doi: 10.1016/j.bbadis.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santoriello C., Zon L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez-Rivera E., Liu Y.P., Verbitsky M., Anderson B.R., Capone V.P., Otto E.A., Yan Z., Mitrotti A., Martino J., Steers N.J. Genetic drivers of kidney defects in the DiGeorge syndrome. N. Engl. J. Med. 2017;376:742–754. doi: 10.1056/NEJMoa1609009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abe S., Nagasaka K., Hirayama Y., Kozuka-Hata H., Oyama M., Aoyagi Y., Obuse C., Hirota T. The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 2011;25:863–874. doi: 10.1101/gad.2016411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katsanis N. The continuum of causality in human genetic disorders. Genome Biol. 2016;17:233. doi: 10.1186/s13059-016-1107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beales P.L., Badano J.L., Ross A.J., Ansley S.J., Hoskins B.E., Kirsten B., Mein C.A., Froguel P., Scambler P.J., Lewis R.A. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am. J. Hum. Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Posey J.E., Harel T., Liu P., Rosenfeld J.A., James R.A., Coban Akdemir Z.H., Walkiewicz M., Bi W., Xiao R., Ding Y. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.