Abstract
Intestinal bacteria play an important role in bile acid metabolism and in the regulation of multiple host metabolic pathways (e.g., lipid and glucose homeostasis) through modulation of intestinal farnesoid X receptor (FXR) activity. Here, we examined the effect of berberine (BBR), a natural plant alkaloid, on intestinal bacteria using in vitro and in vivo models. In vivo, the metabolomic response and changes in mouse intestinal bacterial communities treated with BBR (100 mg/kg) for 5 days were assessed using NMR- and mass spectrometry–based metabolomics coupled with multivariate data analysis. Short-term BBR exposure altered intestinal bacteria by reducing Clostridium cluster XIVa and IV and their bile salt hydrolase (BSH) activity, which resulted in the accumulation of taurocholic acid (TCA). The accumulation of TCA was associated with activation of intestinal FXR, which can mediate bile acid, lipid, and glucose metabolism. In vitro, isolated mouse cecal bacteria were incubated with three doses of BBR (0.1, 1, and 10 mg/ml) for 4 hours in an anaerobic chamber. NMR-based metabolomics combined with flow cytometry was used to evaluate the direct physiologic and metabolic effect of BBR on the bacteria. In vitro, BBR exposure not only altered bacterial physiology but also changed bacterial community composition and function, especially reducing BSH-expressing bacteria like Clostridium spp. These data suggest that BBR directly affects bacteria to alter bile acid metabolism and activate FXR signaling. These data provide new insights into the link between intestinal bacteria, nuclear receptor signaling, and xenobiotics.
Introduction
In addition to their well established role in regulating the metabolism of vitamins, lipids, and other hydrophobic dietary constituents, bile acids activate nuclear receptors and membrane G protein–coupled receptors to mediate lipid, glucose, and energy homeostasis (Kawamata et al., 2003; Li and Chiang, 2013). Notably, the nuclear receptor, farnesoid X receptor (FXR), has gained increasing consideration as a druggable receptor (Lambert et al., 2003; Duran-Sandoval et al., 2004). FXR is a ligand-activated receptor expressed in many tissues, including the liver, intestinal epithelium, adipose tissue, kidney, pancreas, stomach, gall bladder, and macrophage (Forman et al., 1995). FXR activation can protect against obesity, diabetes, and fatty liver disease and can improve hyperlipidemia and hyperglycemia (Zhang et al., 2006; Ali et al., 2015; Arab et al., 2017). Recent studies also suggest that intestinal FXR agonists inhibit bacterial overgrowth and prevent bacterial translocation and epithelial deterioration (Inagaki et al., 2006). Paradoxically, similar beneficial effects toward the improvement of obesity, insulin resistance, and nonalcoholic fatty liver disease are also seen with FXR antagonists (Li et al., 2013; Jiang et al., 2015a,b). Thus, the contextual role and tissue-dependent mechanism for the beneficial effects of FXR is important.
More recent studies indicated that intestinal bacteria play a key role in obesity and related metabolic diseases, especially by mediating bile acid biosynthesis and FXR signaling (Sayin et al., 2013; Zhang et al., 2016). Previous evidence indicated that the intestinal bacteria regulate bile acid metabolism, thus resulting in FXR activation in the liver and gut (Ridlon et al., 2014). A recent study indicated a novel mechanism for the intestine-restricted FXR agonist fexaramine, which can activate intestinal FXR and Takeda G protein-coupled receptor 5 (TGR5)/glucagon-like peptide GLP-1 signaling through intestinal microbiota activity to protect against obesity and related metabolic disease (Pathak et al., 2018).
Berberine (BBR), a natural plant alkaloid, is the major pharmacologic component in the Chinese herbal medicine Coptis chinensis (Huang-Lian) (Tang et al., 2009). As a traditional Chinese medicine, BBR has been used to treat diarrhea in China (Chen et al., 2015). Accumulating evidence suggests that BBR can improve metabolic syndrome by regulating glucose and lipid metabolism and attenuating insulin resistance (Kong et al., 2004; Zhang et al., 2014; Xu et al., 2017). Since BBR has low bioavailability, modulation of gut bacteria might be one possible mechanism for its antidiabetic and antiobesity effects (Han et al., 2011; Xie et al., 2011). Recent evidence suggested that the lipid-lowering effect of BBR involves modulation of bile acid composition and activation of intestinal FXR signaling (Guo et al., 2016; Sun et al., 2017). However, the underlying mechanisms by which BBR modulates the intestinal bacteria community leading to microbially induced signals have not been explored.
In this study, effects of BBR on bacteria were investigated using both in vitro and in vivo models. We studied the direct effect of BBR on bacteria in vitro, so as to determine mechanisms of action in vivo. We demonstrate that BBR directly altered the gut microbiota by reducing Clostridium spp. and subsequently altered intestinal FXR signaling. These data provide new insights for studying the link between the intestinal bacteria, nuclear receptor signaling, and xenobiotics.
Materials and Methods
Chemicals.
BBR chloride and brain heart infusion broth were purchased from Sigma-Aldrich (St. Louis, MO). Transgenic dough was purchased from Bio-Serve (Flemington, NJ). Bile acid standards and deuterated internal standards were obtained from Sigma-Aldrich and Cayman Chemical (Ann Arbor, MI). Four dyes, including SYBR Green, propidium iodide (PI), bis-(1,3-dibutylbarbituric acid) trimethine oxonol (DiBAC4), and carboxyfluorescein diacetate (CFDA), were purchased from Invitrogen (Carlsbad, CA). Sodium 3-(trimethylsilyl)[2,2,3,3-2H4] propionate and D2O (99.9% in D) were purchased from Cambridge Isotope Laboratories (Miami, FL). Reduced (1×) phosphate-buffered saline (PBS) solution containing 1 g/l l-cysteine was prepared and stored anaerobically for bacterial culture.
Animals.
Animal procedures were performed using protocols approved by the Pennsylvania State University Institutional Animal Care and Use Committee. Twelve male C57BL/6 wild-type mice were purchased from Jackson Laboratories (Bar Harbor, MN). Five-week-old mice were trained to eat bacon-flavored dough pills for 1 week. Dough pills containing BBR were made using tablet molds as previously described (Zhang et al., 2015a, 2016), and one pill contained 2.3 mg BBR (100 mg/kg as final dose). Mice were fed pills containing BBR or vehicle for 5 days (Fig. 1A). Mice were housed singly in an empty cage and monitored to ensure the pill was consumed. Urine and feces were collected after 0, 1, 3, and 5 days of BBR treatment. Blood, liver, cecal content, and intestinal tissue samples were collected and saved at −80°C immediately after sacrifice.
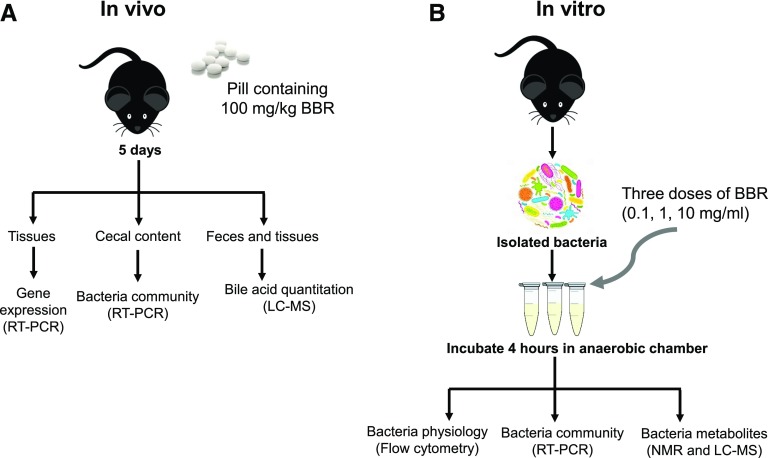
Fig. 1.
Scheme for determining the effects of BBR in vivo (A) and vitro (B). LC, liquid chromatography; MS, mass spectrometry; RT, real time.
Histopathology and Clinical Biochemistry.
Liver tissues embedded in paraffin wax were stained with hematoxylin and eosin. Liver injury markers, including serum alanine transaminase and alkaline phosphatase, were assessed using the VetScan VS2 Chemistry Analyzer (Abaxis Inc., Union City, CA).
Bile Acid Quantitation by Ultra-Performance Liquid Chromatography–Tandem Mass Spectrometry.
Quantitative analysis of bile acids in liver, feces, and ileum tissue was performed with an Acquity ultra-performance liquid chromatograpy (UPLC) system coupled to a Waters Xevo TQS mass spectrometer with an Acquity C8 BEH (2.1 × 100 mm, 1.7 µm) UPLC column (all from Waters, Milford, MA). Tissues (50 mg) and feces (25 mg) were added to 1 ml methanol containing 0.5 μM deuterated internal standards, followed by homogenization. After centrifugation, analytes were detected by multiple reaction monitoring and normalized by their respective internal deuterated standard. The results were quantified by comparing integrated peak areas against a standard curve.
Bile Salt Hydrolase Activity.
Bile salt hydrolase (BSH) activity was measured using a published protocol (Li et al., 2013) with minor modifications. Protein was prepared from fecal samples (50 mg) with 500 µl PBS containing 1 mM phenylmethylsulfonyl fluoride. Incubations were performed at 37°C in 3 mM sodium acetate buffer containing 0.4 mg/ml fecal protein and 5 µM taurocholic acid (TCA)-d4. After 30 minutes of incubation, an equal volume of methanol was added to the reaction. After centrifugation, the supernatant was analyzed by an Acquity UPLC system coupled with a Waters Xevo TQS mass spectrometer.
Tissue RNA Extraction and Quantitative Polymerase Chain Reaction.
Liver and ileum RNA was extracted using TRIzol reagent (Invitrogen). cDNA was synthesized from 1 μg RNA using qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD). Quantitative polymerase chain reaction (qPCR) was carried out using SYBR Green qPCR Master Mix with an ABI Prism 7900HT Fast Real-Time PCR sequence detection system (Applied Biosystems, Waltham, MA). qPCR primers are shown in Supplemental Table 1, and data were normalized to Actb mRNA levels.
In Vitro Bacterial Culture and Flow Cytometry.
In vitro bacterial culture with BBR treatment was performed using a previously published protocol (Maurice and Turnbaugh, 2013) with minor modifications. Briefly, cecal contents from twelve 7-week-old male C57BL/6J mice were mixed and diluted in sterile brain heart infusion broth (1:10 g/ml). After vortexing, 1.8 ml bacterial mixture was transferred to new tubes and divided into four groups. Three groups were treated with three doses of BBR, including high (10 mg/ml), middle (1.0 mg/ml), and low (0.1 mg/ml) doses. After vortexing, incubation of the bacterial mixtures was carried out at 37°C for 4 hours. After incubation, bacterial samples were stained for flow cytometry analysis or kept at −80°C for metabolite and bacteria DNA analysis (Fig. 1B). All experiments were performed in a monitored anaerobic chamber.
After centrifugation of the bacterial mixtures, the supernatants were washed and diluted 120 times with 1× reduced PBS. Diluted samples stained by four different dyes, including SYBR Green, PI, DiBAC4, and CFDA, were analyzed on a BD Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ), and data were analyzed using FlowJo software (version 10; Tree Star, Ashland, OR). The percentages of PI, DiBAC, CFDA, low nucleic acid (LNA), and high nucleic acid (HNA) cells were normalized to SYBR Green.
Bacteria DNA Extraction and qPCR.
DNA was extracted from cecal contents and in vivo bacterial suspension using the E.Z.N.A. Stool DNA Kit (Omega Bio-tek Inc., Norcross, GA). qPCR was carried out with SYBR Green qPCR Master Mix on an ABI Prism 7900HT Fast Real-Time PCR sequence detection system (Applied Biosystems, Waltham, MA). The primers are shown in Supplemental Table 2, and the data were normalized to 16S ribosomal DNA sequences.
1H NMR–Based Metabolomics Experiments.
Urine and feces sample preparation for NMR analysis was carried out as described (Jiang et al., 2012). Bacteria suspension samples were extracted twice with 1 ml methanol/water mix [2:1 (v/v)], followed by three consecutive freeze-thaws. After evaporation via a SpeedVac vacuum concentrator (Thermo Fisher Scientific, Waltham, MA), the extracts were resuspended in 600 μl PBS [0.1 M, 50% (v/v) D2O, and 0.005% 3-(trimethylsilyl)[2,2,3,3-2H4] propionate] and analyzed with a Bruker Avance III 600 MHz NMR spectrometer equipped with an inverse cryogenic probe (Bruker Biospin, Rheinstetten, Germany). A typical one-dimensional NMR spectrum was acquired using the first increment of nuclear overhauser effect spectroscopy sequence with presaturation (NOESYPR1D), and principal component analysis (PCA) and orthogonal projection to latent structure-discriminant analysis were performed with SIMCA-P+ software (Umetrics, Umeå, Sweden). The color-coded correlation coefficient plots from the orthogonal projection to latent structure-discriminant analysis models were carried out with MATLAB (MathWorks Inc., Natick, MA).
FXR Reporter Assay.
Agonist activities of taurine-conjugated bile acids and BBR were measured with the human FXR reporter assay (INDIGO Biosciences Inc., State College, PA). Briefly, reporter cells were incubated with taurine-conjugated bile acids or BBR for 6 hours, after which luminescence was measured by a plate reader.
Statistical Analysis.
Values are represented by means ± S.D. or medians and interquartile ranges. Statistical analyses were performed using unpaired t test analysis for the in vivo experiment and one-way analysis of variance for the in vitro experiment by GraphPad Prism software (version 6.0; GraphPad Software Inc., La Jolla, CA).
Results
BBR Treatment Increases Taurine-Conjugated Bile Acids.
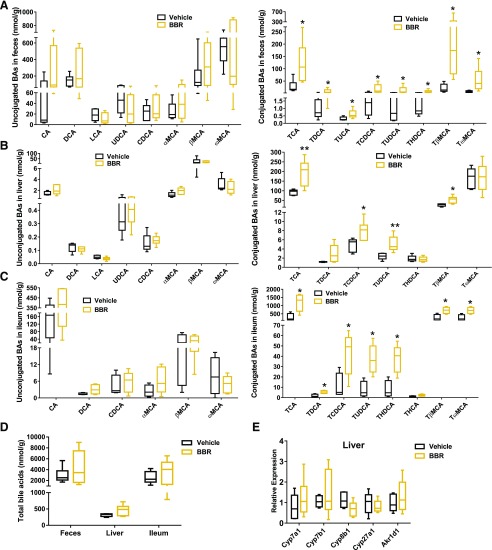
No significant changes in food intake, body weight, or liver histopathology were observed in mice after BBR exposure (Supplemental Fig. 1, A and B). BBR had no effect on serum alanine transaminase and alkaline phosphatase levels (Supplemental Fig. 1C). Previous studies indicated that 2 or 8 weeks of BBR treatment affected bile acid biosynthesis (Guo et al., 2016; Sun et al., 2017). Analysis of bile acid composition showed that 5 days of BBR treatment significantly increased taurine-conjugated bile acid levels in the feces, liver, and ileum (Fig. 2, A–C). BBR treatment markedly increased TCA, tauroursodeoxycholic acid (TUDCA), taurochenodeoxycholic acid (TCDCA), and tauro-β-muricholic (TβMCA) in the feces, liver, and ileum (Fig. 2, A–C). No significant changes in unconjugated bile acids (Fig. 2, A–C) or total bile acids (Fig. 2D) were observed after 5 days of BBR treatment. mRNA expression of genes involved in bile acid synthesis in the liver was unchanged (Fig. 2E). These results suggest that BBR might act as an intestine-restricted FXR activator that does not alter the expression of FXR target genes involved in bile acid synthesis in the liver.
Fig. 2.
Quantification of bile acids in the feces (A), liver (B), ileum (C), and total (D) from mice after 5 days vehicle or BBR treatment. (E) qPCR analysis of mRNA encoding bile acid synthesis in the liver from mice after 5 days vehicle or BBR treatment. Values are medians and interquartile ranges (n = 6 per group). *P < 0.05; **P < 0.01. Hyocholic acid, ursocholanic acid, lithocholenic acid, isolithocholenic acid, allolithocholenic acid, hyodexycholic acid, isodexycholic acid, tauro-ursocholanic acid, 3,7,12 tauro-dehydrocholic acid, TαMCA, TLCA, and THCA were measured that under detection limit in this study. BA, bile acid; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; HCA, hyocholic acid; LCA, lithocholic acid; MCA, muricholic acid; T, taurine-conjugated species; TαMCA, tauro-α-muricholic acid; TDCA, taurodeoxycholic acid; THCA, taurohyocholic acid; THDCA, taurohyodeoxycholic acid; TLCA, taurolithocholic acid; TUCA, tauroursocholanic acid; UDCA, ursodeoxycholic acid.
TCA Activates the FXR Signaling Pathway.
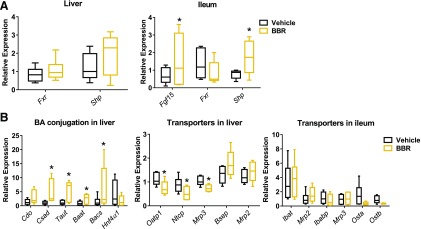
Altered bile acid signaling is associated with FXR signaling (Lefebvre et al., 2009). Five days of BBR treatment activated intestinal FXR target gene expression, including small heterodimer partner (Shp) and fibroblast growth factor 15 (Fgf15), but remained unchanged in the liver (Fig. 3A). These data confirm that BBR promotes intestine-restricted FXR activation. BBR treatment also increased mRNA expression of the taurine transporter (Taut) and the relate-limiting enzyme for the conversation of cysteine to taurine (Csd) in the liver (Fig. 3B). mRNA expression of bile acid–conjugated enzymes (Bacs and Baat) was also significantly higher in the liver after BBR treatment (Fig. 3B), which was consistent with the increased levels of hepatic taurine-conjugated bile acids. We also observed decreased expression of genes involved in bile acid transport (Ntcp and Oatp1) in the liver after BBR treatment (Fig. 3B). These results are consistent with data showing that suppression of these transporters by FXR involves a cascade including FXR-SHP activation (Geier et al., 2007; Dawson et al., 2009).
Fig. 3.
(A) qPCR analysis of Fgf15, Fxr, and Shp mRNAs in the ileum and Fxr and Shp mRNA expression in the liver from mice after 5 days of vehicle or BBR treatment. (B) qPCR analysis of mRNA encoding bile acid conjugation and transporters in the liver and ileum from mice after 5 days of vehicle or BBR treatment. Values are medians and interquartile ranges (n = 6 per group). *P < 0.05. Baat, bile acid-CoA: amino acid N-acyltransferase; Bacs, bile acid-CoA synthetase; Cdo, cysteine dioxygenase; Csad, cysteine sulfinic acid decarboxylase; Hnf4α1, hepatic nuclear factor 4α1; Ibabp, ileal bile acid-binding protein; Ibat, ileal bile acid transporter; Mrp, multidrug resistance-associated protein; Ntcp, Na+/taurocholate cotransporter Oatp, organic anion transporting protein; Ost, organic solute transporter.
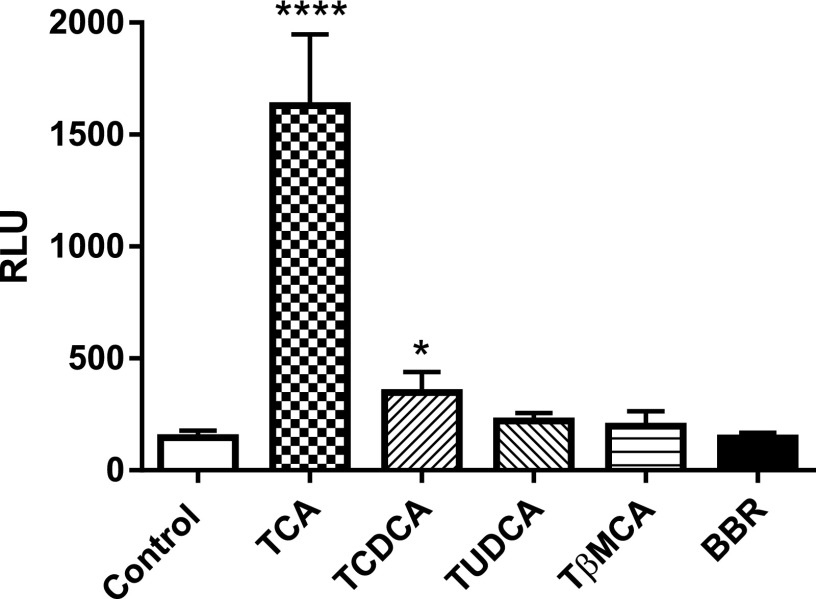
Previous evidence demonstrated that specific taurine-conjugated bile acids had different effects on the FXR signaling pathway (Cyphert et al., 2012). To investigate how altered taurine-conjugated bile acid profiles and BBR can directly affect the FXR signaling pathway, we used an FXR reporter assay to evaluate the FXR agonist activities of taurine-conjugated bile acids (TCA, TCDCA, TUDCA, and TβMCA) and BBR. In vitro, 100 μM TCA and TCDCA effectively activated FXR (increased 7.9-fold and 2.4-fold, respectively), whereas 100 μM TUDCA, TβMCA, and BBR had no effect (Fig. 4). These results indicate that the activation of intestinal FXR signaling by BBR treatment is mainly due to the accumulation of TCA.
Fig. 4.
Luciferase assays of the activation of FXR by 100 μM TCA, TCDCA, TUDCA, TβMCA, and BBR. Values are means ± S.D. (n = 3 per group). *P < 0.05, ****P < 0.0001. RLU, relative light unit.
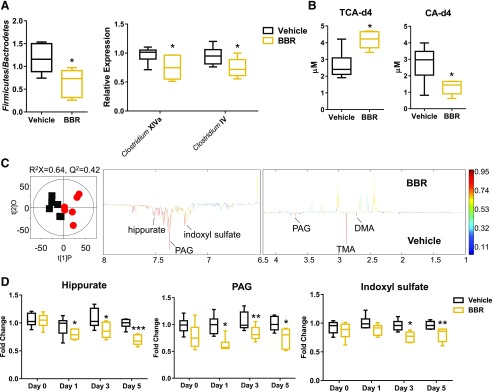
BBR Treatment Decreased Clostridium and BSH Activity in Mice.
Earlier studies reported a significant decrease in bacterial diversity after long-term BBR treatment administered to rats and mice fed a high-fat diet (Xie et al., 2011; Zhang et al., 2012, 2015b). No significant changes were observed in the major phyla of mice that consumed a normal chow diet after short-term BBR treatment (Supplemental Fig. 2). However, significant shifts in the Firmicutes/Bacteroidetes ratio were observed after 5 days of BBR treatment (Fig. 5A). Moreover, BBR treatment decreased levels of Clostridium clusters XIVa and IV (Fig. 5A), which is consistent with significantly lower fecal BSH activity (Fig. 5B). In addition, three-dimensional PCA scores plotted from urine global analysis showed distinct, time-dependent clustering of metabolites after BBR treatment (Supplemental Fig. 3). Urine metabolomics analysis revealed that BBR treatment was associated with decreased levels of common microbial metabolites, including hippurate, indoxyl sulfate, and phenylacetylglycine (Fig. 5, C and D). These data are consistent with the observation of decreased Clostridium, which is involved in the production of microbial metabolites often found in urine (Lord and Bralley, 2008; Lees et al., 2013). However, fecal changes were not as significant as those found in urine, although a trend of decreased butyrate and propionate and a significant increase in glucose after 5 days of BBR treatment was observed (Supplemental Fig. 4).
Fig. 5.
(A) qPCR analysis of Firmicutes/Bacteroidetes, Clostridium XIVa, and Clostridium IV in the cecal contents of mice after 5 days of vehicle or BBR treatment. (B) Fecal BSH enzyme activity after BBR 5 days of vehicle or BBR treatment. (C) Orthogonal projection to latent structure-discriminant analysis scores plot (left) and coefficient plot (right) derived from 1H NMR spectra of urine samples from mice after 5 days of vehicle (squares) or BBR (circles) treatment. The model was evaluated with cross-validated analysis of variance (P = 0.021). (D) Relative abundance of urine bacterial metabolites measured by 1H NMR data from mice after 0, 1, 3, and 5 days of BBR treatment. Values are medians and interquartile ranges (n = 6 per group). *P < 0.05, **P < 0.01, ***P < 0.001. DMA, dimethylamine; PAG, phenylacetylglycine; TMA, trimethylamine.
The Antibiotic Effect of BBR Treatment on Cecal Bacteria In Vitro.
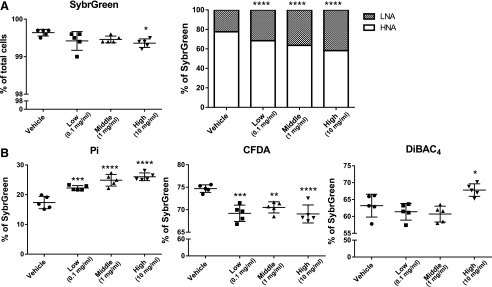
BBR is well known as an effective antimicrobial compound used in the treatment of microbial infections (Yu et al., 2005; Bandyopadhyay et al., 2013). To investigate the direct effect of BBR on the gut microbiota, we used an in vitro model to assess metabolic activity or membrane damage in bacteria using three doses of BBR. We determined the state of bacterial physiology using four different dyes: a dye that stains all cells (SYBR Green I), an impermeable dye that stains only dead or damaged cells (PI), a cell-permeable dye for cellular enzymatic activity (CFDA), and an oxonol dye for loss of membrane polarity (DiBAC4) (Joux and Lebaron, 2000; Maurice and Turnbaugh, 2013; Maurice et al., 2013). BBR caused a significant dose-dependent increase in PI-positive bacteria (from 17.4% ± 2.1% to 26.0% ± 1.3%) and significantly decreased SYBR Green–stained bacteria with a high dose of BBR treatment (Fig. 6). Furthermore, a significant dose-dependent increase in bacteria with LNA content and decreased HNA content was observed with BBR treatment (Fig. 6A). Three doses of BBR treatment all resulted in significant decreases in CFDA-stained bacteria (Fig. 6B), which indicated decreased metabolic activity of bacterial cells with BBR treatment. Moreover, a significant increase in DiBAC4-stained bacteria was observed with a high dose of BBR treatment (Fig. 6B).
Fig. 6.
Flow cytometric analyses of proportions of SYBR Green I, LNA, and HNA (A) and PI, DiBAC4, and CFDA (B) cells from cecal bacteria with vehicle or three doses of BBR treatment. Values are means ± S.D. (n = 5 per group). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. LNA, low nucleic acid; HNA, high nucleic acid.
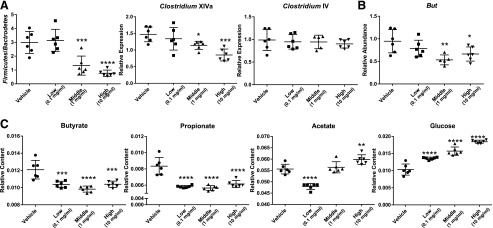
To investigate changes in the composition of bacteria by BBR directly, qPCR bacterial quantification was performed on isolated bacterial DNA. BBR treatment resulted in a dose-dependent decrease in the number of bacteria from Firmicutes and Clostridium XIVa and an increase in the Firmicutes/Bacteroidetes ratio (Fig. 7A; Supplemental Fig. 5). No significant changes in other phyla levels including Bacteroidetes or Actinobacteria were observed after BBR treatment (Supplemental Fig. 5).
Fig. 7.
qPCR analysis of Firmicutes/Bacteroidetes, Clostridium XIVa, and Clostridium IV (A) and bacterially produced butyryl-CoA:acetate CoA-transferase (but) (B) from cecal bacteria with vehicle or three doses BBR treatment. (C) Acetate, propionate, butyrate, and glucose levels in the cecal bacteria with vehicle or three doses of BBR treatment by 1H NMR analysis. Values are means ± S.D. (n = 5 per group, flow cytometry; n = 6 per group, qPCR and NMR). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
1H NMR–based metabolomics was employed to evaluate the direct metabolic effect of BBR on the intestinal microbiota. The three-dimensional PCA score plot showed distinct clustering of bacteria with three doses of BBR treatment (Supplemental Fig. 6). Acetate, propionate, and butyrate are the end metabolites of fermentation of dietary fiber by the gut microbiota (Den Besten et al., 2013). BBR treatment resulted in significantly lower butyrate and propionate levels but higher acetate and glucose levels in bacteria (Fig. 7C). Moreover, levels of bacterial gene butyryl-CoA:acetate CoA-transferase (but) associated with butyrate production were significantly lower with middle and high doses of BBR treatment (Fig. 7B). Taken together, these observations indicate that BBR affects bacteria directly.
Discussion
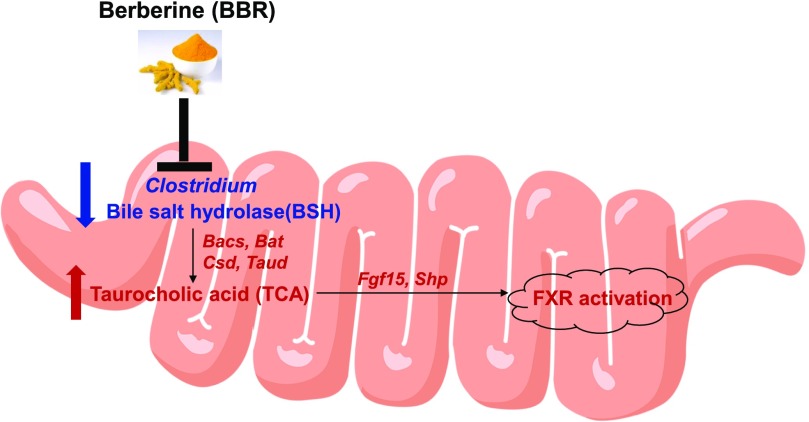
This study demonstrates that remodeling of the intestinal microbiota by BBR leads to alteration of bile acid levels and activation of FXR signaling (Fig. 8). The effect of BBR on the gut microbiota of rats fed a high-fat diet identified a marked decrease in gut microbiota diversity and a phylum shift of gut bacteria in rats (Zhang et al., 2012, 2015b). In this study, 5 days of oral BBR treatment of normal-weight mice led to a significant change in the Firmicutes/Bacteroidetes ratio and reduced Clostridium. These data indicated that the effects of BBR on the gut microbiota are dependent on diet and length of dosing. Furthermore, BBR treatment decreased levels of mouse urine microbial metabolites, including hippurate, phenylacetylglycine, and indoxyl sulfate, which is in agreement with the observation of reduced Clostridium spp. (Lord and Bralley, 2008; Tian et al., 2018).
Fig. 8.
Remodeling of the gut microbiota by BBR leads to a change in the composition of bile acids and associated activation of FXR signaling. The metabolite, bacteria, enzyme, or mRNA in red or blue represents a higher or lower level in the intestines obtained from BBR-treated mice compared with vehicle.
The direct activity of BBR on the intestinal bacteria was observed in vitro using NMR-based metabolomics combined with flow cytometry. In vitro, BBR treatment led to a dose-dependent increase in cell damage and decreased metabolic activity in bacteria, which is consistent with previous studies that showed the effective antibacterial activity of BBR in vitro and in vivo (Yu et al., 2005; Bandyopadhyay et al., 2013). Furthermore, BBR treatment led to decreased HNA cells with higher metabolic activity and growth rates (Wang et al., 2009). Moreover, BBR treatment resulted in a dose-dependent decrease in Firmicutes and Clostridium XIVa but no significant change in Bacteroidetes or Actinobacteria was observed, which supports our observation of reduced Clostridium by BBR treatment in vivo. This is consistent with reports showing that active and damaged subsets in bacteria belong to Firmicutes and 90% of the affected operation taxonomic units were Firmicutes after antibiotics exposure (Maurice et al., 2013). Moreover, BBR treatment decreased levels of the bacterial butyrate-producing gene but and altered bacterial metabolite levels, including reduced levels of butyrate and propionate and increased acetate and glucose levels in vitro. These results indicated inhibition of bacterial fermentation by BBR treatment (Morrison et al., 2006; Vital et al., 2014). Collectively, the data suggest that BBR influences microbial metabolism.
Increased levels of taurine-conjugated bile acids with BBR treatment are likely a combination of both host and bacteria changes. Significantly higher levels of taurine-conjugated bile acids were observed in the liver, ileum, and feces after BBR treatment. Consistently, previous studies also indicated increased levels of taurine-conjugated bile acids in the colon, ileum, and serum by intestine-restricted FXR agonist fexaramine (Pathak et al., 2018). An increase in gene expression involved in conjugation enzymes was observed in the host after BBR treatment, which indicated that BBR increased host bile acid conjugation pathways (Guo et al., 2016). In addition, the increased level of taurine-conjugated bile acids with BBR treatment is also associated with reducing Clostridium and its BSH activity. BSH is a bacterial enzyme that catalyzes the deconjugation of conjugated bile acids (Kumar et al., 2006). Among the Firmicutes, BSH activity has been detected in Lactobacillus and Clostridium (Begley et al., 2006). A recent study indicated that tempol reduced Lactobacillus and its BSH activity, resulting in increased levels of taurine-conjugated bile acids (Li et al., 2013). On the basis of these findings, the decreased BSH activity may result from the reduction or direct killing of Clostridium by BBR treatment. Moreover, the lower BSH activity contributed to the accumulation of taurine-conjugated bile acids in the BBR treatment group.
Activation of intestinal FXR by BBR exposure was associated with the accumulation of TCA. Previous studies showed that bile acid can activate ileal FXR, which simulated FGF15 in the liver and elevated serum FGF15 (Li et al., 2018). One possible explanation for our results is that activation of intestinal FXR by BBR treatment was associated with a marked accumulation of TCA. This is consistent with studies showing that both BBR and TCA treatment activated FXR signaling in the intestine and exerted its lipid-lowering effect (Sun et al., 2017). FXR is a bile acid receptor and plays an key role in the regulation of bile acid metabolism (Forman et al., 1995), and tissue-restricted FXR activation was reported as a new therapeutic angle for obesity and related metabolic diseases (Fang et al., 2015; Pathak et al., 2018). Future work is still needed to investigate whether the same effect of BBR on microbiota occurs in obese or diabetic mice.
In summary, the effect of BBR on gut bacteria was studied using both in vitro and in vivo models. BBR reduced Clostridium and its BSH activity, leading to the accumulation of TCA. The accumulation of TCA was associated with the activation of intestinal FXR. Our data provide new insights for studying the link between the microbiota, nuclear receptor signaling, and xenobiotics.
Acknowledgments
We thank Nelson Thomas Peterson for technical help and Dr. Frank J. Gonzalez for critical discussion of the data.
Abbreviations
- BBR
berberine
- BSH
bile salt hydrolase
- CFDA
carboxyfluorescein diacetate
- DiBAC4
bis-(1,3-dibutylbarbituric acid) trimethine oxonol
- FGF
fibroblast growth factor
- FXR
farnesoid X receptor
- HNA
high nucleic acid
- PBS
phosphate-buffered saline
- PCA
principal component analysis
- PI
propidium iodide
- qPCR
quantitative polymerase chain reaction
- TβMCA
tauro-β-muricholic acid
- TCA
taurocholic acid
- TCDCA
taurochenodeoxycholic acid
- TUDCA
tauroursodeoxycholic acid
- UPLC
ultra-performance liquid chromatography
Authorship Contributions
Participated in research design: Tian, Patterson.
Conducted experiments: Tian, Cai, Gui, Nichols, Zhang, Anitha.
Performed data analysis: Tian, Koo, Patterson.
Wrote or contributed to the writing of the manuscript: Tian, Patterson.
Footnotes
This work was supported in part by the Pennsylvania Department of Health [Tobacco Commonwealth Universal Research Enhancement Funds], the National Institutes of Health National Library of Medicine [Grant T32LM12415-1], and the U.S. Department of Agriculture National Institute of Food and Agriculture [Award 2914-38420-21822].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ali AH, Carey EJ, Lindor KD. (2015) Recent advances in the development of farnesoid X receptor agonists. Ann Transl Med 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab JP, Karpen SJ, Dawson PA, Arrese M, Trauner M. (2017) Bile acids and nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives. Hepatology 65:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Patra PH, Mahanti A, Mondal DK, Dandapat P, Bandyopadhyay S, Samanta I, Lodh C, Bera AK, Bhattacharyya D, et al. (2013) Potential antibacterial activity of berberine against multi drug resistant enterovirulent Escherichia coli isolated from yaks (Poephagus grunniens) with haemorrhagic diarrhoea. Asian Pac J Trop Med 6:315–319. [DOI] [PubMed] [Google Scholar]
- Begley M, Hill C, Gahan CGM. (2006) Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72:1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tao C, Liu Z, Lu M, Pan Q, Zheng L, Li Q, Song Z, Fichna J. (2015) A randomized clinical trial of berberine hydrochloride in patients with diarrhea-predominant irritable bowel syndrome. Phytother Res 29:1822–1827. [DOI] [PubMed] [Google Scholar]
- Cyphert HA, Ge X, Kohan AB, Salati LM, Zhang Y, Hillgartner FB. (2012) Activation of the farnesoid X receptor induces hepatic expression and secretion of fibroblast growth factor 21. J Biol Chem 287:25123–25138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA, Lan T, Rao A. (2009) Bile acid transporters. J Lipid Res 50:2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Sandoval D, Mautino G, Martin G, Percevault F, Barbier O, Fruchart JC, Kuipers F, Staels B. (2004) Glucose regulates the expression of the farnesoid X receptor in liver. Diabetes 53:890–898. [DOI] [PubMed] [Google Scholar]
- Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, Yoshihara E, Perino A, Jacinto S, Lukasheva Y, et al. (2015) Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med 21:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. (1995) Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687–693. [DOI] [PubMed] [Google Scholar]
- Geier A, Wagner M, Dietrich CG, Trauner M. (2007) Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim Biophys Acta 1773:283–308. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zhang Y, Huang W, Selwyn FP, Klaassen CD. (2016) Dose-response effect of berberine on bile acid profile and gut microbiota in mice. BMC Complement Altern Med 16:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lin H, Huang W. (2011) Modulating gut microbiota as an anti-diabetic mechanism of berberine. Med Sci Monit 17:RA164–RA167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. (2006) Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA 103:3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, Cai J, Qi Y, Fang ZZ, Takahashi S, et al. (2015a) Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest 125:386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Xie C, Lv Y, Li J, Krausz KW, Shi J, Brocker CN, Desai D, Amin SG, Bisson WH, et al. (2015b) Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun 6:10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Huang J, Wang Y, Tang H. (2012) Eliminating the dication-induced intersample chemical-shift variations for NMR-based biofluid metabonomic analysis. Analyst (Lond) 137:4209–4219. [DOI] [PubMed] [Google Scholar]
- Joux F, Lebaron P. (2000) Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect 2:1523–1535. [DOI] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, et al. (2003) A G protein-coupled receptor responsive to bile acids. J Biol Chem 278:9435–9440. [DOI] [PubMed] [Google Scholar]
- Kong W, Wei J, Abidi P, Lin M, Inaba S, Li C, Wang Y, Wang Z, Si S, Pan H, et al. (2004) Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med 10:1344–1351. [DOI] [PubMed] [Google Scholar]
- Kumar RS, Brannigan JA, Prabhune AA, Pundle AV, Dodson GG, Dodson EJ, Suresh CG. (2006) Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J Biol Chem 281:32516–32525. [DOI] [PubMed] [Google Scholar]
- Lambert G, Amar MJA, Guo G, Brewer HB, Jr, Gonzalez FJ, Sinal CJ. (2003) The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J Biol Chem 278:2563–2570. [DOI] [PubMed] [Google Scholar]
- Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. (2013) Hippurate: the natural history of a mammalian-microbial cometabolite. J Proteome Res 12:1527–1546. [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. (2009) Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 89:147–191. [DOI] [PubMed] [Google Scholar]
- Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. (2013) Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun 4:2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhao Q, Zhang C, Zhang P, Hu A, Zhang L, Schroder PM, Ma Y, Guo Z, Zhu X, et al. (2018) The ileal FGF15/19 to hepatic FGFR4 axis regulates liver regeneration after partial hepatectomy in mice. J Physiol Biochem 74:247–260. [DOI] [PubMed] [Google Scholar]
- Li T, Chiang JYL. (2013) Nuclear receptors in bile acid metabolism. Drug Metab Rev 45:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord RS, Bralley JA. (2008) Clinical applications of urinary organic acids. Part 2. Dysbiosis markers. Altern Med Rev 13:292–306. [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. (2013) Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF and Turnbaugh PJ (2013) Quantifying and identifying the active and damaged subsets of indigenous microbial communities, in Microbial Metagenomics, Metatranscriptomics, and Metaproteomics (DeLong EF ed) pp 91–107, Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- Morrison DJ, Mackay WG, Edwards CA, Preston T, Dodson B, Weaver LT. (2006) Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br J Nutr 96:570–577. [PubMed] [Google Scholar]
- Pathak P, Xie C, Nichols RG, Ferrell JM, Boehme S, Krausz KW, Patterson AD, Gonzalez FJ, Chiang JYL. (2018) Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 68:1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. (2014) Bile acids and the gut microbiome. Curr Opin Gastroenterol 30:332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. (2013) Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. [DOI] [PubMed] [Google Scholar]
- Sun R, Yang N, Kong B, Cao B, Feng D, Yu X, Ge C, Huang J, Shen J, Wang P, et al. (2017) Orally administered berberine modulates hepatic lipid metabolism by altering microbial bile acid metabolism and the intestinal FXR signaling pathway. Mol Pharmacol 91:110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Feng Y, Tsao S, Wang N, Curtain R, Wang Y. (2009) Berberine and Coptidis rhizoma as novel antineoplastic agents: a review of traditional use and biomedical investigations. J Ethnopharmacol 126:5–17. [DOI] [PubMed] [Google Scholar]
- Tian Y, Nichols RG, Cai J, Patterson AD, Cantorna MT. (2018) Vitamin A deficiency in mice alters host and gut microbial metabolism leading to altered energy homeostasis. J Nutr Biochem 54:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Howe AC, Tiedje JM. (2014) Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hammes F, Boon N, Chami M, Egli T. (2009) Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J 3:889–902. [DOI] [PubMed] [Google Scholar]
- Xie W, Gu D, Li J, Cui K, Zhang Y. (2011) Effects and action mechanisms of berberine and Rhizoma coptidis on gut microbes and obesity in high-fat diet-fed C57BL/6J mice. PLoS One 6:e24520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JH, Liu XZ, Pan W, Zou DJ. (2017) Berberine protects against diet-induced obesity through regulating metabolic endotoxemia and gut hormone levels. Mol Med Rep 15:2765–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Kim KJ, Cha JD, Kim HK, Lee YE, Choi NY, You YO. (2005) Antimicrobial activity of berberine alone and in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. J Med Food 8:454–461. [DOI] [PubMed] [Google Scholar]
- Zhang L, Nichols RG, Correll J, Murray IA, Tanaka N, Smith PB, Hubbard TD, Sebastian A, Albert I, Hatzakis E, et al. (2015a) Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ Health Perspect 123:679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xie C, Nichols RG, Chan SH, Jiang C, Hao R, Smith PB, Cai J, Simons MN, Hatzakis E, et al. (2016) Farnesoid X receptor signaling shapes the gut microbiota and controls hepatic lipid metabolism. mSystems 1:e00070-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, Zhang X, Zhao L. (2015b) Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep 5:14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, Li M, Zhang C, Zhang Z, Zhang Y, et al. (2012) Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One 7:e42529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. (2006) Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci USA 103:1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang H, Li B, Meng X, Wang J, Zhang Y, Yao S, Ma Q, Jin L, Yang J, et al. (2014) Berberine activates thermogenesis in white and brown adipose tissue. Nat Commun 5:5493. [DOI] [PubMed] [Google Scholar]