Abstract
Cellular protein homeostasis (proteostasis) is maintained by a broad network of proteins involved in synthesis, folding, triage, repair and degradation. Chief among these are molecular chaperones and their cofactors that act as powerful protein remodelers. The growing realization that many human pathologies are fundamentally diseases of protein misfolding (proteopathies) has generated interest in understanding how the proteostasis network impacts onset and progression of these diseases. In this minireview, we highlight recent progress in understanding the enigmatic Hsp110 class of heat shock protein that acts as both a potent nucleotide exchange factor to regulate activity of the foldase Hsp70, and as a passive chaperone capable of recognizing and binding cellular substrates on its own, and its integration into the proteostasis network.
Keywords: Hsp70, Hsp110, proteostasis, chaperone, folding, nucleotide exchange factor, holdase, proteopathy, neurodegenerative disease
Introduction
Molecular chaperones aid in the proper folding of nascent polypeptide chains into their functionally active confirmations during synthesis and additionally recognize misfolded proteins to refold or target for degradation (Hartl et al., 2011). Proteotoxic cellular stress activates unfolded protein responses resulting in the coordinated expression of chaperones and other so-called heat shock proteins to maintain protein homeostasis (proteostasis). Misfolded proteins have the propensity to aggregate and interfere with normal functions of the proteome; therefore, safeguarding cellular proteostasis is essential for cell survival. Numerous neurodegenerative disorders such as Alzheimer’s (AD), Parkinson’s (PD), and Huntington’s (HD) diseases are linked to the alternative folding and aggregation of key proteins and have thus been termed proteopathies (Soto, 2003). The heat shock protein 70 (Hsp70) class of molecular chaperones are ancient and ubiquitous modulators of cellular proteostasis that operate by iterative cycles of substrate binding and release allosterically coupled to ATP binding and hydrolysis. These linked actions are carried out by nucleotide binding (NBD) and substrate binding (SBD) domains highly conserved throughout evolution (Fig. 1A). Two highly related but distinct sub-groups of Hsp70 possess additional sequence elements and insertions in the SBD and have been classified as Hsp110 and Grp170 (glucose-related protein) (Easton et al., 2000). The last fifteen years has witnessed an explosion of interest and advancement in our understanding of these previously underappreciated members of the proteostasis network. For simplicity this mini-review will largely focus on current trends in thinking regarding the multiple roles that Hsp110s play in cellular biology, considering relevant features of the Grp170 chaperones where appropriate.
Figure 1. A, Domain architecture of Hsp70 superfamily.
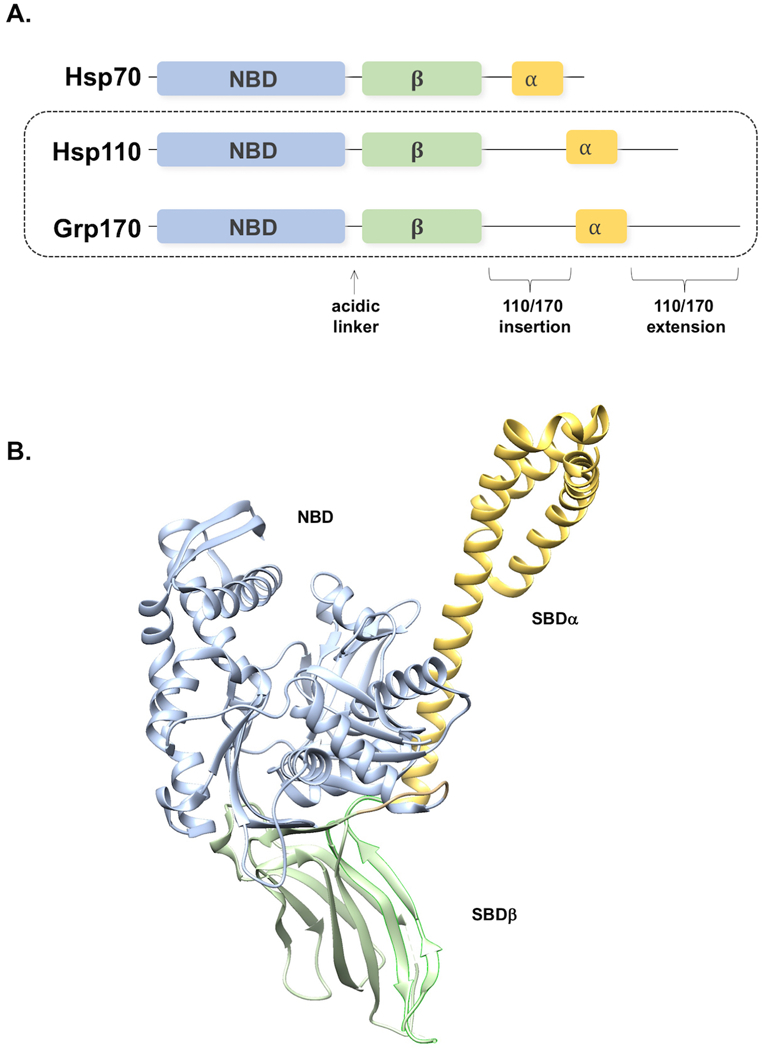
NBD, nucleotide binding domain. β, β-sandwich peptide binding domain. α, α-helical bundle. Locations of additional sequences in the Hsp110 and Grp170 chaperone classes relative to Hsp70 are indicated, as is the acidic linker. B, Ribbon structure of the yeast Hsp110 Sse1 (PDB file 2QXL), domains colored as in A and labeled as indicated.
Hsp110s are unique from Hsp70
Hsp110 is encoded by two highly homologous genes, SSE1 and SSE2, in Saccharomyces cerevisiae, which together are required for viability (Trott et al., 2005). However, deletion of SSE1 alone results in significant slow growth and other phenotypes as described below (Hideyuki et al., 1993). Despite strong homology (27% identity overall and 36% in the NBD with yeast Hsp70 Ssa1), several findings in budding yeast support the notion that Hsp110 is functionally distinct from Hsp70. First, mutation of several key residues in the Sse1 NBD known to be required for ATP hydrolysis in Hsp70 did not compromise growth, suggesting that ATP hydrolysis is not required for normal functions (Shaner et al., 2004). Second, Sse1 was found to exist in stable heterodimeric complexes with cytoplasmic Hsp70 molecules (Shaner et al., 2005; Yam et al., 2005). Third, the Sse1 and other Hsp110 SBDs, characterized in Hsp70 chaperones by β-sandwich and α-helical bundle domains that work together to bind peptide substrates or extended regions of polypeptides, show low sequence homology (~15%) to the Hsp70 SBD and contain an extra “loop” sequence of unknown function. A seminal crystal structure of Sse1 revealed that the SBDα is stably docked against the NBD, and therefore does not participate in high-affinity substrate binding by SBDβ (Fig. 1B) (Liu and Hendrickson, 2007). Additionally, the interdomain linker between the NBD and SBD in Hsp110 is a charged rather than hydrophobic sequence, as it is in Hsp70s, a distinction that would be expected to disrupt interdomain allosteric communication (Kityk et al., 2015). Suspicions that Hsp110 had indeed diverged from Hsp70 were confirmed when yeast and human Hsp110s were shown to act as potent nucleotide exchange factors (NEF) for Hsp70 by several groups (Dragovic et al., 2006; Raviol et al., 2006; Shaner et al., 2006). Hsp70 ATP hydrolysis is coupled with substrate binding and a NEF is required to mediate ADP dissociation from Hsp70, allowing ATP recharging and substrate release (Dragovic et al., 2006; Raviol et al., 2006). Finally, two groups solved the co-crystal structure of Sse1 with Hsp70, revealing an elegant heterodimer with substantial buried interface in the two NBDs, coupled with an “embrace” of the Hsp70 NBD by the Hsp110 α-helical bundle in an extended conformation (Schuermann et al., 2008; Polier et al., 2008). Interaction details were further supported by hydrogen-deuterium and chemical crosslinking experiments for Sse1/Ssa1 as well as the ER-localized pair Lhs1/Kar2 (Andréasson et al., 2008; Andréasson et al., 2010). In keeping with the earlier yeast genetic findings, ATP binding in the Hsp110 NBD, but not hydrolysis, is needed for association with Hsp70 and resultant nucleotide exchange activity (Shaner et al., 2006). In turn, the binding of ATP to Hsp70 leads to temporary dissociation of the Hsp70-Hsp110 complex (Andréasson et al., 2008). Mutations that disrupt ATP binding and/or Hsp70 association in yeast result in loss of function phenotypes in yeast, supporting the notion that NEF activity is a core Hsp110 property (Polier et al., 2008; Shaner et al., 2004). At least one report claims that Hsp110 can act as an ATP-powered foldase in concert with Hsp40/J-proteins, but this finding has not been corroborated to date (Mattoo et al., 2013). Additionally, the recently defined interaction site for J-domain proteins in Hsp70 chaperones is lacking in the Hsp110 family, further complicating this functional model (Kityk et al., 2018).
Biochemical experiments with purified proteins demonstrated that Hsp110 homologs in several species are potent passive chaperones, or “holdases,” defined by the capacity to hold unfolded substrates in a folding-competent state without energy input, but lacking the ability to accelerate substrate refolding (Fig. 2A) (Brodsky et al., 1999; Oh et al., 1999). This is in contrast to Hsp70 chaperones, which are weak holdases, but in the presence of J-proteins and ATP are potent “foldases,” helping to accelerate the rate of substrate refolding. Correspondingly, the core β-sandwich element of the SBD is very poorly conserved between Hsp70 and Hsp110, and human and yeast Hsp110 only exhibit 38% sequence identity despite retaining the same peptide binding capacity and function (Polier et al., 2010). This divergence in sequence suggests Hsp70 and Hsp110 may exhibit different substrate binding preferences, an assertion supported by findings that a known Hsp70 high affinity-binding peptide, FITC-ala-p5, does not demonstrably associate with Hsp110 (Goeckeler et al., 2008). More recent studies corroborated this result and further established that Hsp110 and Grp170 chaperones exhibit a preference for peptide substrates rich in aromatic residues (Behnke and Hendershot, 2014; Behnke et al., 2016; Xu et al., 2012). Residues within loops connecting β-strands in the β-sandwich domain have been shown to mediate differential peptide selectivity by Hsp70 and Hsp110, although interestingly, swapping loop residues from Hsp110 into the bacterial Hsp70 DnaK SBD increased recognition of an Hsp110-preferred peptide, but the converse was not true (Xu et al., 2012). Additionally, in a recent study, four amino acid substitutions predicted to negatively impact substrate binding were introduced into the L3,4 loop of the Sse1 SBD. However, Sse1 holdase activity for two different unfolded protein substrates was subsequently reduced by only 50% and resulted in minor impact to cellular proteostasis in yeast (Garcia, et al., 2017).
Figure 2. Models of Hsp110 function.
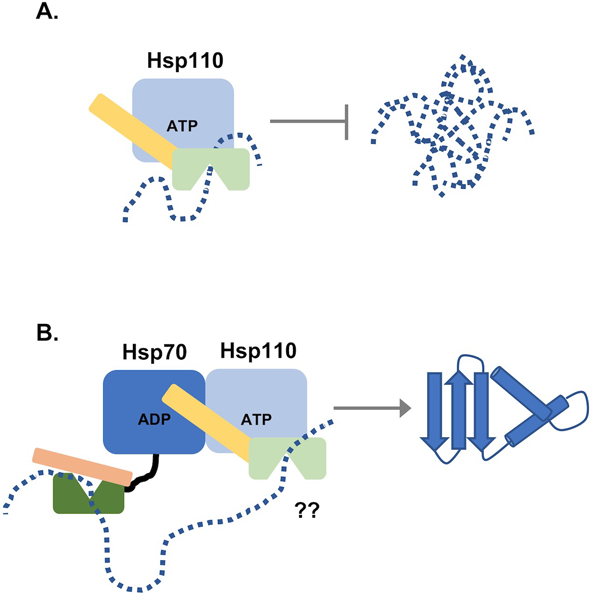
A, Hsp110 can bind and hold unfolded or partially folded polypeptides, sequestering them from aggregation. This activity is independent of nucleotide state in the NBD but it is generally assumed that ATP is bound within the NBD in cells. B, Hsp110 interacts with Hsp70 primarily via contacts in both NBDs, leaving the respective SBDs exposed and available for binding the same extended polypeptide, likely at different regions defined by SBD preference as described in the text. However, this arrangement has not been conclusively demonstrated. Dashed line represents unfolded polypeptide.
Grp170, the organellar Hsp110
The ER-localized Grp170 (Lhs1) has been demonstrated to be an NEF for the luminal Hsp70 (BiP/Kar2) as well as a passive chaperone, similar to cytosolic Hsp110 (Andréasson et al., 2010; Steel et al., 2004). Grp170 and Hsp110 are present in all major eukaryotic taxa, indicating the two proteins have a shared ancestral lineage (Easton et al., 2000). Grp170 binds aggregation-prone regions of ER substrates, specifically sequences enriched for aromatic residues akin to Hsp110 (Behnke et al., 2016). It has been suggested that π stacking, the transient interactions between aromatic rings, might mediate association of aromatic residues in Hsp110/Grp170 SBDs and those in non-native polypeptides (Behnke et al., 2016; McGaughey et al., 1998). Like Hsp110, Grp170 also possesses large insertions in the SBDα and an unstructured loop in SBDβ that both modulate substrate binding (Wang et al., 2015). Deletion of the unstructured SBD β-domain loop increased Grp170 binding to the unfolded peptide NS-1 LC, whereas deletion of the carboxyl-terminal alpha helical domain greatly reduced Grp170 NC-1 LC interaction (Behnke et al., 2016). Together, these findings indicate a possible alternative peptide binding mechanism for different Hsp110s, and certainly a lack of full understanding of peptide recognition by this class of chaperones.
Integration into the Proteostasis Network
Hsp110 and other Hsp70 NEFs
Three distinct classes of mammalian Hsp70 nucleotide exchange factors have been described, all with homologs in budding yeast (indicated in parentheses) where their relative contributions to proteostasis have been most investigated: Hsp110 (Sse1/2), BAG-1 (Snl1), and HspBP1 (Fes1). A comprehensive analysis of their respective structures and biochemical characteristics has been recently published (Bracher and Verghese, 2015). All three types of Hsp70 NEF perform the same fundamental function of inducing conformational shifts (through lobe displacement or rotation) in the Hsp70 NBD upon binding that reduce affinity for ADP, allowing exchange for ATP. The existence of three distinct classes of cytosolic Hsp70 NEF (and two classes, Grp170 and the HspBP1 homolog BAP/Sil1, in the ER) raises several questions. 1) What selection pressure with regard to protein quality control resulted in generation and retention of three different proteins with the same apparent function? 2) Are they indeed redundant or do they have specialized functions in vivo? and 3) What features dictate their differential partnerships with Hsp70 in cellular activities? Although these questions remain incompletely answered, progress has been made. As stated earlier, loss of both SSE genes in yeast is lethal, establishing that Hsp110 is essential for life in the presence of Fes1 and the ER membrane-tethered Snl1. However, overexpression of either Fes1 or truncated Snl1 lacking the membrane anchor partially suppresses sse1Δsse2Δ lethality, suggesting that at least part of the explanation may include lower cellular abundance of both proteins (Fes1, 5-fold lower than Sse1; Snl1, 20-fold lower than Sse1) (Ghaemmaghami et al., 2003; Raviol et al., 2006; Sadlish et al., 2008). To date, no phenotypes or functional consequences have been revealed in snl1Δ cells. Snl1 appears to exist in a complex with assembled ribosomes on the ER in both budding yeast and the related pathogenic yeast Candida albicans, but the biological significance of this interaction is unknown (Verghese and Morano, 2012). Cells lacking Sse1 or Fes1 accumulate terminally misfolded proteins, and loss of Sse1 specifically impairs folding of nascent polypeptides (Abrams et al., 2014; Gowda et al., 2013). Snl1 therefore apparently plays a far more restricted role in proteostasis, likely due to both low levels of expression and its subcellular localization. Cells lacking Sse1, Fes1 and Snl1 remain competent to refold thermally aggregated firefly luciferase, but loss of all Hsp110 activity brought about by inactivation of a temperature-sensitive SSE1 allele abolished this activity (Abrams et al., 2014; Kaimal et al., 2017).
While Hsp110 clearly demonstrates the ability to recognize unfolded or misfolded polypeptides, it is unclear how or why Fes1 might be targeted to terminally misfolded substrates. However, in a pair of fascinating recent discoveries, an amino-terminal domain in HspBP1 family members Fes1 and the human ER-localized BAP was shown to mimic unfolded proteins to ensure the release of substrates from Hsp70 (Gowda et al., 2018; Rosam et al., 2018). Some BAG-domain proteins also possess unstructured domains that mimic Hsp70 substrates to promote substrate release while performing NEF activity (Rauch et al., 2016). These findings suggest substrate mimicry may be a general mechanism for NEFs to promote Hsp70 substrate release that is not shared by the Hsp110/Grp170 family. Additionally, whereas Sse1/2 strongly interacts with both Ssa and Ssb Hsp70 isoforms in the yeast cytosol, Fes1 appears to be restricted to interaction with Ssa in vivo, effectively removing it from consideration as an important NEF for nascent protein synthesis on the ribosome (Abrams et al., 2014).
Hsp110 and recruitment of the disaggregase Hsp104
Hsp110 collaborates with the disaggregase Hsp104 to re-solubilize misfolded protein aggregates as well as amyloid deposits in yeast cells. A recent study demonstrated that Sse1 expression accelerated the rate of Hsp104-mediated disaggregation of chemically aggregated firefly luciferase (Kaimal et al., 2017). Hsp110/Hsp70 complexes are recruited early to aggregates independently of Hsp104, but Hsp104 requires Hsp110/Hsp70 to efficiently localize and resolubilize aggregates (Kaimal et al., 2017). Importantly, this activity requires that Hsp110 associate with Hsp70, suggesting either that Hsp104 cannot recognize Hsp110 as it does Hsp70 through the M-domain, or that the Hsp110 SBD is not competent to recognize protein aggregates in the absence of Hsp70 (Kaimal et al., 2017). On the other hand, Hsp110 has also been shown to counteract the effects of Hsp104 on prion propagation by promoting the nucleation of [PSI+] precursors (Sadlish et al., 2008). It is therefore possible that the properties of amyloid and aggregate structures dictate differential roles for Hsp110. Because Hsp110/Hsp70 (and Hsp40) complexes are also involved in the refolding of substrates extracted from aggregates, cells lacking Hsp110 are therefore deficient in several steps of the protein rescue pathway - recruitment, disaggregation and refolding - sharply underscoring the importance of this chaperone co-factor in proteostasis.
Hsp110 powers disaggregation in a chaperone triad
Maximal protein refolding by Hsp70 in vitro has been demonstrated to require both Hsp40 as an activator of the ATP hydrolysis step as well as a NEF to facilitate efficient nucleotide exchange and promote rapid cycling between low- and high-affinity substrate binding states in the Hsp70 SBD. Prior to the discovery of eukaryotic NEFs, refolding experiments were limited to the Hsp70/Hsp40 machinery, and to substrates that had been chemically denatured and maintained in an unfolded state. Aggregated substrates were viewed as recalcitrant to refolding unless the fungal disaggregase Hsp104 was included in reactions (Glover and Lindquist, 1998). These findings raised the question of how metazoans deal with protein aggregation, as no Hsp104 homologs are known outside of plant, fungal and bacterial lineages. It was revealed that inclusion of Hsp110 with Hsp70/Hsp40 facilitated a slower but still efficient disaggregation reaction (Rampelt et al., 2012; Shorter, 2011). This holds true for mammalian and yeast chaperone homologs, demonstrating conservation of this remarkable activity. Moreover, the Hsp110/Hsp70/Hsp40 disaggregase requires Hsp110 NEF activity but is not replaceable with other Hsp70 NEFs, indicative of an as yet unexplained specificity (Nillegoda et al., 2015; Rauch and Gestwicki, 2014; Tzankov et al., 2008). It is likely not contributed by Hsp110 substrate binding, as partial inactivation of the Sse1 SBD had no detrimental effect on either refolding or disaggregation by the chaperone triad (Garcia et al., 2017). Further investigation indicates that maximal disaggregase activity from this system is achieved with an equimolar mix of both class A and class B Hsp40 proteins, which despite a high degree of similarity play incompletely understood complementary and even synergistic roles (Nillegoda et al., 2015). The in vivo relevance of this disaggregase activity was established in Caenorhabditis elegans, where knockdown of Hsp110, but not the Bag homolog, resulted in failure to clear heat shock-induced luciferase aggregates (Rampelt et al., 2012). However, unlike Hsp104, addition of Hsp110 does not rapidly accelerate disaggregation of high order amyloids or prions (Shorter, 2011), suggesting that the two disaggregase machines are not interchangeable.
Implications for Hsp110 involvement in diseases of protein misfolding
Protein misfolding and aggregation is at the heart of neurodegenerative disorders such as Amyotrophic lateral sclerosis (ALS) and Parkinson’s (PD), Huntington’s (HD), and Alzheimer’s (AD) diseases (Soto, 2003). Loss of Hsp110 specifically has been shown to modulate amyloid formation and exacerbate disease phenotypes in models of ALS, HD, and AD. In a mouse AD model, Hsp110−/− mice exhibited age-dependent increase and accumulation of hyper-phosphorylated tau protein, a hallmark of disease progression, along with AD-related behavioral defects (Eroglu et al., 2010). In a Drosophila HD model, loss of Hsp110 led to age-dependent accumulation of Huntingtin (Htt) protein amyloids and acceleration in disease related phenotypes. In this same study, it was demonstrated that over-expression of Hsp110 had a remarkable protective effect, preventing Huntingtin aggregation and onset of disease phenotypes (Zhang et al., 2010). Further evidence for a major role for Hsp110 in HD is provided by the finding that Hsp110 and Hsp40 cooperatively prevent aggregation of polyQ-expanded Htt and associated neurodegeneration (Kuo et al., 2013).
The aggregation of mutant superoxide dismutase 1 (SOD1) has been linked to motor neuron degeneration in the inherited form of ALS. Addition of the ALS-associated SOD1 variant G85R (SOD1G85R) to giant squid axoplasm results in inhibition of axonal transport in this classic model (Song et al., 2013). Co-addition of Hsp110 rescued this transport defect, although it is not clear if the observed outcome was due to partnering with endogenous squid Hsp70 or passive chaperoning by the added Hsp110. In a corroborating mouse study, Hsp110−/− animals expressing SOD1G85R showed enhanced aggregation-dependent onset of ALS characteristics such as neural aggregates and reduced lifespan. Correspondingly, overexpression of Hsp110 resulted in reduced SOD1G85R aggregation and increased lifespan (Nagy et al., 2016). Despite strong evidence supporting involvement of Hsp110 in neurodegenerative disease progression, it is worth noting that humans express three distinct Hsp110 genes: Apg-1, Apg-2, and Hsp105α. The presence of multiple Hsp110s in neuromuscular tissues may complicate a full understanding of Hsp110 function in neurodegenerative disease due to functional redundancy. While the default interpretation may be to assume that Hsp110 is acting in the context of the disaggregase activity, it is difficult to reconcile this model with the fact that overexpression of Hsp110 alone is sufficient to inhibit aggregation and subsequent disease progression, likely in a super-stoichiometric manner. However, Hsp110 may be playing multiple, non-mutually exclusive roles in these scenarios: it may power disaggregation that is lost upon genetic deletion and passively sequester toxic or pre-toxic oligomers of disease-linked proteins when highly overexpressed (Fig. 2A, B). The generation of Hsp110 mutants that specifically abrogate NEF or substrate binding activity is required to deconvolute these two models.
Conclusions and Future Directions
The biological importance of Hsp110 as a key eukaryotic Hsp70 NEF is now well established. However, the in vivo relevance of its chaperone properties requires further investigation. Furthermore, details of substrate recognition by the Hsp110 SBD are lacking. Namely, more than 20 years after establishing that Hsp110 binds substrates, no structural details have been forthcoming regarding how and where peptide substrates bind. An X-ray or high-resolution cryoelectron microscopy structure would provide important insight into the Hsp110 chaperoning mechanism. It is generally accepted that Hsp110-class chaperones are better holdases than Hsp70s, and do so in an energy-independent manner. The recent findings that they also exhibit slightly altered peptide specificity relative to Hsp70 suggests both complementary biological functions as well as the possibility of novel binding characteristics and unique substrate profiles in vivo. It has been established that the protein quality control network plays a significant role in the onset and progression of various human diseases, notably neurodegenerative disorders of protein misfolding. To date, few studies have focused on delineating the roles of Hsp70 NEFs, and Hsp110 specifically, in proteopathies, but it is becoming abundantly clear that they will turn out to be major players in many of these diseases. Further work must determine whether nucleotide exchange activity is paramount and what role, if any, substrate binding plays in disease progression. To date, disease-linked mutations in Hsp110 have not been identified, in contrast to the case with the ER-localized NEF Sil1, where amino acid substitutions have been found that result in Marinesco-Sjögren syndrome (Senderek et al., 2005). As mentioned previously, this may be due to the presence of multiple Hsp110 genes in humans. Given the remarkable pace of discovery and generation of new understanding of the biological roles of the Hsp110 chaperone in recent years, it is anticipated that these outstanding questions will be answered soon and the information used to drive therapeutic intervention that exploits this powerful protein remodeler.
Acknowledgments
We apologize to the many investigators whose contributions we were unable to cite due to the brevity of this minireview. Work in the authors’ laboratory was supported by NIH grants GM074696, GM127287 and AG051046.
REFERENCES
- Abrams JL, Verghese J, Gibney PA, and Morano KA (2014). Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. J. Biol. Chem. 289, 13155–13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Fiaux J, Rampelt H, Druffel-Augustin S, and Bukau B (2008). Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc. Natl. Acad. Sci. U. S. A. 105, 16519–16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Rampelt H, Fiaux J, Druffel-Augustin S, and Bukau B (2010). The endoplasmic reticulum Grp170 acts as a nucleotide exchange factor of Hsp70 via a mechanism similar to that of the cytosolic Hsp110. J. Biol. Chem. 285, 12445–12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, and Hendershot LM (2014). The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s. J. Biol. Chem. 289, 2899–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Mann MJ, Scruggs F-L, Feige MJ, and Hendershot LM (2016). Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Mol. Cell 63, 739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher A, and Verghese J (2015). The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, and McCracken AA (1999). The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274, 3453–3460. [DOI] [PubMed] [Google Scholar]
- Dragovic Z, Broadley S Shomura Y., Bracher A., and Hartl FU. (2006). Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, and Subjeck JR (2000). The Hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones 5, 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu B, Moskophidis D, and Mivechi NF (2010). Loss of Hsp110 leads to age-dependent tau hyperphosphorylation and early accumulation of insoluble amyloid. Mol. Cell. Biol. 30, 4626–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia VM, Nillegoda NB, Bukau B, and Morano KA (2017). Substrate binding by the yeast Hsp110 nucleotide exchange factor and molecular chaperone Sse1 is not obligate for its biological activities. Mol. Biol. Cell 28, 2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, and Weissman JS (2003). Global analysis of protein expression in yeast. Nature 425, 737–741. [DOI] [PubMed] [Google Scholar]
- Glover JR, and Lindquist S (1998). Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82. [DOI] [PubMed] [Google Scholar]
- Goeckeler JL, Petruso AP, Aguirre J, Clement CC, Chiosis G, and Brodsky JL (2008). The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 582, 2393–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda NKC, Kandasamy G, Froehlich MS, Dohmen RJ, and Andréasson C (2013). Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc. Natl. Acad. Sci. U. S. A. 110, 5975–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda NKC, Kaimal JM, Kityk R, Daniel C, Liebau J, Öhman M, Mayer MP, and Andréasson C (2018). Nucleotide exchange factors Fes1 and HspBP1 mimic substrate to release misfolded proteins from Hsp70. Nat. Struct. Mol. Biol. 25, 83–89. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, and Hayer-Hartl M (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. [DOI] [PubMed] [Google Scholar]
- Hideyuki M, Takayoshi K, Hozumi T, Dai H, Tokichi M, and Chikako T (1993). Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene 132, 57–66. [DOI] [PubMed] [Google Scholar]
- Kaimal JM, Kandasamy G, Gasser F, and Andréasson C (2017). Coordinated Hsp110 and Hsp104 activities power protein disaggregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 37, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kityk R, Vogel M, Schlecht R, Bukau B, and Mayer MP (2015). Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 6, 8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kityk R, Kopp J, and Mayer MP (2018). Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69, 227–237.e4. [DOI] [PubMed] [Google Scholar]
- Kuo Y, Ren S, Lao U, Edgar BA, and Wang T (2013). Suppression of polyglutamine protein toxicity by co-expression of a heat-shock protein 40 and a heat-shock protein 110. Cell Death Dis. 4, e833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, and Hendrickson WA (2007). Insights into Hsp70 Chaperone Activity from a Crystal Structure of the Yeast Hsp110 Sse1. Cell 131, 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo RUH, Sharma SK, Priya S, Finka A, and Goloubinoff P (2013). Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J. Biol. Chem. 288, 21399–21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey GB, Gagné M, and Rappé AK (1998). π-Stacking Interactions. J. Biol. Chem. 273, 15458–15463. [DOI] [PubMed] [Google Scholar]
- Nagy M, Fenton WA, Li D, Furtak K, and Horwich AL (2016). Extended survival of misfolded G85R SOD1-linked ALS mice by transgenic expression of chaperone Hsp110. Proc. Natl. Acad. Sci. 113, 5424–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, et al. (2015). Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HJ, Easton DP, Murawski M, Kaneko Y, and Subjeck J (1999). The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J. Biol. Chem. 274, 15712–15718. [DOI] [PubMed] [Google Scholar]
- Polier S, Dragovic Z, Hartl FU, and Bracher A (2008). Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079. [DOI] [PubMed] [Google Scholar]
- Polier S, Hartl FU, and Bracher A (2010). Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J. Mol. Biol. 401, 696–707. [DOI] [PubMed] [Google Scholar]
- Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, and Bukau B (2012). Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31, 4221–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch JN, and Gestwicki JE (2014). Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 289, 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch JN, Zuiderweg ERP, and Gestwicki JE (2016). Non-canonical interactions between heat shock cognate protein 70 (Hsc70) and Bcl2-associated anthanogene (BAG) co-chaperones are important for client release. J. Biol. Chem. 291, 19848–19857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, and Bukau B (2006). Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosam M, Krader D, Nickels C, Hochmair J, Back KC, Agam G, Barth A, Zeymer C, Hendrix J, Schneider M, et al. (2018). Bap (Sil1) regulates the molecular chaperone BiP by coupling release of nucleotide and substrate. Nat. Struct. Mol. Biol. 25, 90–100. [DOI] [PubMed] [Google Scholar]
- Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andréasson C, Lindquist S, and Bukau B (2008). Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS One 3, e1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenz LE, Jin S, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R. (2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31(2):232–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, Breitbach-Faller N, Rudnik-Schöneborn S, Blaschek A, Wolf NI, Harting I, et al. (2005). Mutations in SIL1 cause Marinesco-Sjögren syndrome, a cerebellar ataxia with cataract and myopathy. Nat. Genet. 37, 1312–1314. [DOI] [PubMed] [Google Scholar]
- Shaner L, Trott A, Goeckeler JL, Brodsky JL, and Morano KA (2004). The function of the yeast molecular chaperone Sse1 Is mechanistically distinct from the closely related Hsp70 family. J. Biol. Chem. 279, 21992–22001. [DOI] [PubMed] [Google Scholar]
- Shaner L, Wegele H, Buchner J, and Morano KA (2005). The Yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 280, 41262–41269. [DOI] [PubMed] [Google Scholar]
- Shaner L, Sousa R, and Morano KA (2006). Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry 45, 15075–15084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J (2011). The Mammalian Disaggregase Machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 6, e26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Nagy M, Ni W, Tyagi NK, Fenton WA, Lopez-Giraldez F, Overton JD, Horwich AL, and Brady ST (2013). Molecular chaperone Hsp110 rescues a vesicle transport defect produced by an ALS-associated mutant SOD1 protein in squid axoplasm. Proc. Natl. Acad. Sci. 110, 5428–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C (2003). Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4, 49–60. [DOI] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, and Stirling CJ (2004). Coordinated activation of Hsp70 chaperones. Science 303, 98–101. [DOI] [PubMed] [Google Scholar]
- Trott A, Shaner L, and Morano KA (2005). The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics 170, 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzankov S, Wong MJH, Shi K, Nassif C, and Young JC (2008). Functional divergence between co-chaperones of Hsc70. J. Biol. Chem. 283, 27100–27109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, and Morano KA (2012). A lysine-rich region within fungal BAG domain-containing proteins mediates a novel association with ribosomes. Eukaryot. Cell 11, 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pezeshki AM, Yu X, Guo C, Subjeck JR, and Wang X-Y (2015). The endoplasmic reticulum chaperone GRP170: from immunobiology to cancer therapeutics. Front. Oncol. 4, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Sarbeng EB, Vorvis C, Kumar DP, Zhou L, and Liu Q (2012). Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J. Biol. Chem. 287, 5661–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam AY-W, Albanèse V, Lin H-TJ, and Frydman J (2005). Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280, 41252–41261. [DOI] [PubMed] [Google Scholar]
- Zhang S, Binari R, Zhou R, and Perrimon N (2010). A Genomewide RNA interference screen for modifiers of aggregates formation by mutant huntingtin in Drosophila. Genetics 184, 1165–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]


