Abstract
Background
Multiple interrelated pathways contribute to the pathogenesis of preeclampsia, and variants in susceptibility genes may play a role among Filipinos, an ethnically distinct group with high prevalence of the disease. The objective of this study was to examine the association between variants in maternal candidate genes and the development of preeclampsia in a Philippine population.
Methods
A case-control study involving 29 single nucleotide polymorphisms (SNPs) in 21 candidate genes was conducted in 150 patients with preeclampsia (cases) and 175 women with uncomplicated normal pregnancies (controls). Genotyping for the GRK4 and DRD1 gene variants was carried out using the TaqMan Assay, and all other variants were assayed using the Sequenom MassARRAY Iplex Platform. PLINK was used for SNP association testing. Multilocus association analysis was performed using multifactor dimensionality reduction (MDR) analysis.
Results
Among the clinical factors, older age (P < 1 × 10–4), higher BMI (P < 1 × 10–4), having a new partner (P = 0.006), and increased time interval from previous pregnancy (P = 0.018) associated with preeclampsia. The MDR algorithm identified the genetic variant ACVR2A rs1014064 as interacting with age and BMI in association with preeclampsia among Filipino women.
Conclusions
The MDR algorithm identified an interaction between age, BMI and ACVR2A rs1014064, indicating that context among genetic variants and demographic/clinical factors may be crucial to understanding the pathogenesis of preeclampsia among Filipino women.
Electronic supplementary material
The online version of this article (10.1186/s12884-018-2152-z) contains supplementary material, which is available to authorized users.
Keywords: Association study, Multifactor dimensionality reduction, Philippines, Preeclampsia, Single nucleotide polymorphism
Background
Hypertensive disorders of pregnancy account for 36.7% of all maternal deaths in the Philippines [1], which is much higher than the worldwide rate of 18% [2]. Included among these hypertensive diseases affecting pregnant women is preeclampsia, a severe and diverse disorder that is associated with life-threatening multi-organ maternal complications and which causes serious feto-placental problems. It accounted for 22.5% of hypertensive patient admissions at the hospital where this study was conducted [3].
Preeclampsia is a multifactorial disease, with both genetic and environmental factors contributing to its development. Multiple interrelated pathways have been suggested to contribute to its pathogenesis. Previous studies have tested genes with potential biological relevance in specific pathways to ascertain whether certain variants influence the disease process. The biological pathways impacted by preeclampsia include but are not limited to aberrant placental development and dysfunctional hemodynamic and renal functions, impaired immune function, free radical dysregulation and lipid peroxidation, and defects in coagulation and fibrinolysis. We have previously found variants of the VEGF-A and VEGFR1 genes to associate with preeclampsia among Filipinos, an ethnically distinct group with high prevalence [4]. These genes are important in angiogenesis, a critical process in the establishment of normal pregnancy and in preeclampsia.
The effect of single gene variation will likely be contingent on other genetic variations (gene-gene interaction, or epistasis) and environmental factors (gene-environment interaction). Since many genes and environmental factors interact to cause multifactor and polygenic diseases, including preeclampsia, the effect of any single gene may be too small to be detected using traditional statistical methods, which do not take these interactions into account. The multifactor dimensionality reduction (MDR) algorithm has been designed as an alternative to traditional statistical methods to deal with high-order gene/factor interactions [5, 6]. MDR has many other advantages over traditional methods. It is model-free, i.e., it does not assume any particular genetic model, and requires only a small sample size that can be used for case-control studies [5–7]. MDR has been successfully applied in detecting gene-gene interactions for a number of clinical phenotypes, which include bronchial asthma [8], autism [9], essential hypertension [10, 11], and type II diabetes [12].
In this study, we evaluated the role of contextual effects in the development of preeclampsia among Filipino women by analyzing 29 previously reported susceptibility SNPs found in 21 genes. The 21 genes are involved in various pathways that regulate the processes implicated in the development of preeclampsia and are as follows: endothelial/angiogenesis (VEGFA, VEGFC, VEGFR1, VEGFR3); dopaminergic system (GRK4, DRD1); renin-angiotensin system (AGT); immunity and inflammation (ERAP2, CTLA4, IL1A, TNSF13B); lipid metabolism (LPL); oxidative stress and detoxification (eNOS, CYP1A2, PON1, EPHX1, GSTP1); key signaling proteins (ACVR2A), hormone and neurotransmitter regulation (COMT) and protein biosynthesis (MTR and MTRR).
Methods
This is a case-control study that included 381 individuals. Of these, 56 were removed in the final analysis because they had more than 2 genotypes missing. Of the 325 that remained, 150 were patients with preeclampsia (cases) and 175 were women with uncomplicated normal pregnancies (controls). Subjects were recruited upon admission to the hospital and were followed up until 6 weeks after delivery. Subjects included in the normal pregnancy control group had blood pressures ≤120/80 mmHg, consistent with the latest guidelines on hypertension [13]. Blood pressure was measured according to the Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure [14] and verified twice with at least a 4-h interval. Exclusion criteria for the control group were a history of hypertension and pregnancy-induced hypertension, multiple pregnancy, molar pregnancy, personal and family history of diabetes mellitus, ischemic heart disease, cerebrovascular accident, and renal disease.
Included in the preeclampsia group were patients who had a resting systolic blood pressure ≥ 140 mmHg and/or a diastolic blood pressure ≥ 90 mmHg and had proteinuria after 20 weeks of gestation. Proteinuria was defined as ≥300 mg protein in a 24-h urine collection or a urine protein dipstick of ≥2+. Exclusion criteria for the preeclampsia group were a history of hypertension, renal disease, proteinuria before the 20th week of pregnancy, multiple pregnancy, diabetes mellitus, ischemic heart disease, cerebrovascular accident, and renal disease.
This study was undertaken in accordance with the Declaration of Helsinki at the Department of Obstetrics and Gynecology of the University of the Philippines, Philippine General Hospital (UP-PGH) in Manila. All subjects provided informed written consent. The study was approved by the UP-PGH Ethics Review Board.
SNP selection, blood sampling, DNA extraction, and genotyping
The candidate genes and respective variants were selected from different pathophysiological pathways involved in the development of preeclampsia and based on previous association studies. The ACVR2A SNPs in particular have been shown to be associated with preeclampsia in a study involving Brazilian [15] and Norwegian women [16]. Criteria for SNP selection included SNPs for which an association with preeclampsia has been reported in other ethnic populations and those that are also supported by the platform used for genotyping. The gene variants in relevant pathways include: endothelial/angiogenesis (VEGFA [17–19], VEGFC [20], VEGFR1 [20–22]; renin-angiotensin system (AGT [23–25]); immunity and inflammation (ERAP2 [26], CTLA4 [27, 28], IL1A [29, 30], TNSF13B [31]); lipid metabolism (LPL [27, 32]); oxidative stress and detoxification (eNOS [33, 34], CYP1A2 [35], PON1 [36–39], EPHX1 [40, 41], GSTP1 [42]); key signaling proteins (ACVR2A [15, 16]), hormone and neurotransmitter regulation (COMT [43, 44]) and protein biosynthesis (MTR and MTRR) [45–48]. Also included are SNPs in the dopaminergic system (GRK4 [49–52] and DRD1 [52]), which have been established to be associated strongly with hypertension.
Venous blood samples for DNA extraction and genotyping were collected after a definitive diagnosis of preeclampsia or normal pregnancy. Three milliliters of venous blood were extracted at the time of hospital admission by venipuncture at the antecubital fossa, collected in a vacutainer with EDTA, and stored at 4 °C until DNA extraction. DNA was extracted from the peripheral blood mononuclear cells using the QIAamp® DNA Mini Kit. DNA purity and quantity were determined using a Nanodrop 2000 spectrometer (Thermo Scientific, Waltham, MA, USA). The DNA was stored at 4 °C until genetic profiling. Genotyping of the GRK4 and DRD1 variants was carried out using the TaqMan Assay at the University of Maryland Biopolymer-Genomics Core Facility. Genotyping of the other genes was carried out using the Sequenom MassARRAY Iplex Platform at the Center for Genomic Sciences of the University of Hong Kong. Repeat genotyping for 16 samples was performed for quality control.
Statistical analysis
Statistical analysis for the clinical parameters was performed using STATA software, version 14.1 [53]. All values were expressed as mean/median ± standard error, while the association of known categorical risk factors was analyzed using Pearson’s chi-square test. Odds ratios (OR) were calculated to determine the odds of developing preeclampsia when the individual had the clinical factor of interest. OR was used for binary logistic regression and multinomial logistic regression. Minor allele frequencies and Hardy-Weinberg Equilibrium were calculated for each SNP using PLINK 1.9. Of the 29 SNPs, 6 were removed from further analysis (AGT, LPL, FLT1, ERAP1, FLT4 and TNFSF13B) because these SNPs were monomorphic, or had a minor allele frequency below 5%. Accumulated/average Cross Validation testing, training, consistency and permutation P values were calculated using MDR [6, 54]. The MDR algorithm and ViSEN software [55] were applied to the genetic data to enable the detection and characterization of epistatic SNP-SNP interactions and SNP-clinical factor (age and BMI) interaction. To identify a correct multi-locus model, the Acc. CV testing (Accumulated/average Cross Validation testing) and CV Consistency (Cross Validation Consistency) were calculated for each model.
Results
Normal pregnancies and preeclampsia outcomes differed in several demographic and clinical characteristics. Mothers with normal pregnancies were significantly younger (25.2 vs. 30.3; P = < 1 × 10− 4) (Table 1) and had lower BMI (21.75 vs. 23.5; P = < 1 × 10− 4) (Table 1). The mean interval from last pregnancy was shorter in normal outcomes (2.03 years vs. 2.87 years; P = 0.018). Having a new partner also associated with preeclampsia (P = 0.006). However, neither smoking (P = 0.957) nor gravidity (P = 0.435) associated with preeclampsia (Table 2).
Table 1.
Age, BMI, and interval year are risk factors for preeclampsia
| Variable | Category | Mean/Median | P value |
|---|---|---|---|
| Age | NP | Mean: 25.2 ± 0.45 Median: 24 | |
| PE | Mean: 30.3 ± 0.53 Median: 31 | < 1 × 10−4* | |
| BMI3 | NP | Mean: 21.75 ± 0.19 Median: 21.39 | |
| PE | Mean: 23.5 ± 0.21 Median: 23.31 | < 1 × 10−4* | |
| Interval Year | NP | Mean: 2.03 ± 0.21 Median: 1 | |
| PE | Mean: 2.87 ± 0.28 Median: 1 | 0.018* |
Data are expressed as mean ± SEM and median; odds ratio (OR) used for binary logistic regression
NP Normal pregnancy, PE Preeclampsia, BMI Body mass index
*P values < 0.05 are statistically significant
The table summarizes the demographic, clinical characteristics, and risk factors of preeclampsia patients and controls with normal pregnancy. The preeclampsia patients were generally older, had higher BMI, and longer interval year from previous pregnancy compared with control subjects.
Table 2.
A new partner is a risk factor for preeclampsia
| Variable | Category | No | Yes | Pearson’s correlation | P value | OR |
|---|---|---|---|---|---|---|
| Smoking | NP | 181 | 17 | |||
| PE | 167 | 16 | 0.003 | 0.957 | 1.020077 | |
| Alcohol | NP | 184 | 14 | |||
| PE | 165 | 18 | 1.737 | 0.333 | 1.433766 | |
| Nulliparous | NP | 109 | 89 | |||
| PE | 108 | 75 | 0.61 | 0.435 | 0.8504994 | |
| Previous | New | |||||
| Partner | NP | 173 | 25 | |||
| PE2 | 140 | 43 | 7.67 | 0.006* | 2.125 |
Data used odds ratio (OR) for binary logistic regression and for multinomial logistic regression
NP Normal pregnancy, PE Preeclampsia
*P values < 0.05 are statistically significant
Among the risk factors analyzed (smoking, alcoholic beverage consumption, nulliparity, and having a new partner) in preeclampsia patients and controls with normal pregnancy, only having a new partner was associated with increased risk of preeclampsia.
No SNPs associated with preeclampsia in the unadjusted analyses (Table 3). Association of single SNPs was also run adjusting for age, BMI, interval between pregnancies, and new partner (Additional file 1: Table S1). After adjusting for these covariates, one SNP reached nominal statistical significance (VEGF-A rs3025039; P = 0.022). This was not significant after adjusting for multiple testing (Bonferroni threshold P = 0.0023).
Table 3.
Gene variants included in the study
| Chr | SNP | Gene | Product (processes involved in) | MAF | OR | Allelic P value |
Genotypic P value |
|---|---|---|---|---|---|---|---|
| 1 | rs1051740 | EPHX1 | Epoxide hydrolase (xenobiotic metabolism) | 0.4547 | 1.039 | 0.8109 | 0.6148 |
| 1 | rs699 | AGT | Angiotensinogen (products elicit vasoconstriction) | 0.1277 | 1.098 | 0.6899 | NA |
| 1 | rs1805087 | MTR | 5-Methyltetrahydrofolate-Homocysteine Methyltransferase (methionine biosynthesis) | 0.1357 | 1.013 | 0.9568 | NA |
| 2 | rs3783550 | IL1A | Interleukin 1-alpha (immunity, inflammation, hematopoiesis) | 0.2031 | 1.003 | 0.988 | 0.7627 |
| 2 | rs1014064 | ACVR2A | Activin A Receptor Type 2A (growth and differentiation) | 0.4414 | 0.8699 | 0.3805 | 0.556 |
| 2 | rs2161983 | ACVR2A | 0.4385 | 0.8921 | 0.4717 | 0.5188 | |
| 2 | rs231775 | CTLA-4 | Cytotoxic T-Lymphocyte Associated Protein 4 (inhibition of immune responses) | 0.4462 | 1.22 | 0.216 | 0.4626 |
| 4 | rs2960306 | GRK4 | G protein-coupled receptor kinase 4 (receptor desensitization) | 0.0679 | 1.763 | 0.0709 | NA |
| 4 | rs1024323 | GRK4 | 0.1354 | 1.26 | 0.3133 | NA | |
| 4 | rs1801058 | GRK4 | 0.4599 | 1.024 | 0.8797 | 0.8192 | |
| 4 | rs7664413 | VEGF-C | Vascular endothelial growth factor C (lymphangiogenesis) | 0.2436 | 1.316 | 0.1412 | 0.181 |
| 5 | rs1801394 | MTRR | Methionine synthase reductase (methionine biosynthesis) | 0.2754 | 0.9219 | 0.645 | 0.8572 |
| 5 | rs2549782 | ERAP 2 | Endoplasmic reticulum aminopeptidase 2 (antigen processing) | 0.4046 | 1.097 | 0.5622 | 0.8404 |
| 5 | rs4532 | DRD1 | Dopamine D1 receptor (sodium transport, blood pressure regulation) | 0.2105 | 1.01 | 0.9594 | 0.9148 |
| 6 | rs2010963 | VEGF-A | Vascular endothelial growth factor A (angiogenesis, vasculogenesis, endothelial cell growth & migration) | 0.223 | 0.8585 | 0.4495 | NA |
| 6 | rs3025039 | VEGF-A | 0.1188 | 0.6282 | 0.0626 | NA | |
| 7 | rs662 | PON1 | Paraoxonase 1 (inactivation of organophosphates, inhibition of atherosclerosis formation) | 0.4306 | 0.9923 | 0.9612 | 0.9841 |
| 7 | rs1799983 | NOS3 | Endothelial NOS (vascular relaxation, antioxidant activity) | 0.1813 | 0.9324 | 0.7342 | 0.829 |
| 11 | rs1695 | GSTP1 | Glutathione S-Transferase Pi 1 (xenobiotic metabolism) | 0.2954 | 1.073 | 0.6809 | 0.8688 |
| 13 | rs12584067 | VEGFR-1 | Vascular endothelial growth factor receptor 1 (cell proliferation and differentiation | 0.0776 | 1.282 | 0.3978 | NA |
| 13 | rs722503 | VEGFR-1 | Vascular endothelial growth factor receptor 1 (cell proliferation and differentiation | 0.1660 | 0.8019 | 0.3593 | NA |
| 15 | rs2470890 | CYP1A2 | Cytochrome P450 1A2 (xenobiotic metabolism) | 0.0923 | 0.8182 | 0.4643 | NA |
| 22 | rs4633 | COMT | Catechol-O-methyltransferase (degrades catecholamines) | 0.1692 | 1.056 | 0.7962 | NA |
Chr Chromosome, SNP Single nucleotide polymorphism, MAF Minor allele frequencies, OR Odds ratio, NA Not appplicable
The alleles and genotypes of the SNPs are not associated with preeclampsia when analyzed using PLINK. NA corresponds to the inability to determine a genotypic P value, due to a small number of counts in at least 1 of the 3 genotypes (i.e., AA/AT/TT not present, only AA/AT), thus, only allelic P value is given.
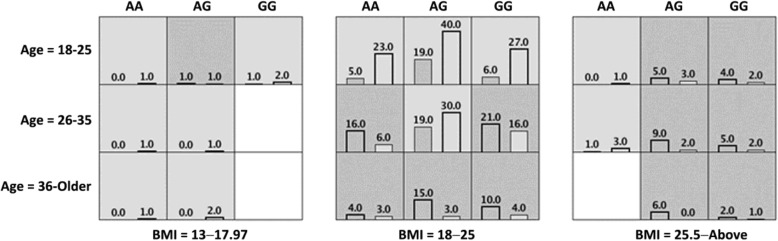
Context-dependent effects were assessed using MDR. Only one model that included a genetic variant, as well as the significant demographic and clinical variables, was statistically significant; it included maternal age as three categories (18–25; 26–35; 36-Older); maternal BMI as three categories (13–17.97; 18–25; 25.1-Above) and rs1014064 (cross validation testing prediction = 60.95%; permutation P = 0.005 and cross validation consistency = 7/10) (Table 4). Upon examining the model, it was evident that the genetic effect was present primarily in the middle range of BMI (18–25) (Fig. 1). In low BMI ranges (13–17.97), the outcome was dominated by a protective effect and in high BMI ranges (25.1-above) by a risk-increasing effect of body mass. When re-analyzing only the middle range of BMI (18–25), genetics appeared to play a significant non-additive role (cross validation testing prediction = 64.88%; permutation p = < 1 × 10− 4 and cross validation consistency = 10/10; Fig. 1). There was no statistical epistasis detected using ViSEN.
Table 4.
MDR analysis of genetic variants, adjusting for age and BMI
| SNP rs1014064 | GENE ACVR2A | ||
|---|---|---|---|
| INTERACTION | Acc. CV Testing | CV Consistency | |
| Age | 0.6457 | 10/10 | |
| Age, BMI | 06729 | 10/10 | |
| rs1014064, Age, BMI | 0.6095 | 7/10 | P = 0.005* |
There is an interaction between ACVR2A rs1014064, age, and BMI
MDR Multifactor dimensionality reduction, SNP Single nucleotide polymorphism, BMI Body mass index
*P value < 0.05 is statistically significant
Fig. 1.
MDR model for interaction of rs1014064, age, and BMI. Each cell shows counts of preeclampsia on the left and normal pregnancy on the right. When re-analyzing only the middle range of BMI (18–25), genetics appeared to play a significant non-additive role in predicting preeclampsia, P < 1 × 10− 4, cross validation testing prediction = 64.88%, cross validation consistency = 10/10; MDR, multifactor dimensionality reduction; BMI, body mass index
The genetic data were analyzed for epistasis using MDR and ViSEN statistical software. Table 3 summarizes the different gene variants that were included in the analysis, as well as pertinent information for each, including the gene product and the processes in which it is involved, chromosome location, minor allele frequencies (MAF), and odds ratio (OR). Monomorphic SNPs (VEGFR1 rs7335588, AGT rs41271499, and LPL rs268) and with MAF < 0.05 (VEGFR3 rs307826, ERAP2 rs17408150, and TNFSF13B rs16972194) were excluded from analyses. Genotype frequencies for all the SNPs were in Hardy-Weinberg equilibrium.
Discussion
Non-linear interactions among multiple genetic and environmental or clinical factors are now understood to be important components in understanding the underlying pathogenesis, especially when considering the genetic bases of complex diseases such as preeclampsia. The MDR algorithm is a well-known data mining strategy that provides an improved representation of the genotypic and phenotypic data and enables better detection of higher-order interactions, such as epistatic interactions [6]. In the current study, MDR identified a four-locus model that underscores a possible interaction among rs1014064 (ACVR2A), rs7664413 (VEGF-C), rs2549782 (ERAP2), and rs662 (PON1) variants when un-adjusted for age or BMI (Additional file 2: Table S2). However, when age and BMI were adjusted for, these effects disappeared. A significant interaction was found in a model involving the genetic variant rs1014064 (ACVR2A) and the demographic and clinical variables, age and BMI.
The significant gene identified, ACVR2A, encodes the receptor for Activin A. ACVR2A expression in the placenta throughout pregnancy indicates its possible role in the regulation of placental development and function [56]. Initial studies have shown a linkage between preeclampsia and various parts of chromosome 2, where the ACVR2A gene is localized. The first reported locus for preeclampsia that met the criteria for genome-wide association significance was seen in chromosome 2p13 and 2q23 in a study involving Icelandic families, representing 343 affected women [57]. Two other genome-wide association studies identified other loci in chromosome 2 distinct from those seen in the initial study, i.e., 2p25 in 15 families with 49 affected women from Finland [58] and 2p11–12 and 2q22 involving 34 families, representing 121 affected women from Australia and New Zealand [59]. With the reported significant linkage to chromosome 2q22, the same group identified the ACVR2A gene as a strong positional candidate gene [60].
The MDR analysis identified the ACVR2A rs1014064, an intronic variant (A to G), as the only significant variant. This variant associated with preeclampsia in a large Norwegian population-based study (the HUNT study), together with the other ACVR2A variant (rs2161983) [16], which was also evaluated and found not associated with preeclampsia in this study, and with early onset preeclampsia in a Brazilian population [15].
MDR has been used to identify the important role of epistasis in polygenic disorders, such as sporadic breast cancer [6] and essential hypertension. A two-locus model including ACE and GRK4 successfully predicted the blood pressure phenotype 70.5% of the time [10]. A genetic model based on the three common GRK4 SNPs was 94.4% predictive of salt-sensitive hypertension, while a single-locus model with only the GRK4 A142V variant was 78.4% predictive. By contrast, for low-renin hypertension, a two-locus model that includes the GRK4 A142V variant and cytochrome P450 11B2 (CYP11B2) C-344 T was 77.8% predictive [11]. These results reflect the differences in the underlying genetics and the crucial role of epistasis in the development of the different hypertension-related phenotypes. Considering the spectrum of the clinical presentation of preeclampsia, it is conceivable that different phenotypes of the disease may involve specific gene polymorphisms, i.e., locus heterogeniety. In fact, the presence of severe forms of preeclampsia, HELLP syndrome and eclampsia, have been suggested to have their own set of predisposing gene variants. It is therefore important to know which specific genes contribute the most to their development.
We also used ViSEN software, which provides a global interaction map to identify and corroborate risk-associated SNPs, visualize putative gene interactions and generate an interaction or concept map for preeclampsia. Similar to what we observed using MDR, we detected no statistically significant two- and three-way epistatic interactions with ViSEN. Due to its limitation to analyze only up to three-way epistasis, the four-locus model that we observed with MDR was undetected. Moreover, ViSEN as a statistical tool has its own limitations, e.g., the statistical epistasis quantifications in ViSEN only consider discrete traits and cannot incorporate measures on continuous traits like age and BMI [55].
In the analysis of genetic datasets, an important consideration is the power of analytical methods to identify accurate predictive models of disease. The MDR approach overcomes the common setbacks found in other methods. It is non-parametric, model-free, and can identify high-order gene-gene interactions [6, 54, 61]. It retains its power to analyze in the presence of genotyping error and missing data (up to 5%). However, it has its own limitations, including a decrease in power in the presence of phenocopies and genetic heterogeneity [61].
With respect to marginal effects, one of the SNPs we genotyped in VEGF-A, rs3025039, that was previously identified as associating with preeclampsia in the Philippines [4] also showed a marginal association in our data set when adjusting for covariates (p = 0.022). Although this result would not stand after adjusting for multiple testing, as a replication it provided additional evidence that this variant confers pre-eclampsia risk especially since the direction of effect was the same in the present and the previous studies. A second SNP in this gene, rs722503, that was previously reported to be associated with preeclampsia, but only in pregnancies with women over 40, did not show evidence for significance in the current study (Additional file 1: Table S1). Of note, in the age and BMI adjusted model for rs722503 the p value did get smaller as compared to the unadjusted model, which is consistent with an age-related effect. This, however, was not surprising as very few pregnancies involved women over 40 (4). In addition, this SNP did not appear in our MDR analyses when all other SNPs were included.
A notable limitation of this study is the non-inclusion of other SNPs that have been shown to be associated with preeclampsia in specific ethnic groups, or more importantly in multi-gene meta-analysis studies involving different ethnic groups [25, 27, 62, 63] and in multigene association studies [64]. These include the gene variants of FV, F2, ACE, SERPINE1, AGTR1, MTHFR, and MMP-9. The ACE gene variant is an insertion/deletion polymorphism, which cannot be detected by the method used for genotyping in our studies. The other genes, although included in the initial list for analysis, were eventually dropped from the analysis due to technical problems. These genes, however, should be included in future studies.
Conclusions
Preeclampsia is a multifactorial disease, with both genetic and environmental factors contributing to its development. Genetic variants from multiple, interrelated pathways have been suggested to contribute to the pathogenesis of the disease. The MDR algorithm enabled the analysis of high-order gene/factor interactions and identified ACVR2A rs1014064 as important in modulating preeclampsia risk among older Filipino women with a middle-range BMI.
Additional files
Table S1. Association of SNPs, adjusting for statistically significant risk factors. (DOCX 16 kb)
Table S2. MDR analysis of genetic variants, without adjusting for age and BMI. (DOCX 14 kb)
Acknowledgements
Not Applicable.
Funding
This study was supported by a grant from University of the Philippines Emerging Interdisciplinary Research (EIDR) Program and the Philippine Genome Center; and in part by the authors’ respective grants: NIH CTSA grant TL1 TR000441 and the Gilliam Fellowship from the Howard Hughes Medical Institute (GRT); a minigrant from the National Kidney Foundation of Maryland (VMV); NIH grants R37HL023081, HL092196 (PAJ): NIH grant R01LM010098 and a grant from March of Dimes (SMW). Funding bodies had no role in the design of the study and collection, analysis, interpretation of data, and in writing the manuscript.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ACVR2A
Activin A Receptor Type 2A
- AGT
Angiotensinogen
- COMT
Catechol-O-methyltransferase
- CTLA4
Cytotoxic T-Lymphocyte Associated Protein 4
- CYP1A2
Cytochrome P450 family 1 subfamily A member 2
- DRD1
Dopamine D1 receptor
- eNOS
Endothelial nitric oxide synthase
- EPHX1
Epoxide hydrolase
- ERAP2
Endoplasmic reticulum aminopeptidase 2
- GRK4
G protein-coupled receptor kinase 4
- GSTP1
Glutathione S-Transferase Pi 1
- IL1A
Interleukin 1-alpha
- LPL
Lipoprotein lipase
- MDR
Multifactor dimensionality reduction
- MTR
5-Methyltetrahydrofolate-Homocysteine Methyltransferase
- MTRR
Methionine synthase reductase
- PON1
Paraoxonase 1
- SNPs
Single nucleotide polymorphisms
- TNSF13B
Tumor necrosis factor ligand superfamily member 13B
- VEGFA
Vascular endothelial growth factor A
- VEGFC
Vascular endothelial growth factor C
- VEGFR1
Vascular endothelial growth factor receptor 1
- VEGFR3
Vascular endothelial growth factor receptor 3
Authors’ contributions
MDA, VMV PAJ, CPS conceived and designed the study; MDA, JAN performed the experiments; LDB clinical input; JAN contributed in the initial analysis, interpretation of results, and manuscript preparation; GRT, SMW analyzed the data; MDA, GRT wrote the final paper; VMV, CPS, PAJ, SMW, review and editing; CPS, PAJ, SMW, VMV, funding acquisition. All authors read and approved the final draft of the manuscript.
Ethics approval and consent to participate
This study was approved by the University of the Philippines, Philippine General Hospital (UP-PGH) Ethics Review Board. All subjects provided informed written consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Melissa D. Amosco, Email: mdamosco@up.edu.ph
Gloria R. Tavera, Email: gloria.tavera@case.edu
Van Anthony M. Villar, vvillar@email.gwu.edu
Justin Michael A. Naniong, Email: jananiong@mbb.upd.edu.ph
Lara Marie G. David-Bustamante, Email: laraob@yahoo.com
Scott M. Williams, Email: smw154@case.edu
Cynthia P. Palmes-Saloma, Email: cpsaloma@up.edu.ph
References
- 1.Department of Health - National Epidemiology Center. The 2013 Philippine health statistics. http://www.dohgovph/sites/default/files/publications/2013PHScompressed_0pdf. Accessed 5 Apr 2018.
- 2.Khan KS, Wojdyla D, Say L, Gulmezoglu AM, Van Look PF. WHO analysis of causes of maternal death: a systematic review. Lancet. 2006;367:1066–1074. doi: 10.1016/S0140-6736(06)68397-9. [DOI] [PubMed] [Google Scholar]
- 3.David-Ona D, De Castro DM, Baltazar AC. The distribution of hypertension in the Philippine general hospital after 4 decades (a comparative study) Acta Med Philipp. 2013;47(3):49–52. [Google Scholar]
- 4.Amosco M, Villar VA, Naniong JM, David-Bustamante LM, Jose PA, Palmes-Saloma CP. VEGF-A and VEGFR1 SNPs associate with preeclampsia in a Philippine population. Clin Exp Hypertens. 2016;38(7):578–585. doi: 10.3109/10641963.2016.1174252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19(3):376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 6.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards TL, Lewis K, Velez DR, Dudek S, Ritchie MD. Exploring the performance of multifactor dimensionality reduction in large scale SNP studies and in the presence of genetic heterogeneity among epistatic disease models. Hum Hered. 2009;67:183–192. doi: 10.1159/000181157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan IH, Leung TF, Tang NL, Li CY, Sung YM, Wong GW, et al. Gene-gene interactions for asthma and plasma total IgE concentration in Chinese children. J Allergy Clin Immunol. 2006;117:127–133. doi: 10.1016/j.jaci.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Ma DQ, Whitehead PL, Menold MM, Martin ER, Ashley-Koch AE, Mei H, et al. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am J Hum Genet. 2005;77:377–388. doi: 10.1086/433195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams SM, Ritchie MD, Phillips JA, Dawson E, Prince M, Dzhura E, et al. Multilocus analysis of hypertension: a hierarchical approach. Hum Hered. 2004;57:28–38. doi: 10.1159/000077387. [DOI] [PubMed] [Google Scholar]
- 11.Sanada H, Yatabe J, Midorikawa S, Hashimoto S, Watanabe T, Moore JH, et al. Single-nucleotide polymorphisms for diagnosis of salt-sensitive hypertension. Clin Chem. 2006;52:352–360. doi: 10.1373/clinchem.2005.059139. [DOI] [PubMed] [Google Scholar]
- 12.Cho YM, Ritchie MD, Moore JH, Park JY, Lee KU, Shin HD, et al. Multifactor-dimensionality reduction shows a two-locus interaction associated with type 2 diabetes mellitus. Diabetologia. 2004;47:549–554. doi: 10.1007/s00125-003-1321-3. [DOI] [PubMed] [Google Scholar]
- 13.Whelton PK, Carey RM. The 2017 clinical practice guideline for high blood pressure. JAMA. 2017;318(21):2073–2074. doi: 10.1001/jama.2017.18209. [DOI] [PubMed] [Google Scholar]
- 14.Chobanian AV, Bakris GL, Black HR, Cushman CC, Green LA, Izzo JL, et al. The National High Blood Pressure Education Program Coordinating Committee. Seventh report of the joint National Committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira LC, Gomes CE, Araújo AC, Bezerra PF, Duggal P, Jeronimo SM. Association between ACVR2A and early-onset preeclampsia: replication study in a northeastern Brazilian population. Placenta. 2015;36(2):186–190. doi: 10.1016/j.placenta.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Roten LT, Johnson MP, Forsmo S, Fitzpatrick E, Dyer TD, Brennecke SP, et al. Association between the candidate susceptibility gene ACVR2A on chromosome 2q22 and pre-eclampsia in a large Norwegian population-based study (the HUNT study) Eur J Hum Genet. 2009;17:250–257. doi: 10.1038/ejhg.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res. 2000;37:443–448. doi: 10.1159/000054076. [DOI] [PubMed] [Google Scholar]
- 18.Shim JY, Jun JK, Jung BK, Kim SH, Won HS, Lee PR, et al. Vascular endothelial growth factor gene +936 C/T polymorphism is associated with preeclampsia in Korean women. Am J Obstet Gynecol. 2007;197:271–274. doi: 10.1016/j.ajog.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 19.Bányász I, Szabo S, Bokodi G, Vannay A, Vasarhelyi B, Szabo A, et al. Genetic polymorphisms of vascular endothelial growth factor in severe pre-eclampsia. Mol Hum Reprod. 2006;12:233–236. doi: 10.1093/molehr/gal024. [DOI] [PubMed] [Google Scholar]
- 20.Srinivas SK, Morrison AC, Andrela CM, Elovitz MA. Allelic variations in angiogenic pathway genes are associated with preeclampsia. Am J Obstet Gynecol. 2010;202:445.e1–445.11. doi: 10.1016/j.ajog.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 21.Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt-1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003;111:649–658. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergmann A, Ahmad S, Cudmore M, Gruber AD, Wittschen P, Lindenmaier W, et al. Reduction of circulating soluble Flt-1 alleviates preeclampsia-like symptoms in a mouse model. J Cell Mol Med. 2010;14:1857–1867. doi: 10.1111/j.1582-4934.2009.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward K, Hata A, Jeunemaitre X, Helin C, Nelson L, Namikawa C, et al. A molecular variant of angiotensinogen associated with preeclampsia. Nat Genet. 1993;4:59–61. doi: 10.1038/ng0593-59. [DOI] [PubMed] [Google Scholar]
- 24.Sethi AA, Nordestgaard BG, Tybjaerg-Hansen A. Angiotensinogen gene polymorphism, plasma angiotensinogen, and risk of hypertension and ischemic heart disease: a meta-analysis. Arterioscler Thromb Vasc Biol. 2003;23:1269–1275. doi: 10.1161/01.ATV.0000079007.40884.5C. [DOI] [PubMed] [Google Scholar]
- 25.Staines-Urias E, Paez MC, Doyle P, Dudbridge F, Serrano NC, Ioannidis JP, et al. Genetic association studies in pre-eclampsia: systematic meta-analyses and field synopsis. Int J Epidemiol. 2012;41:1764–1775. doi: 10.1093/ije/dys162. [DOI] [PubMed] [Google Scholar]
- 26.Johnson MP, Roten LT, Dyer TD, East CE, Forsmo S, Blangero J, et al. The ERAP2 gene is associated with preeclampsia in Australian and Norwegian populations. Hum Genet. 2009;126:655–666. doi: 10.1007/s00439-009-0714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buurma A, Turner R, Driessen A, Mooyaart A, Schoones J, Bruijn JA, et al. Genetic variants in pre-eclampsia: a meta-analysis. Pregnancy Hypertens. 2012;2:204. doi: 10.1016/j.preghy.2012.04.051. [DOI] [PubMed] [Google Scholar]
- 28.Jääskeläinen E, Toivonen S, Keski-Nisula L, Paattiniemi EL, Helisalmi S, Punnonen K, et al. CTLA-4 polymorphism 49A-G is associated with placental abruption and preeclampsia in Finnish women. Clin Chem Lab Med. 2008;46:169–173. doi: 10.1515/CCLM.2008.034. [DOI] [PubMed] [Google Scholar]
- 29.Cotran RS, Pober JS. Cytokine-endothelial interactions in inflammation, immunity, and vascular injury. J Am Soc Nephrol. 1990;1(3):225–235. doi: 10.1681/ASN.V13225. [DOI] [PubMed] [Google Scholar]
- 30.Goddard KA, Tromp G, Romero R, Olson JM, Lu Q, Xu Z, et al. Candidate-gene association study of mothers with preeclampsia and their infants, analyzing 775 SNPs in 190 genes. Hum Hered. 2007;63(1):1–16. doi: 10.1159/000097926. [DOI] [PubMed] [Google Scholar]
- 31.Fenstad MH, Johnson MP, Roten LT, Aas PA, Forsmo S, Klepper K, et al. Genetic and molecular functional characterization of variants within TNFSF13B, a positional candidate preeclampsia susceptibility gene on 13q. PLoS One. 2010;5(9). 10.1371/journal.pone.0012993. [DOI] [PMC free article] [PubMed]
- 32.Yang W, Huang J, Ge D, Yao C, Duan X, Shen Y, et al. Lipoprotein lipase gene is in linkage with blood pressure phenotypes in Chinese pedigrees. Hum Genet. 2004;115:812. doi: 10.1007/s00439-004-1108-8. [DOI] [PubMed] [Google Scholar]
- 33.Savvidou MD, Vallance PJ, Nicolaides KH, Hingorani AD. Endothelial nitric oxide synthase gene polymorphism and maternal vascular adaptation to pregnancy. Hypertension. 2001;38:1289–1293. doi: 10.1161/hy1201.097305. [DOI] [PubMed] [Google Scholar]
- 34.Kobashi G, Yamada H, Ohta K, Kato E, Ebina Y, Fujimoto S. Endothelial nitric oxide synthase gene (NOS3) variant and hypertension in pregnancy. Am J Med Genet. 2001;103:241–244. doi: 10.1002/ajmg.1535. [DOI] [PubMed] [Google Scholar]
- 35.Eichelberger KY, Baker AM, Woodham PC, Haeri S, Strauss RA, Stuebe AM. Second-trimester maternal serum paraxanthine, CYP1A2 activity, and the risk of severe preeclampsia. Obstet Gynecol. 2015;126:725–730. doi: 10.1097/AOG.0000000000001041. [DOI] [PubMed] [Google Scholar]
- 36.Kumru S, Aydin S, Gursu MF, Ozcan Z. Changes of serum paraoxonase (an HDL-cholesterol-associated lipophilic antioxidant) and arylesterase activities in severe preeclamptic women. Eur J Obstet Gynecol Reprod Biol. 2004;114:177–181. doi: 10.1016/j.ejogrb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 37.Serrato M, Marian AJ. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Invest. 1995;96:3005–3008. doi: 10.1172/JCI118373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uzun H, Benian A, Madazli R, Topçuoğlu MA, Aydin S, Albayrak M. Circulating oxidized low-density lipoprotein and paraoxonase activity in preeclampsia. Gynecol Obstet Investig. 2005;60:195–200. doi: 10.1159/000087205. [DOI] [PubMed] [Google Scholar]
- 39.Wheeler JG, Keavney BD, Watkins H, Collins R, Danesh J. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls: meta-analysis of 43 studies. Lancet. 2004;363(9410):689–695. doi: 10.1016/S0140-6736(04)15642-0. [DOI] [PubMed] [Google Scholar]
- 40.Zusterzeel PL, Peters WH, Visser W, Hermsen KJ, Roelofs HM, Steegers EA. A polymorphism in the gene for microsomal epoxide hydrolase is associated with pre-eclampsia. J Med Genet. 2001;38:234–237. doi: 10.1136/jmg.38.4.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laasanen J, Romppanen EL, Hiltunen M, Helisalmi S, Mannermaa A, Punnonen K, et al. Two exonic single nucleotide polymorphisms in the microsomal epoxide hydrolase gene are jointly associated with preeclampsia. Eur J Hum Genet. 2002;10:569–573. doi: 10.1038/sj.ejhg.5200849. [DOI] [PubMed] [Google Scholar]
- 42.Zusterzeel PL, Peters WH, De Bruyn MA, Knapen MF, Merkus HM, Steegers EA. Glutathione S-transferase isoenzymes in decidua and placenta of preeclamptic pregnancies. Obstet Gynecol. 1999;94:1033–1038. doi: 10.1016/s0029-7844(99)00433-0. [DOI] [PubMed] [Google Scholar]
- 43.Hill LD, York TP, Kusanovic JP, Gomez R, Eaves LJ, Romero R, et al. Epistasis between COMT and MTHFR in maternal-fetal dyads increases risk for preeclampsia. PLoS One. 2011;6(1):e16681. doi: 10.1371/journal.pone.0016681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanasaki K, Palmsten K, Sugimoto H, Ahmad S, Hamano Y, Xie L, et al. Deficiency in catechol-O-methyltransferase and 2-methoxyoestradiol is associated with pre-eclampsia. Nature. 2008;453:1117–1121. doi: 10.1038/nature06951. [DOI] [PubMed] [Google Scholar]
- 45.Furness DL, Fenech MF, Khong YT, Romero R, Dekker GA. One-carbon metabolism enzyme polymorphisms and uteroplacental insufficiency. Am J Obstet Gynecol. 2008;199(3):276.e1–276.e8. doi: 10.1016/j.ajog.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 46.Hoque MM, Bulbul T, Mahal M, Islam NA, Ferdausi M. Serum homocysteine in pre-eclampsia and eclampsia. Bangladesh Med Res Counc Bull. 2008;34:16–20. doi: 10.3329/bmrcb.v34i1.1165. [DOI] [PubMed] [Google Scholar]
- 47.Mignini LE, Latthe PM, Villar J, Kilby MD, Carroli G, Khan KS. Mapping the theories of preeclampsia: the role of homocysteine. Obstet Gynecol. 2005;105:411–425. doi: 10.1097/01.AOG.0000151117.52952.b6. [DOI] [PubMed] [Google Scholar]
- 48.Sorensen TK, Malinow MR, Williams MA, King IB, Luthy DA. Elevated second-trimester serum homocysteine levels and subsequent risk of preeclampsia. Gynecol Obstet Investig. 1999;48:98–103. doi: 10.1159/000010148. [DOI] [PubMed] [Google Scholar]
- 49.Felder RA, Sanada H, Xu J, Yu PY, Wang Z, Watanabe H, et al. G protein-coupled receptor kinase 4 gene variants in human essential hypertension. Proc Natl Acad Sci U S A. 2002;99:3872–3877. doi: 10.1073/pnas.062694599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu C, Xi B. Pooled analyses of the associations of polymorphisms in the GRK4 and EMILIN1 genes with hypertension risk. Int J Med Sci. 2012;9:274–279. doi: 10.7150/ijms.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Premont RT, Macrae AD, Stoffel RH, Chung N, Pitcher JA, Ambrose C, et al. Characterization of the G protein coupled receptor kinase GRK4. Identification of four splice variants. J Biol Chem. 1996;271:6403–6410. doi: 10.1074/jbc.271.11.6403. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Sun ZQ, Liu SS, Yang LN. Association between GRK4 and DRD1 gene polymorphisms and hypertension: a meta-analysis. Clin Interv Aging. 2015;11:17–27. doi: 10.2147/CIA.S94510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.StataCorp . Stata statistical software: release 14. College Station: StataCorp LP; 2015. [Google Scholar]
- 54.Gui J, Moore JH, Williams SM, Andrews P, Hillege HL, van der Harst P, et al. A simple and computationally efficient approach to multifactor dimensionality reduction analysis of gene-gene interactions for quantitative traits. PLoS One. 2013;8(6):e66545. doi: 10.1371/journal.pone.0066545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu T, Chen Y, Kiralis JW, Moore JH. ViSEN: methodology and software for visualization of statistical epistasis networks. Genet Epidemiol. 2013;37:283–285. doi: 10.1002/gepi.21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muttukrishna S, Child TJ, Groome NP, Ledger WL. Source of circulating levels of inhibin a, pro alpha C- containing inhibins and activin a in early pregnancy. Hum Reprod. 1997;12:1089–1093. doi: 10.1093/humrep/12.5.1089. [DOI] [PubMed] [Google Scholar]
- 57.Arngrímsson R, Sigurardttir S, Frigge ML, Bjarnadóttir RI, Jónsson T, Stefánsson H, et al. A genome-wide scan reveals a maternal susceptibility locus for preeclampsia on chromosome 2p13. Hum Mol Genet. 1999;8:1799–1805. doi: 10.1093/hmg/8.9.1799. [DOI] [PubMed] [Google Scholar]
- 58.Laivuori H, Lahermo P, Ollikainen V, Widen E, Haiva-Mallinen L, Sundström H, et al. Susceptibility loci for preeclampsia on chromosomes 2p25 and 9p13 in Finnish families. Am J Hum Genet. 2003;72:168–177. doi: 10.1086/345311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moses EK, Lade JA, Guo G, Wilton AN, Grehan M, Freed K, et al. A genome scan in families from Australia and New Zealand confirms the presence of a maternal susceptibility locus for pre-eclampsia, on chromosome 2. Am J Hum Genet. 2000;67:1581–1585. doi: 10.1086/316888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moses EK, Fitzpatrick E, Freed KA, Dyer TD, Forrest S, Elliott K, et al. Objective prioritization of positional candidate genes at a quantitative trait locus for pre-eclampsia on 2q22. Mol Hum Reprod. 2006;12:505–512. doi: 10.1093/molehr/gal056. [DOI] [PubMed] [Google Scholar]
- 61.Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 62.Wang X, Bai T, Liu S, Pan H, Wang B. Association between thrombophilia gene polymorphisms and preeclampsia: a Meta-analysis. PLoS One. 2014;9(6):e100789. doi: 10.1371/journal.pone.0100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang B, Fan S, Zhi X, Li Y, Liu Y, Wang D, et al. Associations of MTHFR gene polymorphisms with hypertension and hypertension in pregnancy: a meta analysis from 114 studies with 15411 cases and 21970 controls. PLoS One. 2014;9(2):e87497. doi: 10.1371/journal.pone.0087497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luizon MR, Palei ACT, Sandrim VC. Polymorphisms and haplotypes in candidate genes related to angiogenesis and endothelial dysfunction in preeclampsia. J Pregnancy. 2012;2012:914704. doi: 10.1155/2012/914704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Association of SNPs, adjusting for statistically significant risk factors. (DOCX 16 kb)
Table S2. MDR analysis of genetic variants, without adjusting for age and BMI. (DOCX 14 kb)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.