Abstract
Background
MicroRNAs (miRNAs) have been associated with the Hirschsprung disease (HSCR) pathogenesis, however, the findings are still inconclusive. We aimed to investigate the effect of miRNA-206 and its targets, fibronectin 1 (FN1), serum deprivation response (SDPR), and paired box 3 (PAX3) expressions on multifactorial HSCR in Indonesia, a genetically distinct group within Asia.
Methods
We determined the miRNA-206, FN1, SDPR and PAX3 expressions in both the ganglionic and aganglionic colon of HSCR patients and control colon by quantitative real-time polymerase chain reaction (qRT-PCR).
Results
Twenty-one sporadic HSCR patients and thirteen controls were ascertained in this study. The miRNA-206 expression was up-regulated (2-fold) in the ganglionic colon and down-regulated (0.5-fold) in the aganglionic colon compared to the control group (ΔCT 12.4 ± 3.0 vs. 14.1 ± 3.9 vs. 13.1 ± 2.7), but these differences did not reach significant levels (p = 0.48 and p = 0.46, respectively). Interestingly, the FN1 expression was significantly increased in both the ganglionic (38-fold) and aganglionic colon (18-fold) groups compared to the control group ΔCT 5.7 ± 3.0 vs. 6.8 ± 2.3 vs. 11.0 ± 5.0; p = 0.001 and p = 0.038, respectively). Furthermore, the expressions of SDPR were similar in the ganglionic, aganglionic and control colon groups (ΔCT 2.4 ± 0.6 vs. 2.2 ± 0.4 vs. 2.1 ± 0.6; p = 0.16 and p = 0.39, respectively), while no change was observed in the PAX3 expression between the ganglionic, aganglionic, and control colon groups (ΔCT 3.8 ± 0.8 vs. 4.1 ± 0.8 vs. 3.7 ± 1.1; p = 0.83 and p = 0.44, respectively).
Conclusion
Our study is the first report of aberrant FN1 expressions in the colon of patients with HSCR and supplies further insights into the contribution of aberrant FN1 expression in the HSCR pathogenesis.
Keywords: FN1, Hirschsprung disease, Indonesia, miRNA-206, PAX3, SDPR
Background
Hirschsprung disease (HSCR: MIM# 142623) is a complex genetic disorder characterized by the absence of ganglion cells in the intestines, resulting in a functional obstruction in children. HSCR is classified as follows: short-segment HSCR, long-segment HSCR, and total colonic aganglionosis [1, 2]. The incidence of HSCR varies among ethnic groups with 1.5, 2.1, and 2.8 cases per 10,000 live births in European, African and Asian ancestry cases, respectively [1, 2].
At least 15 genes have been associated with the pathogenesis of HSCR, with the RET gene as primarily responsible for HSCR [1, 2]. However, the majority of those genes make minor contributions to HSCR [3–5]. Recent studies have proposed some microRNAs (miRNAs) targets contribute important roles in the pathogenesis of HSCR, but the findings are still inconclusive [6–8]. miRNA is a small non-coding RNA that deregulates gene expression at the posttranscriptional level. It is stable and easily measureable in the patients’ tissue and blood specimens, including HSCR patients’ colon [6–8].
miRNA-206 has been shown to be down-regulated and targeted three genes, named fibronectin 1 (FN1), serum deprivation response (SDPR), and paired box 3 (PAX3), in HSCR patients in Chinese population [7]. In addition, some genetic differences might exist among Asian population [9] and our previous study revealed that the impact of SEMA3 rs11766001 variant differs among ethnic groups[10]. Therefore, we aimed to investigate the expressions of miRNA-206 and its targets, FN1, SDPR, and PAX3, in HSCR patients in Indonesia, a genetically distinct group within Asia.
Material and methods
Patients
This study was conducted at Dr. Sardjito Hospital, a referral and academic hospital in Yogyakarta, Indonesia. All children with the age of < 18 years old with diagnosis of HSCR according to clinical findings, contrast enema and histopathology were involved in this study, except those that had low quality of total RNA [4, 5, 10–12].
The ganglionic and aganglionic colon of HSCR patients were collected at definitive surgery, while the control colon samples were obtained at stoma closure from anorectal malformation patients [12].
A written informed consent was signed by the HSCR patients’ and control parents to ascertain this study. The Institutional Review Board of the Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada/Dr. Sardjito Hospital gave approval for this study (KE/FK/786/EC/2015).
Total RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
The miRCURY™ RNA Isolation Kit-Tissue (Exiqon A/S, Denmark) was used to extract the total RNA from colon tissue. Subsequently, the total RNA was measured using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Only high quality RNAs with the OD260/280 ratios of 1.8 to 2.0 were utilized for the subsequent experiment.
The qRT-PCR was performed to determine the expression of miRNA-206, FN1, SDPR, and PAX3 using the BioRad CFX Real-Time PCR System (California, USA), the Universal cDNA Synthesis Kit II (Exiqon A/S, Denmark), ExiLENT SYBR® Green Master Mix Kit (Exiqon A/S, Denmark), and miRCURY™ LNA™ Universal RT microRNA PCR System (Exiqon A/S, Denmark). U6 small nuclear RNA (snRNA) served as a control for analysis of miRNA-206 expression, while glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized as a reference gene for analysis of FN1, SDPR, and PAX3 expression. All qRT-PCR reactions were performed in duplicate.
The hsa-miRNA-206 and U6 snRNA primers were 5’-ACGAGTTTAGAGCCGGATAGCCACACAC-3′ (RT), 5’-TGACGAGTTTAGAGCCGGATAG-3′ (forward), and 5’-GCGTTGTCTGGAATGTAAGGAAGT -3′ (reverse); and 5’-CTCGCTTCGGCAGCACA-3′ (forward) and 5’-AACGCTTCACGAATTTGCGT-3′ (reverse), respectively [13], while the primer sequence for FN1, SDPR, PAX3, and GAPDH were 5′-CAAGCCAGATGTCAGAAGC-3′ (forward) and 5′-GGATGGTGCATCAATGGCA-3′ (reverse); 5′-AGTCACGGTGCTCACGCTCC-3′ (forward) and 5′- GTTGCTGGTGGAGGCCTGGT-3′ (reverse); 5'-ACCACCTTCACAGCAGAACA-3' (forward) and 5'-CAGCTTGCTTCCTCCATCTT-3' (reverse); and 5′-GCACCGTCAAGGCTGAGAAC-3′ (forward) and 5′-TGGTGAAGACGCCAGTGGA-3′ (reverse), respectively [12, 14–17].
The Livak (2-ΔΔCT) method was used to analyze the miRNA-206, FN1, SDPR, and PAX3 expression level [18].
Statistical analysis
The miRNA-206, FN1, SDPR, and PAX3 expressions were determined as mean values ± standard deviation (SD) and t-tests were used to determine any statistical differences between the ganglionic and aganglionic colon of HSCR patients and control groups. A p-value < 0.05 was considered statistically significant.
Results
We obtained twenty-one colon samples from sporadic non-syndromic HSCR patients, of whom 12 and 9 were males and females, respectively, and thirteen colon specimens from non-HSCR patients. Most (90%) patients had short-segment HSCR and underwent transanal endorectal pull-through (76%) (Table 1).
Table 1.
Clinical characteristics of Indonesian HSCR patients involved in this study
| Characteristic | n (%); months ± SD |
|---|---|
| Gender | |
| Male | 12 (57) |
| Female | 9 (43) |
| Aganglionosis type | |
| Short | 19 (90) |
| Long | 2 (10) |
| Total colon aganglionosis | 0 |
| Age at diagnosis | 14.3 ± 31.2 |
| Colostomy | 5 (28) |
| Age at definitive surgery | 22.1 ± 34.1 |
| Definitive surgery | |
| Transanal endorectal pull-through | 16 (76) |
| Duhamel | 3 (14) |
| Soave | 2 (10) |
Although the miRNA-206 expression was up-regulated (2-fold) in the ganglionic colon and down-regulated (0.5-fold) (Fig. 1) in the aganglionic colon compared to the control group (ΔCT 12.4 ± 3.0 vs. 14.1 ± 3.9 vs. 13.1 ± 2.7), but these differences did not reach significant levels (p = 0.48 and p = 0.46, respectively) (Table 2).
Fig. 1.
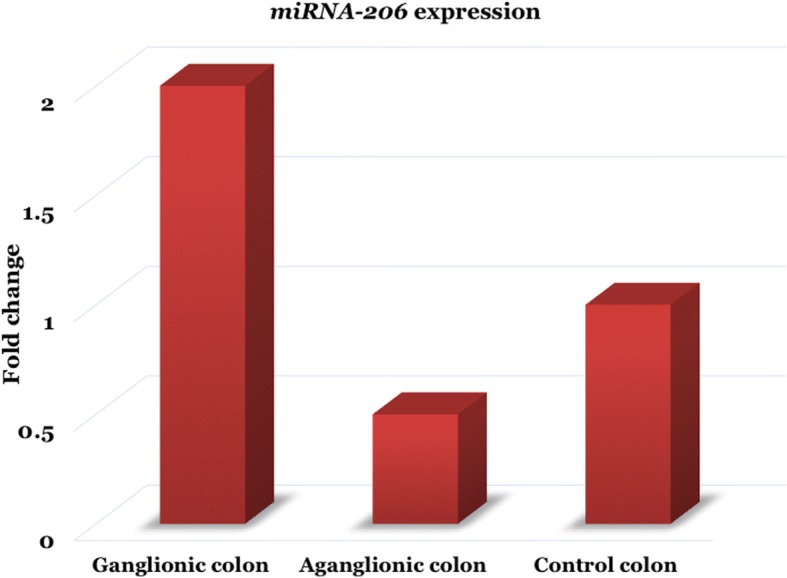
The miRNA-206 expression was up-regulated (2-fold) in the ganglionic colon and down-regulated (0.5-fold) in the aganglionic colon compared to the control group, but these differences did not reach significant level
Table 2.
The miRNA-206 expression in both the ganglionic and aganglionic colon of HSCR patients and control colon
| ΔCT ± SD | ΔΔCT (95% CI) | 2-ΔΔCT (Fold change) | p-value | |
|---|---|---|---|---|
| Ganglionic colon | 12.4 ± 3.0 | −0.8 (−3.0–1.4) | 2.0 | 0.48 |
| Aganglionic colon | 14.1 ± 3.9 | 1.0 (− 1.7–3.6) | 0.5 | 0.46 |
| Control colon | 13.1 ± 2.7 |
Interestingly, the FN1 expression was significantly up-regulated in both the ganglionic (38-fold) and aganglionic colon (18-fold) (Fig. 2) groups compared to the control group (ΔCT 5.7 ± 3.0 vs. 6.8 ± 2.3 vs. 11.0 ± 5.0; p = 0.001 and p = 0.038, respectively) (Table 3).
Fig. 2.
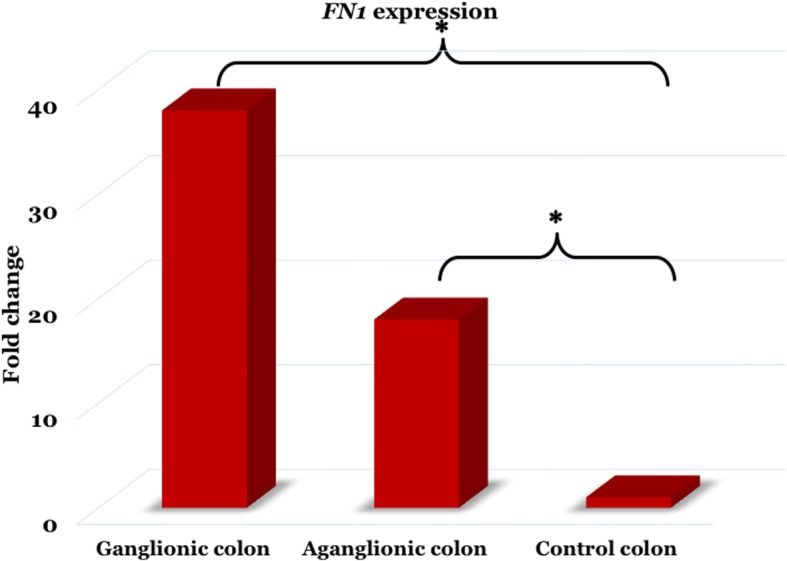
The FN1 expression was increased in both the ganglionic (38-fold) and aganglionic colon (18-fold) groups compared to the control group, with p-value of 0.001 and 0.038, respectively. *, p < 0.05
Table 3.
The FN1 expression in both the ganglionic and aganglionic colon of HSCR patients and control colon
| ΔCT ± SD | ΔΔCT (95% CI) | 2-ΔΔCT (Fold change) | p-value | |
|---|---|---|---|---|
| Ganglionic colon | 5.7 ± 3.0 | −5.3 [− 8.2 – (−)2.3] | 38 | 0.001* |
| Aganglionic colon | 6.8 ± 2.3 | −4.1 [− 8.1 – (−)0.2] | 18 | 0.038* |
| Control colon | 11.0 ± 5.0 |
*, p < 0.05 is considered statistically significant
Furthermore, the expressions of SDPR were similar in the ganglionic, aganglionic and control colon groups (ΔCT 2.4 ± 0.6 vs. 2.2 ± 0.4 vs. 2.1 ± 0.6; p = 0.16 and p = 0.39, respectively) (Table 4), while no change was observed in the PAX3 expression between the ganglionic, aganglionic, and control colon groups (ΔCT 3.8 ± 0.8 vs. 4.1 ± 0.8 vs. 3.7 ± 1.1; p = 0.83 and p = 0.44, respectively) (Table 5).
Table 4.
The SDPR expression in both the ganglionic and aganglionic colon of HSCR patients and control colon
| ΔCT ± SD | ΔΔCT (95% CI) | 2-ΔΔCT (Fold change) | p-value | |
|---|---|---|---|---|
| Ganglionic colon | 2.4 ± 0.6 | 0.3 (−0.1–0.8] | 0.8 | 0.16 |
| Aganglionic colon | 2.2 ± 0.4 | 0.2 (− 0.2–0.6) | 0.9 | 0.39 |
| Control colon | 2.1 ± 0.6 |
Table 5.
The PAX3 expression in both the ganglionic and aganglionic colon of HSCR patients and control colon
| ΔCT ± SD | ΔΔCT (95% CI) | 2-ΔΔCT (Fold change) | p-value | |
|---|---|---|---|---|
| Ganglionic colon | 3.8 ± 0.8 | 0.1 (−0.9–1.1) | 0.9 | 0.83 |
| Aganglionic colon | 4.1 ± 0.8 | 0.4 (− 0.7–1.5) | 0.8 | 0.44 |
| Control colon | 3.7 ± 1.1 |
Discussion
We describe new data on the miRNA-206 expression in Indonesian HSCR patients. We were unable to find evidence of the impact of miRNA-206 in the pathogenesis of HSCR in Indonesian population, although its expression was ~ 2-fold up-regulated and ~ 0.5-fold down-regulated (Fig. 1) in the ganglionic and the aganglionic colon of HSCR patients, respectively, compared to the control colon. These results are different with previous report [7]. It has been shown that the miRNA expression significantly differed between two populations, CEU (Utah residents with northern and western European ancestry) and YRI (Yoruba people from Ibadan, Nigeria) [19]. In addition, miRNA-26a expression was also different between the prostate cancer cell lines derived from African American ancestry and those derived from Caucasian ancestry [20]. Interestingly, the population differences in miRNA expression are affected by genetic variants [19]. Therefore, the miRNA-206 expression differences between previous report and our study might relate to Indonesian genetic structure ethnicity [9, 10].
The down-regulation of miRNA-206 has been hypothesized to be involved in the pathogenesis of HSCR patient through the SDPR up-regulation resulting in the deformation of the caveolae of neural crest cells in the intestines [7]. Our study reveals a new evidence opposing this hypothesis by providing data from a population genetically different from previous study [7]. However, our results should be interpreted with some caution since our study had a different approach from the previous report [7]; we determined the miRNA-206 expression in the colon tissue using RT-PCR only (vs. they also performed in vitro study employing the human 293 T and SH-SY5Y cell lines). Also, it should be noted the main weakness of our study is the small sample size, which suggests that a larger sample size needs to be involved to clarify and confirm our results.
Although several miRNAs have been shown to have a role in HSCR pathogenesis, however, the evidence for actual etiology remains inconclusive [6–8]. Therefore, in the meanwhile, it is always challenging to determine which miRNAs have the strongest impact on the HSCR pathogenesis. Those miRNAs may serve as potential biomarkers and/or molecular therapy for patients with HSCR in the future since the miRNAs are stable and easily measureable in the patients’ tissue and blood specimens.
Moreover, our study showed that the expression of PAX3 did not differ between the HSCR and the control groups. PAX3 has been associated with syndromic HSCR, i.e. Waardenburg syndrome [21]. Our cohort patients are non-syndromic HSCR, therefore, it might be important to conduct a study involving the syndromic HSCR to clarify the results.
Intriguingly, the expression of FN1 was strongly up-regulated in both the ganglionic and aganglionic colon of HSCR patients compared to the control colon. To the best of our knowledge, this report is the first study of aberrant FN1 expressions in the colon of HSCR patients. It has been shown that FN1 is up-regulated by enteric glial cells in the proliferating intestinal epithelial cells [22]. HSCR is a developmental defect of the enteric nervous system (ENS). HSCR pathogenesis might involve the compromised condition of genes responsible for gangliogenesis of the ENS [1–4] and/or their interactions [1, 2, 5, 23]. Furthermore, integration of different pathways synchronizing neurogenesis and gliogenesis is also important for the proper development of ENS and defects in any of these signaling elements might result in HSCR [24, 25]. Gui et al. showed that GDNF stimulates neuronal differentiation, while NRG1 strongly induces the glial differentiation of enteric neural crest cells (ENCCs) [24], whereas Ngan et al. revealed that Ptch1 knockout in mouse ENCCs promotes up-regulated Dll1 expression and stimulates the Notch signaling, resulting in a premature gliogenesis and reduced ENCC progenitors in intestines [25]. Therefore, further in vitro assay of FN1 knockdown in primary culture of ganglion (mixture of neurons and glial cells) are necessary to see the effect of FN1 knockdown on the proliferation, differentiation and survival of both neurons and glial cells, and the balance of neurogenesis and gliogenesis. Unfortunately, we do not have any data on in vitro assay of FN1 knockdown in primary culture of ganglion due to resource limitations in our laboratory.
Conclusion
Our study is the first report of aberrant FN1 expressions in the colon of patients with HSCR and supplies further insights into the contribution of aberrant FN1 expression in the HSCR pathogenesis.
Acknowledgements
We thank the patients and their families who have contributed in these studies. We are thankful for the English Services Center, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, for editing the grammar and proofreading of our manuscript. We are also grateful to the numerous nurses (Dr. Sardjito Hospital), Dr. Dian Nirmala Sirait, and all those who provided excellent technical support and assistance during the study.
Funding
This work was supported by a grant from the Ministry of Research, Technology and Higher Education, Indonesia to G. and A.M. (PUPT 2416/UN1-P.III/DIT-LIT/LT/2017 and PDUPT 192/UN1/DITLIT/DIT-LIT/LT/2018).
Availability of data and materials
All data generated or analyzed during this study are included in the submission. The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Abbreviations
- FN1
Fibronectin 1
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HSCR
Hirschsprung disease
- miRNA
MicroRNA
- PAX3
Paired box 3
- qRT-PCR
Quantitative real-time polymerase chain reaction
- SDPR
Serum deprivation response
- snRNA
Small nuclear RNA
Authors’ contributions
G, AM, and KI conceived the study. G drafted the manuscript, AM and KI critically revised the manuscript for important intellectual content. NYPB, ASK, WS, FDM, and G facilitated all project-related tasks. All authors have read and approved the manuscript, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The Ethical Committee of the Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada/Dr. Sardjito Hospital gave approval for this study (KE/FK/786/EC/2015). The HSCR patients and controls were ascertained for this study after their parents signed a written informed consent form.
Consent for publication
Not applicable.
Competing interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gunadi, Phone: +62-274-631036, Email: drgunadi@ugm.ac.id.
Nova Yuli Prasetyo Budi, Email: novayuliprasetyo@ymail.com.
Alvin Santoso Kalim, Email: alvin_2495@yahoo.com.
Wiwid Santiko, Email: dr.wiwid.santiko@gmail.com.
Fuad Dheni Musthofa, Email: fuaddmusthofa@gmail.com.
Kristy Iskandar, Email: kristy.iskandar@mail.ugm.ac.id.
Akhmad Makhmudi, Email: akhmad_makhmudi@yahoo.com.
References
- 1.Alves MM, Sribudiani Y, Brouwer RW, et al. Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev Biol. 2013;382:320–329. doi: 10.1016/j.ydbio.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 2.Amiel J, Sproat-Emison E, Garcia-Barcelo M, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 3.Emison ES, Garcia-Barcelo M, Grice EA, et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet. 2010;87:60–74. doi: 10.1016/j.ajhg.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunadi BNYP, Sethi R, et al. NRG1 variant effects in patients with Hirschsprung disease. BMC Pediatr. 2018;18:292. doi: 10.1186/s12887-018-1265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunadi KA, Ling AY, et al. Effects of RET and NRG1 polymorphisms in Indonesian patients with Hirschsprung disease. J Pediatr Surg. 2014;49:1614–1618. doi: 10.1016/j.jpedsurg.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sergi CM, Caluseriu O, McColl H, Eisenstat DD. Hirschsprung's disease: clinical dysmorphology, genes, micro-RNAs, and future perspectives. Pediatr Res. 2017;81:177–191. doi: 10.1038/pr.2016.202. [DOI] [PubMed] [Google Scholar]
- 7.Sharan A, Zhu H, Xie H, Li H, Tang J, Tang W, Zhang H, Xia Y. Down-regulation of miR-206 is associated with Hirschsprung disease and suppresses cell migration and proliferation in cell models. Sci Rep. 2015;5:9302. doi: 10.1038/srep09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang W, Li H, Tang J, Wu W, Qin J, Lei H, et al. Specific serum microRNA profile in the molecular diagnosis of Hirschsprung's disease. J Cell Mol Med. 2014;18:1580–1587. doi: 10.1111/jcmm.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tajima A, Pan IH, Fucharoen G, et al. Three major lineages of Asian Y chromosomes: implications for the peopling of east and Southeast Asia. Hum Genet. 2002;110:80–88. doi: 10.1007/s00439-001-0651-9. [DOI] [PubMed] [Google Scholar]
- 10.Gunadi, Makhmudi A, Agustriani N, Rochadi. Effects of SEMA3 polymorphisms in Hirschsprung disease patients. Pediatr Surg Int 2016;32:1025–1028. [DOI] [PubMed]
- 11.Setiadi JA, Dwihantoro A, Iskandar K, Heriyanto DS. Gunadi. The utility of the hematoxylin and eosin staining in patients with suspected Hirschsprung disease. BMC Surg. 2017;17:71. doi: 10.1186/s12893-017-0267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunadi SM, Budi NYP, Kalim AS, Iskandar K, Dwihantoro A. The impact of down-regulated SK3 expressions on Hirschsprung disease. BMC Med Genet. 2018;19:24. doi: 10.1186/s12881-018-0539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu ZB, Han SP, Bai YF, Zhu C, Pan Y, Guo XR. microRNA expression profiling in fetal single ventricle malformation identified by deep sequencing. Int J Mol Med. 2012;29:53–60. doi: 10.3892/ijmm.2012.888. [DOI] [PubMed] [Google Scholar]
- 14.Bluyssen HA, Lolkema MP, van Beest M, et al. Fibronectin is a hypoxia-independent target of the tumor suppressor VHL. FEBS Lett. 2004;556:137–142. doi: 10.1016/S0014-5793(03)01392-9. [DOI] [PubMed] [Google Scholar]
- 15.Ozturk S, Papageorgis P, Wong CK, et al. SDPR functions as a metastasis suppressor in breast cancer by promoting apoptosis. Proc Natl Acad Sci U S A. 2016;113:638–643. doi: 10.1073/pnas.1514663113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen W, Chen G, Dong R, Zhao R, Zheng S. MicroRNA-21/PTEN/Akt axis in the fibrogenesis of biliary atresia. J Pediatr Surg. 2014;49:1738–1741. doi: 10.1016/j.jpedsurg.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Bajpai VK, Kerosuo L, Tseropoulos G, et al. Reprogramming postnatal human epidermal keratinocytes toward functional neural crest fates. Stem Cells. 2017;35:1402–1415. doi: 10.1002/stem.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Huang RS, Gamazon ER, Ziliak D, et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011;8:692–701. doi: 10.4161/rna.8.4.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theodore S, Rhim J, Turner T, Yates C. MiRNA 26a expression in a novel panel of African American prostate cancer cell lines. Ethn Dis. 2010;20:96–100. [PMC free article] [PubMed] [Google Scholar]
- 21.Pingault V, Ente D, Dastot-Le Moal F, et al. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 22.Van Landeghem L, Mahé MM, Teusan R, et al. Regulation of intestinal epithelial cells transcriptome by enteric glial cells: impact on intestinal epithelial barrier functions. BMC Genomics. 2009;10:507. doi: 10.1186/1471-2164-10-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunadi IK, Makhmudi A, Kapoor A. Combined genetic effects of RET and NRG1 susceptibility variants on multifactorial Hirschsprung disease in Indonesia. J Surg Res. 2019;233:96–99. doi: 10.1016/j.jss.2018.07.067. [DOI] [PubMed] [Google Scholar]
- 24.Gui H, Tang WK, So MT, et al. RET and NRG1 interplay in Hirschsprung disease. Hum Genet. 2013;132:591–600. doi: 10.1007/s00439-013-1272-9. [DOI] [PubMed] [Google Scholar]
- 25.Ngan ES, Garcia-Barceló MM, Yip BH, et al. Hedgehog/notch-induced premature gliogenesis represents a new disease mechanism for Hirschsprung disease in mice and humans. J Clin Invest. 2011;121:3467–3478. doi: 10.1172/JCI43737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the submission. The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.


