Abstract
The human genome harbors an abundance of repetitive DNA; however, its function continues to be debated. Microsatellites—a class of short tandem repeat—are established as an important source of genetic variation. Array length variants are common among microsatellites and affect gene expression; but, efforts to understand the role and diversity of microsatellite variation has been hampered by several challenges. Without adequate depth, both long-read and short-read sequencing may not detect the variants present in a sample; additionally, large sample sizes are needed to reveal the degree of population-level polymorphism. To address these challenges we present the Comparative Analysis of Germline Microsatellites (CAGm): a database of germline microsatellites from 2529 individuals in the 1000 genomes project. A key novelty of CAGm is the ability to aggregate microsatellite variation by population, ethnicity (super population) and gender. The database provides advanced searching for microsatellites embedded in genes and functional elements. All data can be downloaded as Microsoft Excel spreadsheets. Two use-case scenarios are presented to demonstrate its utility: a mononucleotide (A) microsatellite at the BAT-26 locus and a dinucleotide (CA) microsatellite in the coding region of FGFRL1. CAGm is freely available at http://www.cagmdb.org/.
INTRODUCTION
Microsatellites are regions of DNA characterized by short, 1–6 bp, motifs repeated in tandem to form an array. Over 600 000 unique microsatellites exist in the human genome embedded in gene introns, gene exons and regulatory regions (1,2). A number of databases provide searchable interfaces to microsatellites within the human reference genome (3–9), but none provide data on actual polymorphism rates among and within human populations. This is important because microsatellite polymorphisms occur frequently due to strand slip replication and heterozygote instability, in fact, mutation rates for microsatellites can be 10 000 times higher than single nucleotide polymorphisms (SNPs), which have estimated mutation rate of 1 × 10−8 (10). Observed array length variations are largely consistent with a stepwise model: a model based on the idea that each mutation adds or subtracts a single repeat unit to the microsatellite array (11). However, microsatellites remain understudied compared to SNPs (12). In addition, the study of microsatellites can face technical limitations. Microsatellites are challenging for most next-generation sequencing (NGS) platforms and alignment algorithms (13,14); without adequate sequencing depth both long-reads (454-sequencing) and short-reads (Illumina) are prone to errors. Gapped alignment algorithms can detect most microsatellite insertions and deletions (indels) (15); but, alignment of large insertion variants remains challenging. Moreover, large sample sizes are needed to understand the degree of population-level polymorphism that exists in these regions.
Even with adequate data, several additional sequencing and bioinformatics challenges remain: polymerase chain reaction (PCR) amplification can introduce ‘stutter’ errors; optimal alignments may not be unique; and the likelihood of various allele combinations can be difficult to quantify (16,17). Bioinformatics tools for microsatellite genotyping use several strategies to address these challenges (17). Many tools are now designed to anticipate PCR errors (17) and some use custom alignment techniques (17). Others have used informed error profiles that incorporate read properties such as unit, length and base quality (18). For example, Repeatseq uses a Bayesian approach guided by an informed error profile; it was developed using data from the 1000 genomes project (18). A variety of statistical approaches are used to assign genotype likelihoods (17).
Microsatellites have the capacity to affect gene expression (1) by inducing Z-DNA and H-DNA folding (19); altering nucleosome positioning (20); and changing the spacing of DNA binding sites (1). These possibilities are made even more intriguing by the non-random distribution of microsatellites in the human genome (2). In particular, the number of microsatellites in gene exons and regulatory regions far exceeds what is expected due to chance alone (1). Typically, microsatellites found in exons have 3 or 6 bp repeat motifs (21). Consequently, mutations result in single amino acid gain or loss; but, do not result in frameshifts. For these reasons microsatellites have been called the ‘tuning knobs’ of gene expression (1,22,23).
The 1000 Genomes Project was launched in 2008 with the aim of creating the world's largest public catalog of human genetic variation (24). Now finished, the complete data collection includes exome and whole genome sequencing data from over 2500 individuals. These individuals in turn come from 26 worldwide populations belonging to 5 ethnicities (super populations). The data are of high quality; in particular, a combination of high-depth exome sequencing and low-depth whole genome sequencing was used to detect low frequency variants (>1%) (24). Indeed, over 88 million variants are now cataloged: 84.7 million SNPs, 3.6 million short indels and 60 000 structural variants. However, the ability to query microsatellite variation within this data is not part of the 1000 genomes project. A catalog of human microsatellite variation is needed for at least three reasons. First, microsatellites are known to affect gene expression. Trinucleotide repeats can have dramatic effects and are known to increase the risk of particular genetic diseases such as Huntington’s disease, spinocerebellar ataxia and myotonic dystrophy (25). Second, recent studies have demonstrated the diagnostic potential of microsatellites for a limited number of cancers: breast cancer (26,27), ovarian cancer (27) and lung cancer (28). Third, germline DNA is easily obtained and therefore a well-suited target for genomic based testing for somatic microsatellite polymorphisms. Additional motives for cataloging microsatellite variants stem from their role in evolutionary biology, forensics, population genetics and kinship analysis, just to name a few.
We introduce the Comparative Analysis of Germline Microsatellites; its acronym—CAGm—coincides with the important class of polyglutamine microsatellites. The database is designed to assist with future studies of germline microsatellites and enhance our understanding of human genetic variation. Analysis is included for germline microsatellites in whole exome sequencing of 2529 individuals in the 1000 genomes project. The database is implemented using MySQL: a popular open source relational database. Samples can be easily grouped by population, ethnicity and gender. Microsatellites can be searched by gene, functional element and location. Users can query genotypes, view multiple sequence alignments and easily download data for further analysis. The database has a wide range of additional capabilities. Database content is fully described with examples and future directions are discussed. The database is freely available at http://www.cagmdb.org/.
MATERIALS AND METHODS
Microsatellite list generation
A list of microsatellites in version 38 of the human reference genome was generated with a custom Perl script ‘searchTandemRepeats.pl’ using default parameters. This script has been used in previous microsatellite studies (29) and is freely available online: http://genotan.sourceforge.net/#_Toc324410847. The initial list generated with this script included 1 671 121 microsatellites. Only microsatellites of 100 bp or less were included for subsequent analysis. This limitation stems from the need to have both 3′ and 5′ flanking regions present to determine a microsatellite genotype; and, the 100 bp read length of Illumina (Solexa) sequencing technology. Briefly, the ‘searchTandemRepeats.pl’ script first searches for pure repetitive stretches: no impurities allowed. Imperfect repeats and compound repeats are then handled using a ‘mergeGap’ parameter with a default value of 10 bp. Essentially, impurities that interrupt stretches of pure repeat sequence are tolerated unless they exceed 10 bp. Likewise, repeats closer than 10 bp are considered compound. The result is that repeats in the CAGm database are highly pure; and, components of compound repeats are highly pure. To mitigate the likelihood of improper read mapping between microsatellites, we removed all subsets of microsatellites possessing the same repeat motif between 5 bp 3′ and 5′ flanking regions. For example, the microsatellites ‘GCTGC(A)34CTTAG’ and ‘GCTGC(A)15CTTAG’ were removed from our initial list of microsatellites. Our filtered list included 625 178 microsatellites unique in the human genome.
Microsatellite genotyping
We used the program Repeatseq to determine the genotype of microsatellites in next generation sequencing reads (18). Methods used by the program itself can be found elsewhere (18). Repeatseq has been used in previous studies of microsatellites and is freely available: https://github.com/adaptivegenome/repeatseq. Repeatseq operates on three input files: a reference genome, a file containing reads aligned to the human reference genome (.bam file) and a list of known microsatellites (see ‘Materials and Methods’ section above). The unique advantage of Repeatseq over other microsatellite genotyping programs is that it realigns each read to the reference genome prior to array length detection. Result files in variant call format (.vcf) for all samples are available on request.
Each microsatellite genotype includes a pair of alleles which are in turn indicated by their base pair length. For example, the genotype ‘14|15’ indicates a heterozygote genotype with 14 and 15 bp alleles, respectively. The genotype ‘15|15’ indicates a homozygote genotype with two copies of the 15 bp allele.
Samples
Samples were downloaded from phase 3 of the 1000 genome project: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/. All samples listed in the phase 3 index — 20130502.phase3.exome.alignment.index—were included for analysis. Metadata for each sample was retrieved from the sample info file provided by the 1000 genomes project: 20130606_sample_info.txt. A summary of samples included in the CAGm database is shown in Table 1.
Table 1.
Summary of samples in the CAGm database
| Ethnicity | Gender | Samples |
|---|---|---|
| AFR | M | 318 |
| F | 349 | |
| EUR | M | 239 |
| F | 263 | |
| AMR | M | 171 |
| F | 181 | |
| EAS | M | 250 |
| F | 264 | |
| SAS | M | 263 |
| F | 231 |
Five ethnicities (super populations) are shown: African (AFR), American (AMR), European (EUR), East Asian (EAS) and South Asian (SAS). Each ethnicity draws from four to seven populations (not shown).
Implementation
The CAGm database is implemented with MySQL: a popular open source relational database. A website designed to interact with the database is implemented with HTML, PHP, CSS and Javascript. Codes written with PHP are adapted from the book, PHP and mysql for dynamic web sites: visual quickpro guide (30).
RESULTS
Database content and usage
The CAGm database contains information on 625 178 microsatellites across 2529 individuals in the 1000 genomes project. These individuals in turn come from 26 worldwide populations belonging to 5 ethnicities (super populations). The database contains 31 645 227 microsatellites genotypes: 12 513 genotypes on average for each sample. Genotypes are easily filtered against their statistical likelihood. Access to the 1 560 636 846 next generation sequencing reads—used for microsatellite genotyping—provides a source of validation and further analysis.
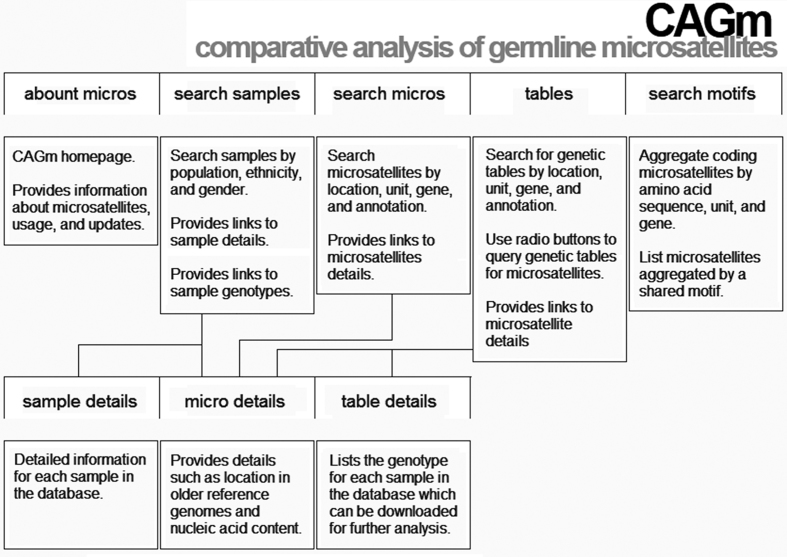
Contents of the CAGm database can be searched and downloaded with a modern web browser. No user registration is required. The website is navigated with assistance of five main pages: about micros; search samples; search micros; tables; and search motifs. Each page displays information in column-wise tables that are easily downloaded in Microsoft Excel spreadsheets. Once downloaded users can perform additional analysis directly in excel or import as a data frame using python, R or perl. In what follows we describe the use and functionality provided by each main page. See Figure 1 for a sitemap. Then usage is demonstrated with respect to a mononucleotide (A) microsatellite at the BAT-26 locus and a dinucleotide (CA) microsatellite in the coding region of FGFRL1.
Figure 1.
A map of the main pages (top row) and partial list of the subordinate pages (bottom row) on the CAGm website.
About micros (index.php)
This page serves as the entry point for the CAGm database. Users are provided a brief description of microsatellites along with instructions for interacting with the database. Users are informed of updates in the version and change log at the bottom of the page.
Search samples (view_samples.php)
This page is used to browse the individual samples in the database. Users can search and sort by ethnicity, population and gender. Ethnicity and population codes are taken directly from the 1000 genomes project. The CAGm database currently contains 2529 samples: 667 Africian (AFR); 352 American (AMR); 514 East Asian (EAS); 502 European (EUR); and 494 South Asian (SAS).
The leftmost columns provide links to the individual sample details and genotypes, respectively. Sample details are taken from the metadata provided by the 1000 genomes project: 20130606_sample_info.txt. The link to genotypes sends users to a separate page listing genetic information for a single sample; once redirected, genetic information is displayed in a column-wise table with each entry corresponding to a single genotype (one row per microsatellite). All genotypes are supported by six or more next generation sequencing reads; however, the number of genotypes available is not the same for every sample. Essentially, the sequenced read depth varies for each sample which in turn affects the number of available genotypes. A link to alignments provides even finer grained resolution allowing users to view the individual NGS reads that overlap a particular microsatellite. Alignments can be used to verify the genetic information (genotypes) provided by the database or as a starting point for new analysis.
Search micros (view_micros.php)
Microsatellites listed in the CAGm database are browsed using the ‘search micros’ page. The search functionality allows users to find microsatellites based on position in the reference genome (GRCh38), gene, annotation and repeat unit. Searching by gene will find all microsatellites within the introns, exons, coding sequence and untranslated regions (utr). On the other hand, searching against an annotation will find matching microsatellites across all genes. For example, to find all microsatellites in coding sequence (cds) a user searches against annotations: the number of matching microsatellites is displayed at the top of the page. Intergenic microsatellites have no gene or annotation values (NULL). Details for each microsatellite are linked to the leftmost column of the data table. These details include—but are not limited to—location in older reference genomes, flanking sequence and nucleotide percentages. Download links to excel spreadsheets are provided. These details are intended to provide users with sufficient information to construct standard bioinformatics files: bed files and fasta files in particular. These can then be used with standard tools such as CruzDB for retrieving additional information about genomic regions (31); GeneCards for investigating potential links to human disease (32); and RefSeq/LocusLink for investigating homologous genes (33).
Tables (search_tables.php)
A primary aim of the CAGm database is to contribute to what is known about human genetic variation. The ‘tables’ page is the entry point for browsing genetic variation by gender, population, and ethnicity (super population). Search functionality is identical to the aforementioned ‘search micros’ page. The difference is that only microsatellites with available genetic tables are listed in the search results. To display a genetic table users select a microsatellite using the radio buttons in the pick column. A second set of radio buttons is used to aggregate samples by gender, population or ethnicity (super population). A final set of radio buttons chooses how to display the genetic information: by genotype, allele or zygosity. Once submitted, users are redirected to a table summarizing genetic variation for the selected microsatellite (see Table 2 for an example). Users can easily re-group using two sets of radio buttons at the bottom of the page.
Table 2.
A key novelty of CAGm are tables that aggregate microsatellite variation by population, ethnicity (super population) and gender
| genotype | AFR | AMR | EUR | EAS | SAS |
|---|---|---|---|---|---|
| 17|21 | 1 | 1 | 0 | 0 | 2 |
| 19|21 | 33 | 11 | 12 | 11 | 30 |
| 21|21 | 403 | 164 | 238 | 198 | 297 |
| 21|23 | 5 | 2 | 0 | 0 | 4 |
| 21|25 | 0 | 0 | 0 | 0 | 1 |
Variants can be aggregated by genotype (shown), allele and zygosity. The table above shows genotypes by ethnicity (super population) for a CA dinucleotide repeat embedded in exon 6 of the FGFRL1 gene (chr4:1025267-1025287). Five ethnicities are shown: African (AFR), American (AMR), European (EUR), East Asian (EAS) and South Asian (SAS).
Each table of genetic information is accompanied by two links: one downloads the data as a Microsoft Excel spreadsheet; another (show table details) provides genotypes for all samples used to generate the table. The list of genotypes for each sample is easily searched, sorted and downloaded for additional analysis.
In general, microsatellite genotypes are difficult to infer with certainty from next generation sequencing reads. Genotypes in the CAGm database were determined using Bayesian model selection guided by an empirically derived error model (18); and, each is assigned a likelihood that the call made is correct. For serious projects it is recommended that the CAGm genotypes be vetted by examining the multiple sequence alignments available for download on the ‘show table details’ page. However, some vetting is built in to the database with a ‘minimum likelihood’ filter. Genotypes with assigned likelihood below the user adjusted value are excluded from genotype tables. We recommend exploring the database with the minimum likelihood set to 50, before moving to higher values.
Search Motifs (view_motifs.php)
This page shows motifs shared among coding microsatellites within the CAGm database; here, motif broadly refers to an amino acid sequence, unit or gene. Microsatellites sharing a motif are aggregated and sorted by their count. Users can limit aggregation to specific regions of the genome or search for specific motifs using a form at the top of the page. The left-most column provides a link that redirects users to a sub-list of the aggregated microsatellites for the selected motif. Users can search the sub-list of microsatellites by genomic region or motif using a form at the top of the page. The sub-list also provides links to details for each microsatellites and radio buttons for displaying genetic tables (if available).
CAGm recapitulates polymorphic variation at the BAT-26 locus
The BAT-26 locus contains a 12-27-repeat polyadenine tract, located on chromosome 2 within the fifth intron of the MSH2 gene. CAGm shows a total of 15 BAT-26 alleles segregating in human populations, with the highest degree of polymorphism in Africa (14 alleles) and the lowest in East Asia (5 alleles). Allelic variation in the size of the poly-A tract at BAT-26 has previously been found in 13 out of 103 healthy African-Americans (12.6%) (34).
Variation in BAT-26 is commonly associated with microsatellite instability (MSI) in colorectal adenomas and carcinomas (35–41). Racial/ethnic disparities in the incidence of colorectal cancer have been observed in USA, with African Americans having increased risk (42–45), but its direct association with BAT-26 polymorphism and MSI has not been studied. Additionally, BAT-26 variation has been implicated in MSI associated with bladder (46) and gastric carcinomas (47). Data provided by CAGm could be used for hypothesis generating and new analysis.
CAGm builds on what is known about FGFRL1
The fibroblast growth factor receptor-like 1 (FGFRL1) gene encompasses the longest exonic dinucleotide microsatellite repeat in the human genome. The human reference genome indicates the dinucleotide unit (CA) is repeated 10 times in the coding region of FGFRL1 (48). Our findings, displayed in CAGm, indicate that microsatellite alleles vary from 8–12 units (see Table 2). Although non-coding dinucleotide microsatellites are highly mutable (10), the length of exonic dinucleotide microsatellites are tightly conserved, because of the possible damaging effects of protein-altering mutations (49). A previous study demonstrated that the CA-microsatellite in FGFRL1 (chr4:1025267-1025287) has a repeat length of 10 in 400 sampled human chromosomes (48). The CAGm database has built on this result by genotyping the same microsatellite in 1413 samples from the 1000 genome project: 1300 of these samples have a repeat length of 10 CA units (21 bp). However, CAGm reveals that 97 samples may carry a 9 CA unit allele (19 bp) and 11 samples may carry an 11 CA unit allele (23 bp). In fact, CAGm also shows 4 samples with an 8 unit allele and 1 with a 12 unit allele. Still, further investigation is needed. Under stringent filter conditions the more extreme calls of 17 and 25 bp are unsupported while the 19 and 23 bp alleles are supported. These data support the hypothesis that the CA-repeat in FGFRL1 is in mutation-selection balance; in other words, strong purifying selection does not fully purge populations of all deleterious alleles.
Variations in FGFRL1 have been associated with clinical phenotypes such as Wolf–Hirschhorn syndrome, a rare genetic disease involving symptoms of delayed development, intellectual disability, ataxia and seizures (50). The Catalogue of Mutations in Cancer (COSMIC) database indicates that FGFRL1 is altered and overexpressed in multiple cancers subtypes e.g. in ovarian tissue, FGFRL1 was overexpressed in 13.2% of the 266 tested samples (51). Interestingly, the COSMIC database identifies insertion/deletion frameshift mutations in exonic dinucleotide microsatellite loci of FGFRL1 in 13 samples. FGFRL1 has not been thoroughly studied despite the fact that variations in this locus may be clinically relevant.
DISCUSSION
It is now acknowledged that microsatellites play an important role in gene expression. Consequently, a growing number of studies are leveraging microsatellites as genetic markers of complex human disease. Although MSI has been studied for decades, less attention has been given to germline microsatellites. This discrepancy is slowly changing. So far, germline microsatellite markers have been proposed for breast cancer (26,27), ovarian cancer (27) and lung cancer (28). The Comparative Analysis of Germline Microsatellites (CAGm) database presented in this work may expedite the discovery of additional markers by serving as a catalog of germline genotypes in healthy individuals. Thus, studies aimed at microsatellite marker discovery should be able to use CAGm for construction of suitable control groups by drawing samples from particular ethnicities, populations or regions.
The use of our database is not limited to biomarker discovery. Future studies could also use these data to shed light on human genetic variation at large. Exonic (coding) microsatellites directly affect translation and primary protein structure. The effects of microsatellites in other functional units may be less direct but nonetheless important. It is hypothesized that microsatellite variants in 5′-UTRs affect transcription and translation; those in 3′-UTRs may cause transcription slippage or disrupted splicing (52). The database presented in this work will contribute by revealing new microsatellite variants and the extent of genetic variation in 26 worldwide populations belonging to 5 ethnicities. We show that the contents of CAGm are consistent with previous studies of a mononucleotide (A) microsatellite at the BAT-26 locus and a dinucleotide (CA) microsatellite in the coding region of FGFRL1. In the case of BAT-26, CAGm reveals a high degree of polymorphism in Africa (14 alleles). In the case of FGFRL1, CAGm reveals several previously unknown array length variants that may affect its function. Many additional unknown variants undoubtedly remain in the database.
So far we have only analyzed microsatellites with array length variants <100 bp: a limitation that stems from the read length of Illumina (Solexa) sequencing technology. Essentially, detection of a microsatellite variant requires reads spanning the entire tandem repeat array and its unique 3′ and 5′ flanking base pairs (18). Reads that truncate the tandem repeat array are unused in our analysis. Unfortunately, we suspect that some of these unused truncated reads are genetically and clinically relevant. Indeed, the disease range for most trinucleotide expansion disorders exceeds 100 bp. Superficially it seems that the CAGm database can easily be improved by using longer read sequencing technologies: Roche 454, IonTorrent, PacBio and NanoPore. However, these technologies are prone to indel errors at a rate much higher than illumina; in the case of microsatellites this is critical limitation. Nevertheless, rapid improvements in sequencing technology may soon pave the way for expansion of the CAGm database to include insertions in excess of 100 bp.
ACKNOWLEDGEMENTS
We thank our biostatistician Liang Shan for assisting with statistical analysis.
FUNDING
Bradley Engineering Foundation to the Edward Via College of Osteopathic Medicine; Edward Via College of Osteopathic Medicine (to N.K.). Funding for open access charge: Edward Via College of Osteopathic Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Li Y.C., Korol A.B., Fahima T., Nevo E.. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 2004; 21:991–1007. [DOI] [PubMed] [Google Scholar]
- 2. Li Y.C., Korol A.B., Fahima T., Beiles A., Nevo E.. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 2002; 11:2453–2465. [DOI] [PubMed] [Google Scholar]
- 3. Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., Bao W., Smit A.F., Wheeler T.J.. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016; 44:D81–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bao W., Kojima K.K., Kohany O.. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015; 6:doi:10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Du L., Li Y., Zhang X., Yue B.. MSDB: a user-friendly program for reporting distribution and building databases of microsatellites from genome sequences. J. Hered. 2013; 104:154–157. [DOI] [PubMed] [Google Scholar]
- 6. Kumar P., Chaitanya P.S., Nagarajaram H.A.. PSSRdb: a relational database of polymorphic simple sequence repeats extracted from prokaryotic genomes. Nucleic Acids Res. 2011; 39:D601–D605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaturvedi A., Tiwari S., Jesudasan R.A.. RiDs db: repeats in diseases database. Bioinformation. 2011; 7:96–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sokol D., Atagun F.. TRedD—a database for tandem repeats over the edit distance. Database (Oxford). 2010; 2010:doi:10.1093/database/baq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subramanian S., Madgula V.M., George R., Kumar S., Pandit M.W., Singh L.. SSRD: simple sequence repeats database of the human genome. Comp. Funct. Genomics. 2003; 4:342–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun J.X., Helgason A., Masson G., Ebenesersdottir S.S., Li H., Mallick S., Gnerre S., Patterson N., Kong A., Reich D. et al. A direct characterization of human mutation based on microsatellites. Nat. Genet. 2012; 44:1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valdes A.M., Slatkin M., Freimer N.B.. Allele frequencies at microsatellite Loci - the stepwise mutation model revisited. Genetics. 1993; 133:737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Payseur B.A., Jing P.C., Haasl R.J.. A genomic portrait of human microsatellite variation. Mol. Biol. Evol. 2011; 28:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Treangen T.J., Salzberg S.L.. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2011; 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shin S., Park J.. Characterization of sequence-specific errors in various next-generation sequencing systems. Mol. Biosyst. 2016; 12:914–922. [DOI] [PubMed] [Google Scholar]
- 15. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hannan A.J. Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 2018; 19:286–298. [DOI] [PubMed] [Google Scholar]
- 17. Gymrek M. A genomic view of short tandem repeats. Curr. Opin. Genet. Dev. 2017; 44:9–16. [DOI] [PubMed] [Google Scholar]
- 18. Highnam G., Franck C., Martin A., Stephens C., Puthige A., Mittelman D.. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 2013; 41:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hannan A.J. Tandem repeat polymorphisms: Mediators of genetic plasticity, modulators of biological diversity and dynamic sources of disease susceptibility. Adv. Exp. Med. Biol. 2012; 769:1–9. [PubMed] [Google Scholar]
- 20. Vinces M.D., Legendre M., Caldara M., Hagihara M., Verstrepen K.J.. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009; 324:1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wren J.D., Forgacs E., Fondon J.W. 3rd, Pertsemlidis A., Cheng S.Y., Gallardo T., Williams R.S., Shohet R.V., Minna J.D., Garner H.R.. Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet. 2000; 67:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kashi Y., King D.G.. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006; 22:253–259. [DOI] [PubMed] [Google Scholar]
- 23. Sawaya S.M., Bagshaw A.T., Buschiazzo E., Gemmell N.J.. Hannan AJ. Promoter Microsatellites as Modulators of Human Disease. Tandem Repeat Polymorphisms: Genetic Plasticity, Neural Diversity and Disease. 2012; NY: Springer; 41–54. [Google Scholar]
- 24. Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P., Gabriel S.B. et al. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Everett C.M. Trinucleotide repeat disorders. Encyclopedia of Movement Disorders. 2010; 3:Academic Press; 290–296. [Google Scholar]
- 26. Kinney N., Varghese R.T., Anandakrishnan R., Garner H.R.. ZDHHC3 as a Risk and mortality marker for breast cancer in African American women. Cancer Inform. 2017; 16:doi:10.1177/1176935117746644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McIver L.J., Fonville N.C., Karunasena E., Garner H.R.. Microsatellite genotyping reveals a signature in breast cancer exomes. Breast Cancer Res. Treat. 2014; 145:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Velmurugan K.R., Varghese R.T., Fonville N.C., Garner H.R.. High-depth, high-accuracy microsatellite genotyping enables precision lung cancer risk classification. Oncogene. 2017; 36:6383–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tae H., Kim D.Y., McCormick J., Settlage R.E., Garner H.R.. Discretized Gaussian mixture for genotyping of microsatellite loci containing homopolymer runs. Bioinformatics. 2014; 30:652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ullman L. Php and Mysql for Dynamic Web Sites: Visual Quickpro Guide. 2011; Berkeley, CA: Peachpit Press. [Google Scholar]
- 31. Pedersen B.S., Yang I.V., De S.. CruzDB: software for annotation of genomic intervals with UCSC genome-browser database. Bioinformatics. 2013; 29:3003–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H. et al. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010; 2010:doi:10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pruitt K.D., Maglott D.R.. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001; 29:137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pyatt R., Chadwick R.B., Johnson C.K., Adebamowo C., de la Chapelle A., Prior T.W.. Polymorphic variation at the BAT-25 and BAT-26 loci in individuals of African origin. Implications for microsatellite instability testing. Am. J. Pathol. 1999; 155:349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parsons R., Myeroff L.L., Liu B., Willson J.K., Markowitz S.D., Kinzler K.W., Vogelstein B.. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995; 55:5548–5550. [PubMed] [Google Scholar]
- 36. Hoang J.M., Cottu P.H., Thuille B., Salmon R.J., Thomas G., Hamelin R.. BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines. Cancer Res. 1997; 57:300–303. [PubMed] [Google Scholar]
- 37. Cravo M., Lage P., Albuquerque C., Chaves P., Claro I., Gomes T., Gaspar C., Fidalgo P., Soares J., Nobre-Leitao C.. BAT-26 identifies sporadic colorectal cancers with mutator phenotype: a correlative study with clinico-pathological features and mutations in mismatch repair genes. J. Pathol. 1999; 188:252–257. [DOI] [PubMed] [Google Scholar]
- 38. Zhou X.P., Hoang J.M., Li Y.J., Seruca R., Carneiro F., Sobrinho-Simoes M., Lothe R.A., Gleeson C.M., Russell S.E., Muzeau F. et al. Determination of the replication error phenotype in human tumors without the requirement for matching normal DNA by analysis of mononucleotide repeat microsatellites. Genes Chromosomes Cancer. 1998; 21:101–107. [DOI] [PubMed] [Google Scholar]
- 39. Brennetot C., Buhard O., Jourdan F., Flejou J.F., Duval A., Hamelin R.. Mononucleotide repeats BAT-26 and BAT-25 accurately detect MSI-H tumors and predict tumor content: implications for population screening. Int. J. Cancer. 2005; 113:446–450. [DOI] [PubMed] [Google Scholar]
- 40. Samowitz W.S., Slattery M.L., Potter J.D., Leppert M.F.. BAT-26 and BAT-40 instability in colorectal adenomas and carcinomas and germline polymorphisms. Am. J. Pathol. 1999; 154:1637–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gonzalez M.L., Causada-Calo N., Santino J.P., Dominguez-Valentin M., Ferro F.A., Sammartino I., Kalfayan P.G., Verzura M.A., Pinero T.A., Cajal A.R. et al. Universal determination of microsatellite instability using BAT26 as a single marker in an Argentine colorectal cancer cohort. Fam. Cancer. 2018; 17:395–402. [DOI] [PubMed] [Google Scholar]
- 42. Rim S.H., Seeff L., Ahmed F., King J.B., Coughlin S.S.. Colorectal cancer incidence in the United States, 1999–2004: an updated analysis of data from the National Program of Cancer Registries and the Surveillance, Epidemiology, and End Results Program. Cancer. 2009; 115:1967–1976. [DOI] [PubMed] [Google Scholar]
- 43. Chien C., Morimoto L.M., Tom J., Li C.I.. Differences in colorectal carcinoma stage and survival by race and ethnicity. Cancer. 2005; 104:629–639. [DOI] [PubMed] [Google Scholar]
- 44. Matanoski G., Tao X., Almon L., Adade A.A., Davies-Cole J.O.. Demographics and tumor characteristics of colorectal cancers in the United States, 1998–2001. Cancer. 2006; 107:1112–1120. [DOI] [PubMed] [Google Scholar]
- 45. Ollberding N.J., Nomura A.M., Wilkens L.R., Henderson B.E., Kolonel L.N.. Racial/ethnic differences in colorectal cancer risk: the multiethnic cohort study. Int. J. Cancer. 2011; 129:1899–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vaish M., Mandhani A., Mittal R.D., Mittal B.. Microsatellite instability as prognostic marker in bladder tumors: a clinical significance. BMC Urol. 2005; 5:doi:10.1186/1471-2490-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Halling K.C., Harper J., Moskaluk C.A., Thibodeau S.N., Petroni G.R., Yustein A.S., Tosi P., Minacci C., Roviello F., Piva P. et al. Origin of microsatellite instability in gastric cancer. Am. J. Pathol. 1999; 155:205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haasl R.J., Payseur B.A.. Remarkable selective constraints on exonic dinucleotide repeats. Evolution. 2014; 68:2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y.C., Korol A.B., Fahima T., Nevo E.. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 2004; 21:991–1007. [DOI] [PubMed] [Google Scholar]
- 50. Battaglia A., Carey J.C., South S.T.. Wolf–Hirschhorn syndrome: A review and update. Am. J. Med. Genet. C Semin. Med. Genet. 2015; 169:216–223. [DOI] [PubMed] [Google Scholar]
- 51. Forbes S.A., Beare D., Bindal N., Bamford S., Ward S., Cole C.G., Jia M., Kok C., Boutselakis H., De T. et al. COSMIC: high-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr. Protoc. Hum. Genet. 2016; 91:10.11.1–10.11.37. [DOI] [PubMed] [Google Scholar]
- 52. Vieira M.L.C., Santini L., Diniz A.L., Munhoz C.D.. Microsatellite markers: what they mean and why they are so useful. Genet. Mol. Biol. 2016; 39:312–328. [DOI] [PMC free article] [PubMed] [Google Scholar]