Abstract
DNA methylation, the most intensively studied epigenetic modification, plays an important role in understanding the molecular basis of diseases. Furthermore, epigenome-wide association study (EWAS) provides a systematic approach to identify epigenetic variants underlying common diseases/phenotypes. However, there is no comprehensive database to archive the results of EWASs. To fill this gap, we developed the EWASdb, which is a part of 'The EWAS Project', to store the epigenetic association results of DNA methylation from EWASs. In its current version (v 1.0, up to July 2018), the EWASdb has curated 1319 EWASs associated with 302 diseases/phenotypes. There are three types of EWAS results curated in this database: (i) EWAS for single marker; (ii) EWAS for KEGG pathway and (iii) EWAS for GO (Gene Ontology) category. As the first comprehensive EWAS database, EWASdb has been searched or downloaded by researchers from 43 countries to date. We believe that EWASdb will become a valuable resource and significantly contribute to the epigenetic research of diseases/phenotypes and have potential clinical applications. EWASdb is freely available at http://www.ewas.org.cn/ewasdb or http://www.bioapp.org/ewasdb.
INTRODUCTION
DNA methylation plays a critical role in regulating chromatin structure (1) and gene expression (2,3), and takes part in a variety of key biological processes including human development (4–7), aging (8,9), genomic imprinting (10,11) and the inactivation of tumor suppressor genes (12).
Recently, a systematic method, named epigenome-wide association study (EWAS), has been developed to identify epigenetic variants associated with complex diseases (13–16) or phenotypes (17). EWAS provides an effective means to understand the molecular basis for disease risk. Meanwhile, the development of high throughput technology, including the generation of Illumina HumanMethylation450 Bead Chip data, makes it possible to complete a large scale EWAS scan (13,18). To date, using EWASs, many complex diseases/phenotypes, including body mass index (BMI), adiposity (19), autism spectrum disorder (ASD) (20) and cancer (13,21,22), have been successfully analyzed.
Using EWASs, achievement has been made in identifying risk epigenetic variations of diseases/phenotypes. However, compared with genome-wide association study (GWAS, identifying common genetic variants associated with diseases/phenotypes), EWAS has lagged behind. To this end, we proposed ‘The EWAS Project’ in 2015 to develop EWAS analysis tools and data resources. Previously, we designed EWAS2.0 JAVA software to identify the association between DNA methylation levels and complex diseases (23–25). However, our investigation of EWAS data resources revealed few databases that archive epigenetic association results from EWASs. This has made it difficult for researchers to view associations between diseases/phenotypes and epigenetics. Here, as an important part of ‘The EWAS Project’, we developed the EWASdb (epigenome-wide association study database, http://www.ewas.org.cn/ewasdb or http://www.bioapp.org/ewasdb), to store and utilize the significant epi-markers identified in EWASs.
The EWASdb can be used to query DNA methylation markers, genes, KEGG pathways, and GO categories which have significant associations with some diseases or phenotypes. In addition, the EWASdb can help to reveal the mechanism of complex diseases at the epigenetic level.
DATABASE CONTENT
DNA methylation data
The Illumina HumanMethylation450 Bead Chip (450K) and Illumina Infinium MethylationEPIC Bead Chip (850K) methylation microarray are the most widely used platforms for epigenome-wide association study, including 482 421 CpG sites and 864 935 CpG sites, respectively. To construct a comprehensive and practical database, we downloaded 907 DNA methylation 450K datasets and 64 DNA methylation 850K datasets updated by the end of July 2018, involving 78 180 samples in matrix format, from the GPL13534 and GPL21145 platforms of the GEO (Gene Expression Omnibus) database (26).
Classification of diseases/phenotypes
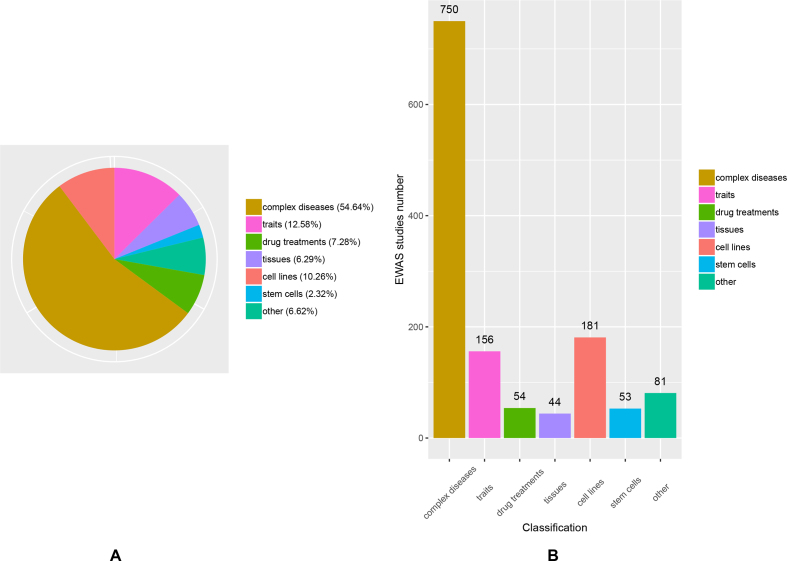
We employed two groups of independent experts to classify the downloaded EWAS datasets. In brief, one group of experts extracted effective information and classified the data based on the published literature corresponding to the EWAS datasets. The other group of experts was responsible for checking the obtained information to ensure the accuracy of the classification. Finally, we curated 1319 (Table 1) EWASs and divided them into seven classifications: ‘disease’, ‘trait’, ‘drug treatment’, ‘tissue’, ‘cell line’, ‘stem cell’, and ‘other’. Among the seven classifications, there are 302 sub-classifications, of which 165 are related to ‘complex disease’, 38 are related to ‘trait’, 22 are related to ‘drug treatment’, 19 are related to ‘tissue’, 31 are related to ‘cell line’, seven are related to ‘stem cell’ and 20 ambiguous sub-classifications were placed in the class of ‘other’ (Figure 1A). The number of the EWASs relevant to each of the seven classifications is shown in Figure 1B.
Table 1.
The summary of the data curated in EWASdb
| Data type | Data number |
|---|---|
| EWAS studies | 1319 |
| EWAS results for single epi-marker (P< 1e–7) | 18 538 029 |
| EWAS results for single epi-marker (P< 1e–3) | 52 292 604 |
| EWAS results for KEGG pathway (P< 1e–3) | 49 967 |
| EWAS results for GO categories (P< 1e–3) | 930 609 |
| EWAS results for BP GO categories (P< 1e–3) | 686 552 |
| EWAS results for MF GO categories (P< 1e–3) | 141 979 |
| EWAS results for CC GO categories (P< 1e–3) | 102 078 |
| Gene | 27 918 |
| Diseases/phenotypes | 302 |
Figure 1.
General statistical situation of the diseases/phenotypes. (A) percentage of sub-classifications relevant to each of the seven classifications; (B) number of the EWASs relevant to each of the seven classifications.
Identification of disease/phenotype related epi-markers
For each EWAS, we identified the association between epigenetic variations and disease/phenotype using EWAS v2.0 software (25). Using a moderate P-value (less than 1.0 × 10−3), we obtained a total of 52 292 604 disease/phenotype related markers (Table 1). Meanwhile, using a strictly significant P-value level (less than 1.0 × 10−7), 18 538 029 CpG loci were obtained (Table 1). In addition, we mapped the gene and chromosomal locations of these significant markers based on the Illumina HumanMethylation450/850 Bead Chip annotation information.
Identification of KEGG pathways
Kyoto Encyclopedia of Genes and Genomes (KEGG) consists of graphical diagrams of biochemical pathways, including metabolic pathways and some known regulatory pathways (27). Gene functions can be systematically analyzed in terms of the networks of genes and molecules. Hence, we performed KEGG pathway analysis of the significant genes to reveal the biochemical pathways they are involved in, and to understand the interactions between these genes. Susceptible biological pathways were identified by performing hypergeometric test analysis. We acquired 49 967 risk KEGG pathways at a significance level with P-values less than 1.0 × 10−3 (Table 1).
Identification of GO (Gene Ontology) Category
To further understand the functional characteristics of disease/phenotype susceptible epigenetic variations we performed Gene Ontology (GO) annotation for each risk gene (28). We used the hypergeometric test method to screen related GO terms for risk CpG loci at a P-value less than 1.0 × 10−3. Finally, we obtained 930 609 disease/phenotype associated GO terms including 686 552 biological processes (BP), 141 979 molecular functions (MF) and 102 078 cellular components (CC) (Table 1).
DATABASE ORGANIZATION AND WEB INTERFACE
Database construction
The EWASdb was constructed based on PHP language, and the web interface was built using HTML and JavaScript. All data of our database are stored in MySQL. The database has been tested using Google Chrome, Firefox, and Internet Explorer web browsers.
Search interface
To make the querying convenient and effective for users, we provide three kinds of search interfaces: (i) EWAS for single marker; (ii) EWAS for KEGG Pathway and (iii) EWAS for GO Category.
EWAS for single marker
In this search module, EWASdb provides five different ways to search for detailed information of single DNA methylation markers associated with diseases/phenotypes: (i) Search by disease/phenotype: users can enter a disease/phenotype, like ‘glioma’, to get a list of all the EWASs related to this disease or phenotype; (ii) Search by EWAS ID: users can enter any item from EWAS1 to EWAS1319 to obtain all the significant loci in this EWAS; (iii) Search by gene: users can input their gene of interest, such as ‘TTTY18’ or ‘TMSB4Y’, to acquire the loci mapped into this gene from all the EWASs; (iv) Search by cg#: users can query by CpG locus to obtain all of the information from this CpG locus of each EWAS and (v) Search by region#: users can fill in the chromosome number and the chromosomal starting and ending positions in the search box to view the significant loci in this region from all of the EWASs.
For each EWAS, users can obtain detailed information, such as ‘GSE ID’, ‘EWAS Title’, ‘Disease/Phenotype’, ‘Group name’, ‘Sample size’, ‘Summary of EWAS’, ‘Contributor’, ‘Public date’ and ‘Citation (PubMed ID)’.
For each epi-marker, users can obtain detailed information, such as ‘EWAS ID’, ‘cg#’, ‘chr#’, ‘Position’, ‘Gene’, ‘Disease/Phenotype’, ‘Classification’, ‘Sample size’, ‘Average beta-value in two groups’, ‘T-statistics’ and ‘P-value’.
EWAS for KEGG Pathway
The EWASdb contains interfaces for users to acquire KEGG pathways significantly associated with diseases/phenotypes. Users can input a pathway ID or pathway name, such as ‘hsa04510’ or ‘Galactose metabolism’, to obtain the details of EWASs involving this pathway. In addition, users can enter any item from EWAS1 to EWAS1319 to obtain the pathways associated with the particular EWAS.
For each KEGG pathway, the following information is displayed: ‘EWAS ID’, ‘KEGG ID’, ‘Classification’, ‘Disease/Phenotype’, ‘Sample groups’, ‘KEGG Pathway Name’, ‘Pathway Gene Number’, ‘Gene Number Annotated In The KEGG Pathway’ and ‘P-value’.
EWAS for GO Category
The database allows users to get the GO terms related to the diseases/phenotypes of interest. Similarly, user can input GO term name or GO term ID, such as ‘mitochondrial genome maintenance’ or ‘GO:0000902’, to obtain the EWASs associated with this GO term. Furthermore, any item from EWAS1 to EWAS1319 can be inputted to obtain the significant GO terms associated with that particular EWAS.
For each GO term, the EWASdb provides detailed information for users, such as: ‘EWAS ID’, ‘GO ID’, ‘Classification’, ‘Disease/Phenotype’, ‘Sample groups’, ‘Categories’, ‘GO Term Name’, ‘GO Term Gene Number’, ‘Gene Number Annotated In The GO Term’ and ‘P-value’.
Browse and download
The database allows users to browse data ‘by class’ and ‘by alph’. For ‘browse by class’, seven classifications are shown on the browse tree. Users can choose any one of them to view the disease/phenotype and the associated EWASs. Furthermore, by clicking the EWAS ID, users can achieve detailed information about the specific EWAS. For ‘browse by alph’, users can get an overview of the EWAS results by clicking the alphabet on the browse tree.
All data can be downloaded freely from the ‘Download’ page. In addition, users can also download the EWAS v2.0 (25) to conduct their own EWAS by clicking the ‘EWAS Software’.
SUMMARY AND DISCUSSION
Over the past few years, GWAS has identified a large number of genomic variants associated with diseases/phenotypes, many of which are in meaningful biological pathways (29). Although GWAS has contributed to our understanding of the occurrence and development of diseases and prognosis, they have failed to fully explain the risk factors that contribute to diseases/phenotypes. The epigenome provides some new insight to understand risk factors affecting the diseases/phenotypes by considering both genetics and environmental perspective.
With the rapid growth in DNA methylation data and the maturity of EWAS methods, increasing numbers of epigenetic variants associated with complex diseases/phenotypes have been identified (28,30,31). A useful database resource, which systemically integrates the results of EWASs, will be of great benefit to researchers. Therefore, we collected the available DNA methylation datasets focusing on different diseases/phenotypes to develop the EWASdb. The EWASdb can be queried in a wide range of ways to meet the requirements of different users. The database will provide a means for researchers to explore and understand the pathogenesis of complex disease from the epigenetic level.
FUTURE DIRECTIONS
EWAS is one of the most useful method to identify genome-wide epigenetic variations associated with diseases/phenotypes. However, it still has some limitations. First, robust findings require large-scale samples and rigorous scientific method (32). In subsequent EWASdb versions, we will collect multiple sets of DNA methylation data for a phenotype or disease and use a meta-analysis strategy to identify more stable and reliable epigenetic markers. In addition, EWAS can more powerful for identifying robust epigenetic markers in common variations rather than rare variations (32). The current EWASdb version contains only the analysis results of common variations. For rare epigenetic variations, we will develop some novel analysis methods to improve its analytical performance in the future.
The EWASdb is a most important part of ‘The EWAS Project’ and has been widely used and downloaded by researchers from more than 40 countries. Our team has the ability to continuously update and maintain the database when more 450K or 850K DNA methylation data are available. We believe that the EWASdb, a comprehensive database, will become a valuable resource and a useful tool in the future.
ACKNOWLEDGEMENTS
We extend our sincere thanks to the anonymous reviewers for carefully reviewing our work and constructive comments to strengthen this manuscript. We also thank the contribution for EWAS from the GEO (Gene Expression Omnibus) database and all the researchers studying in DNA methylation.
FUNDING
National Natural Science Foundation of China [91746113, 81601422, 81600403, 81701350, 31501062, in part]; China Postdoctoral Science Foundation [2016M600259]; Heilongjiang Postdoctoral Fund [LBH-Z16145]; Fundamental Research Funds for the Provincial Universities [2017JCZX45]; Heilongjiang Education Department Fund [12531268, 2012-316]. Funding for open access charge: National Natural Science Foundation of China [91746113].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hamidi T., Singh A.K., Chen T.. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015; 7:247–265. [DOI] [PubMed] [Google Scholar]
- 2. Linn F., Heidmann I., Saedler H., Meyer P.. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol. Gen. Genet.: MGG. 1990; 222:329–336. [DOI] [PubMed] [Google Scholar]
- 3. Shirodkar A.V., St Bernard R., Gavryushova A., Kop A., Knight B.J., Yan M.S., Man H.S., Sud M., Hebbel R.P., Oettgen P. et al. . A mechanistic role for DNA methylation in endothelial cell (EC)-enriched gene expression: relationship with DNA replication timing. Blood. 2013; 121:3531–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li E., Bestor T.H., Jaenisch R.. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992; 69:915–926. [DOI] [PubMed] [Google Scholar]
- 5. Okano M., Bell D.W., Haber D.A., Li E.. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99:247–257. [DOI] [PubMed] [Google Scholar]
- 6. Illingworth R., Kerr A., Desousa D., Jorgensen H., Ellis P., Stalker J., Jackson D., Clee C., Plumb R., Rogers J. et al. . A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008; 6:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moore L.D., Le T., Fan G.. DNA methylation and its basic function. Neuropsychopharmacology. 2013; 38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones M.J., Goodman S.J., Kobor M.S.. DNA methylation and healthy human aging. Aging Cell. 2015; 14:924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li E., Beard C., Jaenisch R.. Role for DNA methylation in genomic imprinting. Nature. 1993; 366:362–365. [DOI] [PubMed] [Google Scholar]
- 11. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007; 447:425–432. [DOI] [PubMed] [Google Scholar]
- 12. Xie Q., Bai Q., Zou L.Y., Zhang Q.Y., Zhou Y., Chang H., Yi L., Zhu J.D., Mi M.T.. Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosomes Cancer. 2014; 53:422–431. [DOI] [PubMed] [Google Scholar]
- 13. Verma M. Epigenome-Wide Association Studies (EWAS) in Cancer. Curr. Genomics. 2012; 13:308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel C.J., Bhattacharya J., Butte A.J.. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010; 5:e10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui T., Zhang L., Huang Y., Yi Y., Tan P., Zhao Y., Hu Y., Xu L., Li E., Wang D.. MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018; 46:D371–D374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su J., Huang Y.H., Cui X., Wang X., Zhang X., Lei Y., Xu J., Lin X., Chen K., Lv J. et al. . Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 2018; 19:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Orozco L.D., Morselli M., Rubbi L., Guo W., Go J., Shi H., Lopez D., Furlotte N.A., Bennett B.J., Farber C.R. et al. . Epigenome-wide association of liver methylation patterns and complex metabolic traits in mice. Cell Metab. 2015; 21:905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore K., McKnight A.J., Craig D., O’Neill F.. Epigenome-wide association study for Parkinson's disease. NeuroMol. Med. 2014; 16:845–855. [DOI] [PubMed] [Google Scholar]
- 19. Wahl S., Drong A., Lehne B., Loh M., Scott W.R., Kunze S., Tsai P.C., Ried J.S., Zhang W., Yang Y. et al. . Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017; 541:81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andrews S.V., Sheppard B., Windham G.C., Schieve L.A., Schendel D.E., Croen L.A., Chopra P., Alisch R.S., Newschaffer C.J., Warren S.T. et al. . Case-control meta-analysis of blood DNA methylation and autism spectrum disorder. Mol. Autism. 2018; 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karlsson A., Jonsson M., Lauss M., Brunnstrom H., Jonsson P., Borg A., Jonsson G., Ringner M., Planck M., Staaf J.. Genome-wide DNA methylation analysis of lung carcinoma reveals one neuroendocrine and four adenocarcinoma epitypes associated with patient outcome. Clin. Cancer Res. 2014; 20:6127–6140. [DOI] [PubMed] [Google Scholar]
- 22. Johansson A., Flanagan J.M.. Epigenome-wide association studies for breast cancer risk and risk factors. Trends Cancer Res. 2017; 12:19–28. [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao L., Liu D., Xu J., Wang Z., Chen Y., Lei C., Li Y., Liu G., Jiang Y.. The framework for population epigenetic study. Brief. Bioinform. 2018; 19:89–100. [DOI] [PubMed] [Google Scholar]
- 24. Xu J., Liu D., Zhao L., Li Y., Wang Z., Chen Y., Lei C., Gao L., Kong F., Yuan L. et al. . EWAS: epigenome-wide association studies software 1.0 - identifying the association between combinations of methylation levels and diseases. Sci. Rep. 2016; 6:37951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu J., Zhao L., Liu D., Hu S., Song X., Li J., Lv H., Duan L., Zhang M., Jiang Q. et al. . EWAS: epigenome-wide association study software 2.0. Bioinformatics. 2018; 34:2657–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. . NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M.. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999; 27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gene Ontology, C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015; 43:D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Visscher P.M., Brown M.A., McCarthy M.I., Yang J.. Five years of GWAS discovery. Am. J. Hum. Genet. 2012; 90:7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okugawa Y., Grady W.M., Goel A.. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015; 149:1204–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vedham V., Verma M.. Cancer-associated infectious agents and epigenetic regulation. Methods Mol. Biol. 2015; 1238:333–354. [DOI] [PubMed] [Google Scholar]
- 32. Flanagan J.M. Epigenome-wide association studies (EWAS): past, present, and future. Methods Mol. Biol. 2015; 1238:51–63. [DOI] [PubMed] [Google Scholar]