Abstract
Long non-coding RNAs (lncRNAs) play crucial roles in regulating gene expression, and a growing number of researchers have focused on the identification of target genes of lncRNAs. However, no online repository is available to collect the information on target genes regulated by lncRNAs. To make it convenient for researchers to know what genes are regulated by a lncRNA of interest, we developed a database named lncRNA2Target to provide a comprehensive resource of lncRNA target genes in 2015. To update the database this year, we retrieved all new lncRNA–target relationships from papers published from 1 August 2014 to 30 April 2018 and RNA-seq datasets before and after knockdown or overexpression of a specific lncRNA. LncRNA2Target database v2.0 provides a web interface through which its users can search for the targets of a particular lncRNA or for the lncRNAs that target a particular gene, and is freely accessible at http://123.59.132.21/lncrna2target.
INTRODUCTION
Development of RNA sequencing technology leads to recognition of large amount of novel transcripts (1,2). The corresponding large-scale genomic and transcriptomic analysis has shown that over 90% of the human genome is actively transcribed (2), while only ∼2% of the genome can encode proteins (3). In comparison, a large portion of the transcripts are non-coding RNAs (ncRNAs) (4), such as long ncRNAs (lncRNAs)
lncRNA plays a crucial role in biological processes (5,6) and even in diseases (7–13). In 2014, Sun et al. presented that BANCR could be a biomarker for poor prognosis of non-small cell lung cancer (NSCLC) by investigating its expression in multiple NSCLC tissues (14). In 2016, Zhu et al. observed that lncRNA MEG3 may be a potential target and therapeutic strategy for diabetes since that the upregulation of MEG3 enhances hepatic insulin resistance (15). Many important databases such as lncRNADisease (16), lncRNASNP2 (17), LncRNAWiki (18), MNDR (19), EVLncRNAs (20), NONCODE (21), LncRNA2Function (22) and TF2LncRNA (23) were developed for collecting the sequence information and functional characteristics of lncRNAs (16–23). For example, LncRNA2Function (22) mapped lncRNAs to Gene Ontology (GO) terms and biological process, lncRNADisease (16) documented associations between lncRNAs and diseases.
Accumulating evidence has shown that lncRNA exert its functions by regulating the expression of target genes. Researchers used to infer lncRNA–target relationships by examining whether a candidate gene is differentially expressed after knocking down or overexpressing a specific lncRNA (24). For example, in 2014, Bell et al. found the decreased expression of Myocardin and numerous smooth muscle contractile genes after knocking down the lncRNA SENCR (25). In 2015, LncRNA2Target v1.0 was initially released with these scattered lncRNA–target relationships (24), which was widely used for predicting potential lncRNA–target relationships, lncRNA disease associations, and so on (26–28). Whereas, the differentially expressed genes after knocking down or overexpressing a lncRNA are potential target genes with the lack of direct lncRNA–target interaction evidence. Recent years, some binding experiments such as immunoprecipitation assays, RNA pull-down assays and luciferase reporter assays was used to identify target genes of lncRNAs (29,30). Therefore, lncRNA2Target 2.0 not only collects all differentially expressed genes after knocking down or overexpressing a lncRNA, but also collects all lncRNA–target relationships confirmed by binding experimental technologies such as luciferase reporter assays, immunoprecipitation assays and pull down arrays.
DATA COLLECTION
To update LncRNA2Target, we collected all lncRNA papers and datasets published from 1 August 2014 to 30 April 2018 by searching the PubMed literature database and Gene Expression Omnibus (GEO) with keywords ‘lncRNA’, ‘lincRNA’ and ‘long non-coding RNA’, respectively. Then, over 1500 papers and 140 new available high-throughput microarray or RNA-seq datasets were retrieved. Subsequently, we downloaded all the published papers and manually extracted information about associations between lncRNAs and their target genes. To ensure data quality, lncRNAs were annotated by NCBI GenBank (31), Ensembl (32) and GENCODE (33), and each of these associations was double checked. Besides, we also downloaded all the high-throughput microarray and RNA-seq datasets before and after knocking down or overexpressing a lncRNA of interest and identified all differentially expressed genes.
A unified analysis process was developed to reanalyze public microarray and RNA-sequencing datasets before and after knocking down or overexpressing a lncRNA. First, microarray raw data was preprocessed by oligo package (34). For RNA-seq data, NGS QC Toolkit (35) was used for quality filtering and trimming, TopHat (36) was used to map reads to reference genome, and HTSeq (37) was used to count how many reads map to each gene. Then, limma (38) was selected for gene expression normalization and differential expression analysis, due to it can be applied to both microarray and RNAseq data with very similar pipelines. At last, genes with significant expression change (adjusted P ≤ 0.05) were considered as targets of a specific lncRNA.
DATABASE CONTENT
LncRNA2Target v2.0 contains 152 137 lncRNA–target associations from 1047 papers and 224 datasets. The detailed statistics on lncRNA–target associations is shown in Table 1. The total number of lncRNA–target associations increased significantly compared previous version. For example, lncRNA2target v1.0 contains 278 human lncRNA–target associations between 68 lncRNAs and 216 target genes from low-throughput methods such as RT-qPCR and western blot. While lncRNA2target v2.0 contains 1465 human lncRNA–target associations between 356 lncRNAs and 689 target genes from low-throughput methods such as RT-qPCR, western blot, luciferase reporter assays, immunoprecipitation assays and RNA pull-down assays.
Table 1.
Comparison between v1.0 and v2.0 of LncRNA2Target database
| Species | Methods | No. of lncRNAs (v1.0, v2.0) | No. of target genes (v1.0, v2.0) | No. of lncRNA–target relationships (v1.0, v2.0) |
|---|---|---|---|---|
| Human | Low-throughputa | (68, 356) | (216, 689) | (278, 1465) |
| High-throughputb | (14, 61) | (11 389, 28 865) | (26 133, 72 102) | |
| Mouse | Low-throughputa | (26, 81) | (95, 188) | (118, 210) |
| High-throughputb | (109, 134) | (14 667, 19 973) | (67 034, 78 360) |
aLow-throughput: Immunoprecipitation assays, RNA pull down assays, Luciferase reporter assays and so on.
bHigh-throughput: Microarray, RNA-seq.
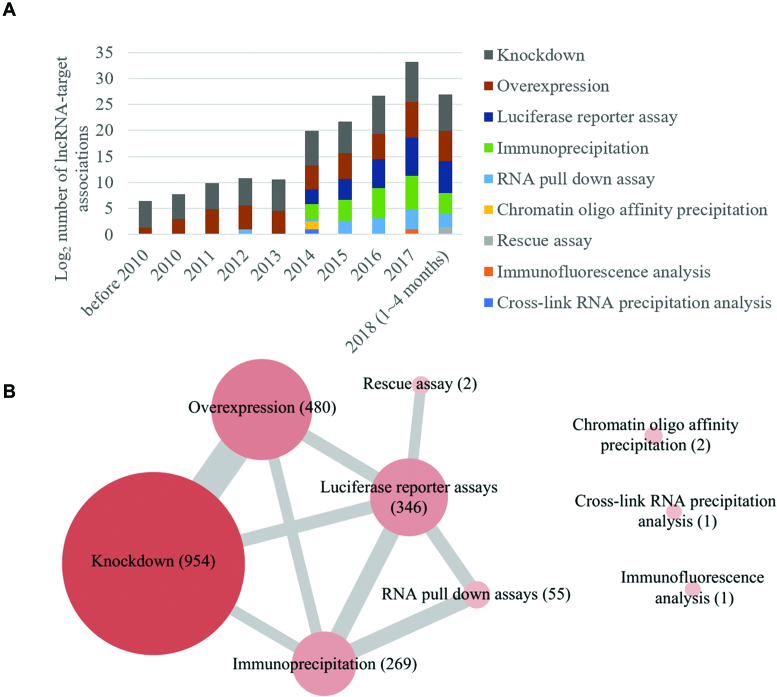
LncRNA2Target v2.0 contains 35 lncRNA–target associations from low-throughput experiments before 2010, and 33, 59, 59, 84, 133, 129, 296, 579 and 268 associations per year from 2010 to 2018, respectively. The number of lncRNA–target associations from low-throughput experiments has been annually increasing since 2010. Especially in 2017, up to 579 lncRNA–target associations were reported. Figure 1 shows that lncRNA knockdown and overexpression were the most common method to infer lncRNA–target relationships, and the number of lncRNA–target associations validated from luciferase reporter assays, immunoprecipitation assays and RNA pull down assays is also increasing rapidly.
Figure 1.
Distribution of lncRNA–target associations identified by low-throughput methods in the LncRNA2Target v2.0. (A) The number distribution of lncRNA–target associations by year. (B) The number distribution of lncRNA–target associations by identified method. Each node represents one low-throughput method. Node size indicates the number of lncRNA–target associations identified by the method. Thickness of the edge represents the number of lncRNA–target associations that is validated by both of the linked methods.
DATABASE ACCESS
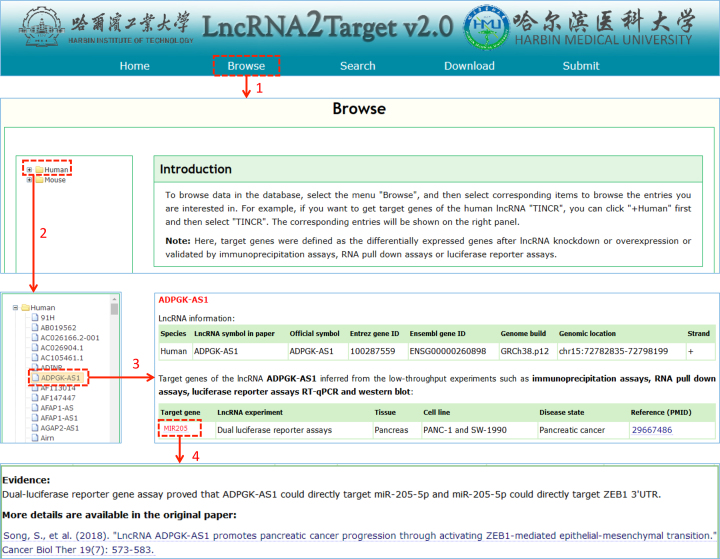
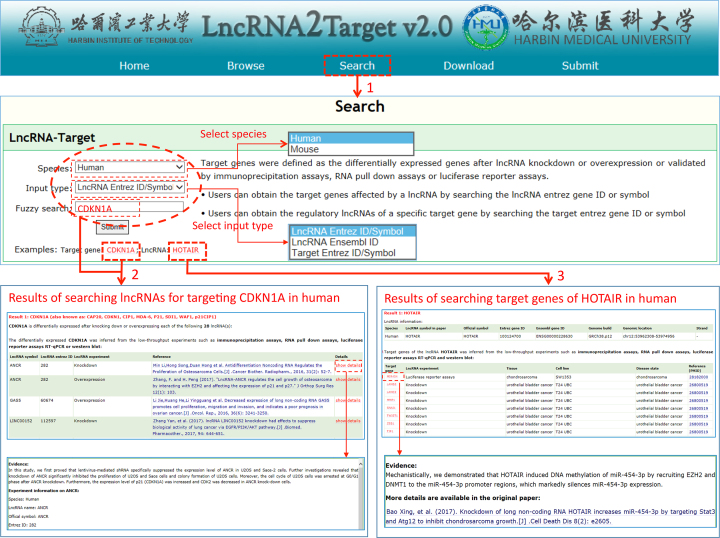
LncRNA2Target v2.0 is publicly available at http://123.59.132.21/lncrna2target. Users can browse, search and download all lncRNA–target relationship data through our web interface. Figure 2 shows the schematic workflow of browsing lncRNA–target associations by lncRNA symbol. lncRNAs of human and mouse will be shown after clicking ‘Human’ and ‘Mouse’ button, respectively. The hyperlink of each lncRNA could be linked to the details of lncRNA information and its target genes. Furthermore, the description of association between lncRNA and gene from the literature could be shown by clicking the hyperlink of the gene. Figure 3 shows the schematic workflow of searching lncRNA–target associations by lncRNA Entrez ID/symbol, lncRNA Ensembl ID and target Entrez ID/symbol. Here, fuzzy search function was provided for retrieving lncRNAs and target genes. The figure gives the searching results of two examples CDKN1A and HOTAIR. In the download page, details of all the lncRNA–target associations could be accessed. In addition, LncRNA2Target web server also provides a submission page, which allows researchers to submit new experimentally verified lncRNA–target associations to the database.
Figure 2.
Schematic workflow of browsing lncRNA–target associations.
Figure 3.
Schematic workflow of searching lncRNA–target associations.
CONCLUSION
LncRNA2Target v2.0 provides a comprehensive resource of lncRNA–target relationships in human and mouse, which aims to facilitate users to browse, search and download all literature-based lncRNA–target associations. The associations based on low-throughput and low-throughput experiments were manually extracted from literature and reanalyzed using a unified analysis process, respectively. Now LncRNA2Target v2.0 contains 152 137 lncRNA–target associations from 1047 papers and 224 datasets. With the development of experimental technologies, more and more relationships between lncRNAs and target genes will be identified. Therefore, to make researchers convenient access the new lncRNA–target associations, we will update lncRNA2Target regularly. We believe that LncRNA2Target v2.0 will be of particular interest to lncRNA community.
FUNDING
National Science and Technology Major Project of China [2016YFC1202302, 2017YFC0907500]; National Nature Science Foundation of China [61571152, 61502125, 81471736, 81671760]; Natural Science Foundation of Heilongjiang Province [F2015006]. Funding for open access charge: National Science and Technology Major Project of China [2016YFC1202302].
Conflict of interest statement. None declared.
REFERENCES
- 1. Li C.H., Chen Y.. Targeting long non-coding RNAs in cancers: progress and prospects. Int. J. Biochem. Cell Biol. 2013; 45:1895–1910. [DOI] [PubMed] [Google Scholar]
- 2. Tehrani S.S., Karimian A., Parsian H., Majidinia M., Yousefi B.. Multiple functions of long non-coding RNAs in oxidative stress, DNA damage response and cancer progression. J. Cell Biochem. 2018; 119:223–236. [DOI] [PubMed] [Google Scholar]
- 3. Sun T., Ye H., Wu C.L., Lee G.S., Kantoff P.W.. Emerging players in prostate cancer: long non-coding RNAs. Am. J. Clin. Exp. Urol. 2014; 2:294–299. [PMC free article] [PubMed] [Google Scholar]
- 4. Mouraviev V., Lee B., Patel V., Albala D., Johansen T.E.B., Partin A., Ross A., Perera R.J.. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic. Dis. 2016; 19:14–20. [DOI] [PubMed] [Google Scholar]
- 5. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ulitsky I., Bartel D.P.. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013; 154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chung S., Nakagawa H., Uemura M., Piao L., Ashikawa K., Hosono N., Takata R., Akamatsu S., Kawaguchi T., Morizono T. et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011; 102:245–252. [DOI] [PubMed] [Google Scholar]
- 9. Cui Z., Ren S., Lu J., Wang F., Xu W., Sun Y., Wei M., Chen J., Gao X., Xu C. et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol. Oncol. 2013; 31:1117–1123. [DOI] [PubMed] [Google Scholar]
- 10. Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G. 3rd, Kenny P.J., Wahlestedt C.. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008; 14:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis. 2012; 46:245–254. [DOI] [PubMed] [Google Scholar]
- 12. Congrains A., Kamide K., Oguro R., Yasuda O., Miyata K., Yamamoto E., Kawai T., Kusunoki H., Yamamoto H., Takeya Y. et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012; 220:449–455. [DOI] [PubMed] [Google Scholar]
- 13. Alvarez M.L., DiStefano J.K.. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One. 2011; 6:e18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun M., Liu X.H., Wang K.M., Nie F.Q., Kong R., Yang J.S., Xia R., Xu T.P., Jin F.Y., Liu Z.J. et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol. Cancer. 2014; 13:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu X., Wu Y.B., Zhou J., Kang D.M.. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem. Biophys. Res. Commun. 2016; 469:319–325. [DOI] [PubMed] [Google Scholar]
- 16. Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q.. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013; 41:D983–D986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miao Y.R., Liu W., Zhang Q., Guo A.Y.. lncRNASNP2: An updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2018; 46:D276–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma L., Li A., Zou D., Xu X., Xia L., Yu J., Bajic V.B., Zhang Z.. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015; 43:D187–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cui T., Zhang L., Huang Y., Yi Y., Tan P., Zhao Y., Hu Y., Xu L., Li E., Wang D.. MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018; 46:D371–D374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou B., Zhao H., Yu J., Guo C., Dou X., Song F., Hu G., Cao Z., Qu Y., Yang Y. et al. EVLncRNAs: A manually curated database for long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 2018; 46:D100–D105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., Zhao L., Li X., Teng X., Sun X. et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018; 46:D308–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang Q., Ma R., Wang J., Wu X., Jin S., Peng J., Tan R., Zhang T., Li Y., Wang Y.. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics. 2015; 16:doi:10.1186/1471-2164-16-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang Q., Wang J., Wang Y., Ma R., Wu X., Li Y.. TF2LncRNA: identifying common transcription factors for a list of lncRNA genes from ChIP-Seq data. Biomed. Res. Int. 2014; 2014:317642–317646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang Q., Wang J., Wu X., Ma R., Zhang T., Jin S., Han Z., Tan R., Peng J., Liu G. et al. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2015; 43:D193–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bell R.D., Long X., Lin M., Bergmann J.H., Nanda V., Cowan S.L., Zhou Q., Han Y., Spector D.L., Zheng D. et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler. Thromb. Vasc. Biol. 2014; 34:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng J., Bai K., Shang X., Wang G., Xue H., Jin S., Cheng L., Wang Y., Chen J.. Predicting disease-related genes using integrated biomedical networks. BMC Genomics. 2017; 18:1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J., Le T.D., Liu L., Li J.. Inferring miRNA sponge co-regulation of protein-protein interactions in human breast cancer. BMC Bioinformatics. 2017; 18:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng L., Shi H., Wang Z., Hu Y., Yang H., Zhou C., Sun J., Zhou M.. IntNetLncSim: an integrative network analysis method to infer human lncRNA functional similarity. Oncotarget. 2016; 7:47864–47874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mohanty V., Gokmen-Polar Y., Badve S., Janga S.C.. Role of lncRNAs in health and disease-size and shape matter. Brief. Funct. Genomics. 2015; 14:115–129. [DOI] [PubMed] [Google Scholar]
- 30. Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Blass G., Kellis M., Werber M. et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013; 24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benson D.A., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2015; 43:D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Giron C.G. et al. Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carvalho B.S., Irizarry R.A.. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26:2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patel R.K., Jain M.. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012; 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]