Abstract
Extracellular vesicles (EVs) are membranous vesicles that are released by both prokaryotic and eukaryotic cells into the extracellular microenvironment. EVs can be categorised as exosomes, ectosomes or shedding microvesicles and apoptotic bodies based on the mode of biogenesis. EVs contain biologically active cargo of nucleic acids, proteins, lipids and metabolites that can be altered based on the precise state of the cell. Vesiclepedia (http://www.microvesicles.org) is a web-based compendium of RNA, proteins, lipids and metabolites that are identified in EVs from both published and unpublished studies. Currently, Vesiclepedia contains data obtained from 1254 EV studies, 38 146 RNA entries, 349 988 protein entries and 639 lipid/metabolite entries. Vesiclepedia is publicly available and allows users to query and download EV cargo based on different search criteria. The mode of EV isolation and characterization, the biophysical and molecular properties and EV-METRIC are listed in the database aiding biomedical scientists in assessing the quality of the EV preparation and the corresponding data obtained. In addition, FunRich-based Vesiclepedia plugin is incorporated aiding users in data analysis.
INTRODUCTION
Extracellular vesicles (EVs) are membranous vesicles that are released by both prokaryotic and eukaryotic cells into the extracellular microenvironment (1,2). EVs contain a lipid bilayer that protects the luminal cargo of nucleic acids, proteins, lipids and metabolites from the harsh environmental conditions (3,4). Based on the mode of biogenesis and release, EVs can be categorised as exosomes, ectosomes or shedding microvesicles and apoptotic bodies (5,6). Exosomes originate from the endosomes whilst ectosomes and apoptotic bodies are released by membrane blebbing of live and apoptotic cells, respectively. The cargo packaged within or associated with the EVs reflect the pathophysiological state of the host cell. For instance, EVs secreted by glioblastoma cells contain mutant EGFR (7) while EVs secreted by PI3K/Akt activated colorectal cancer cells contain phosphorylated AKT (8). Similarly, the cargo of the apoptotic bodies can be altered based on the stimulus for cell death (1). As the cargo of EVs are perturbed based on the precise state of the cell (e.g. stressed, activated or diseased), there has been significant interest in utilizing EVs in biomarker analysis (9). Furthermore, the possibility of EVs mediating both short and long range-based cell-cell communication has significantly advanced our understanding on how cellular communications are orchestrated (10). Owing to the functional role of EVs in intercellular communication and its utility in biomarker analysis, several studies are undertaken with the primary focus of characterizing the cargo of EVs in various conditions.
A compendium of RNA, proteins, metabolites and lipids identified in EVs may aid in identifying subsets of molecular cargo that are specific to the cell type, stimulus, activation state or disease condition. Such class of molecular cargo can be critical in identifying the underlying mechanism by which the biogenesis of EVs occur as well as provide clues for the packaging of the molecules in EVs. A compendium can also aid in the identification of cell type specific EV membrane proteins that could be utilized for immunoaffinity based pull down in patient samples. Here, we describe, Vesiclepedia (http://www.microvesicles.org), a web-based compendium of proteins, RNA, lipids and metabolites that are identified in EVs. Vesiclepedia receives data submissions through continuous community annotation from the biomedical community. In addition to the community participation, Vesiclepedia also contains manually curated data from the published literature. The cargo annotations are complemented with the EV isolation procedures, meta-annotations pertaining to the experiment and EV-METRIC (wherever applicable) which will inform the users on the quality of the EV preparation.
Vesiclepedia
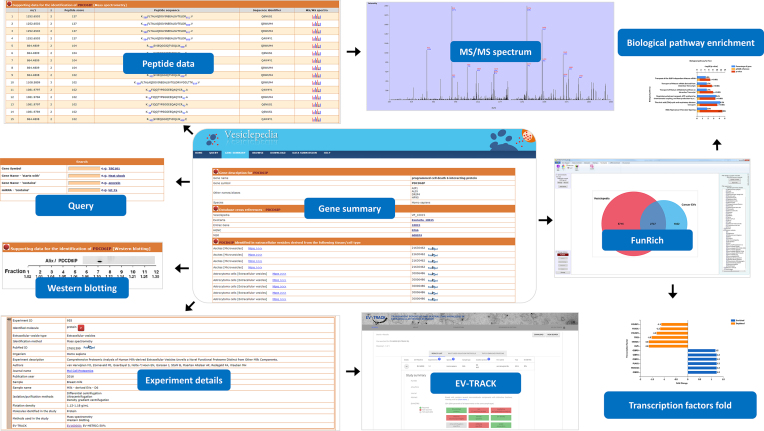
Compared to the exosomal database ExoCarta (11), Vesiclepedia (http://www.microvesicles.org) is a compendium that catalogues proteins, RNAs, lipids and metabolites that are identified in all classes of EVs (12). Vesiclepedia was built using ZOPE, an open source content management system, while Python a portable, interpreted, object oriented programming language was utilised to connect the web interface with the MySQL database. Vesiclepedia provides the users with gene/molecule specific page that contains information on the gene of interest (Figure 1), its external references to other primary databases, meta-annotations pertaining to the experiment description of the study that identified the molecule and gene ontology based annotations that are retrieved from Entrez Gene (13).
Figure 1.
Snapshot of Vesiclepedia pages Vesiclepedia gene summary page of EV enriched protein Alix (PDCD6IP) is displayed. The gene summary page provides data on the molecule of interest, experimental description pertaining to the studies that identified the molecule, Western blotting data to support the identification (if applicable) and mass spectrometry-based peptide details along with the tandem MS spectra is provided wherever possible. Furthermore, the experiment details are supported with EV-METRIC and linked to the EV-TRACK website to provide additional information on the scoring. The entire data from Vesiclepedia can be automatically downloaded through FunRich via the Vesiclepedia plugin and analysed.
DATA ADDITIONS AND NEW FEATURES
Data update
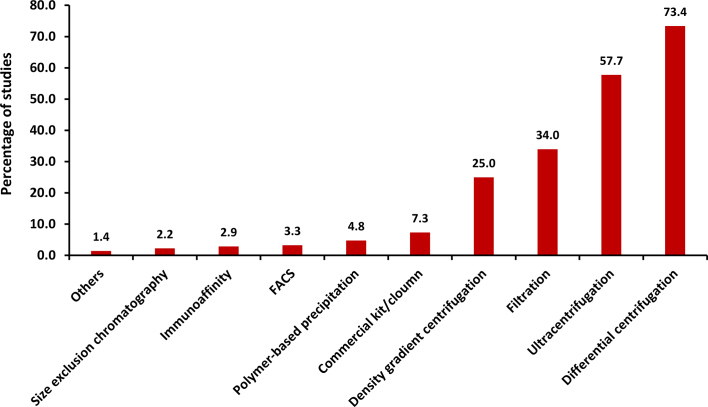
Since the initial description of Vesiclepedia in December 2012, there is more than 3-fold increase in the number of EV studies catalogued in the database. Vesiclepedia currently contains 1254 EV studies, 349 988 protein entries, 38 146 RNA entries and 639 lipid/metabolite entries (Table 1). The data collated in Vesiclepedia were obtained from 41 organisms and 172 sample sources. Among the 1254 studies, 648 studies have been manually annotated whilst the rest were contributed by the EV community. As shown in Figure 2, among the methods employed, ultracentrifugation has been used by 58% of the studies. Although recent trends suggest that authors use a combination of methods (e.g. size exclusion and density gradient centrifugation) to purify EVs, ultracentrifugation has been the most preferred method in the recent past. Due to the significant interest in EV research, multiple companies have developed EV isolation kits/columns which are also used by the biomedical research community. As shown in Figure 2, 7% of the studies have used commercial kits/columns to isolate EVs.
Table 1.
Statistics of data in Vesiclepedia
| 1 | EV studies | 1254 |
| 2 | RNA entries | 38 146 |
| 3 | mRNA entries | 27 646 |
| 4 | Unique mRNA | 17 005 |
| 5 | miRNA entries | 10 520 |
| 6 | Unique miRNA | 2431 |
| 7 | Protein entries | 349 988 |
| 8 | Unique proteins | 32 267 |
| 9 | Lipid/metabolite entries | 639 |
| 10 | Organisms | 41 |
| 11 | Sample sources | 172 |
Figure 2.
Distribution of EV studies based on the isolation method A histogram depicting the distribution of isolation methods employed by the EV studies catalogued in Vesiclepedia is depicted. Differential centrifugation has been used by >73% of the studies to isolate EVs while 57.7% of the studies have used ultracentrifugation. Although the data is depicted as individual methods, most studies have employed a combination of these methods. For instance, filtration accounted for 34% of the EV studies and is always used in conjunction with other procedures to isolate EVs.
EV-TRACK
The EV-TRACK consortium has created an expandable knowledgebase of experimental parameters for publications relating to EVs (14). Based on the EV isolation and characterisation parameters that are reported, each study will receive an EV-METRIC that measures the transparency of reporting experimental meta-annotations. The EV-METRIC is now linked to the experiment details in Vesiclepedia molecule page. Moreover, the EV-METRIC is hyperlinked to the EV-TRACK website to provide additional information as how the EV-METRIC was achieved.
FunRich
An added feature to Vesiclepedia is the incorporation of the entire data into FunRich, a functional enrichment analysis tool (15,16). More importantly, the update of Vesiclepedia data can be performed automatically by clicking the ‘Update’ button. Users can now compare their EV or other datasets with the entire Vesiclepedia data either by Venn diagrams or by using enrichment analysis. Alternatively, users can filter the Vesiclepedia data based on the sample source (e.g. cancer or stem cell), isolation method (e.g. density gradient centrifugation), EV subtype (e.g. ectosomes) and/or species. This new FunRich enabled Vesiclepedia plugin allows the users to analyse the Vesiclepedia data robustly. In addition to data analysis, users can freely download the Vesiclepedia data through FunRich Vesiclepedia plugin. The availability of filters allows the users to customize the downloadable data.
EV community annotation
Although databases are indispensable resources for advancing biomedical research, manual curation of the scientific literature is cumbersome. Hence, in order to constantly update the database, the involvement of the EV scientific community in annotating and contributing datasets is critical. To facilitate the active involvement of the EV scientific community, we have incorporated data submission guidelines. The deposited data can be maintained as private until the manuscript is published or the authors agree to make the data publicly available. The online data submission system can significantly reduce the onus on the curators and aid in the constant maintenance of the database.
Pitfalls of Vesiclepedia
It has been accepted that the terminologies of naming EVs lack global consensus and hence the authors use various terms whilst referring to EVs. The protocols employed also varies between laboratories and hence the comparison between two EV studies is not trivial. Vesiclepedia, similar to other curated databases, collate data from published literature and contributing authors. Hence, the quality of the data is as good as the respective study. To aid users with downstream analyses, Vesiclepedia provides the experiment details as well as the EV-METRIC for assessment. However, we emphasise caution when the data from Vesiclepedia is used for EV based analysis. For instance, Vesiclepedia lists top 100 proteins that were identified in EV studies. The list merely means that these 100 proteins are most often reported in EV studies and does not warrant them to be called as EV markers or enriched proteins. While some of the proteins in the list are bona fide EV enriched proteins, others could be experimental artefacts and could be of low abundance in EVs.
Future directions
Vesiclepedia will be updated with more EV studies through manual curation and community annotation. Furthermore, additional fields such as circular RNA, long non-coding RNA, protein-protein interactions and post-translational modifications will be incorporated in the future. The database provides top 100 proteins that are often identified in EVs and we hope that this list will enable users to compare their respective EV data with most often identified proteins.
DATA AVAILABILITY
Vesiclepedia is a free to use web based database that is available at http://www.microvesicles.org. The data present in Vesiclepedia is available as tab-delimited files through the website or through FunRich enrichment analysis tool (http://www.funrich.org).
ACKNOWLEDGEMENTS
The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
FUNDING
Australian Research Council DP [DP170102312 to S.M.]; Australian Research Council FT [FT180100333] and NHMRC project grant [1122437]. Funding for open access charge: Australian Research Council DP [DP170102312 to S.M.]; Australian Research Council FT [FT180100333];
Conflict of interest statement. None declared.
REFERENCES
- 1. Kalra H., Drummen G.P., Mathivanan S.. Focus on extracellular Vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 2016; 17:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raposo G., Stoorvogel W.. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013; 200:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thery C., Ostrowski M., Segura E.. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009; 9:581–593. [DOI] [PubMed] [Google Scholar]
- 4. Gangoda L., Boukouris S., Liem M., Kalra H., Mathivanan S.. Extracellular vesicles including exosomes are mediators of signal transduction: are they protective or pathogenic. Proteomics. 2015; 15:260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tkach M., Thery C.. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016; 164:1226–1232. [DOI] [PubMed] [Google Scholar]
- 6. Cheung K.H., Keerthikumar S., Roncaglia P., Subramanian S.L., Roth M.E., Samuel M., Anand S., Gangoda L., Gould S., Alexander R. et al. Extending gene ontology in the context of extracellular RNA and vesicle communication. J. Biomed. Semant. 2016; 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J.. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008; 10:619–624. [DOI] [PubMed] [Google Scholar]
- 8. Liem M., Ang C.S., Mathivanan S.. Insulin mediated activation of PI3K/Akt signalling pathway modifies the proteomic cargo of extracellular vesicles. Proteomics. 2017; 17:doi:10.1002/pmic.201600370. [DOI] [PubMed] [Google Scholar]
- 9. Gasecka A., Boing A.N., Filipiak K.J., Nieuwland R.. Platelet extracellular vesicles as biomarkers for arterial thrombosis. Platelets. 2017; 28:228–234. [DOI] [PubMed] [Google Scholar]
- 10. Cossetti C., Iraci N., Mercer T.R., Leonardi T., Alpi E., Drago D., Alfaro-Cervello C., Saini H.K., Davis M.P., Schaeffer J. et al. Extracellular vesicles from neural stem cells transfer IFN-gamma via Ifngr1 to activate Stat1 signaling in target cells. Mol. Cell. 2014; 56:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keerthikumar S., Chisanga D., Ariyaratne D., Saffar H., Anand S., Zhao K.N., Samuel M., Pathan M., Jois M., Chilamkurti N. et al. ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 2016; 428:688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalra H., Simpson R.J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V.C., Borras F.E., Breakefield X., Budnik V. et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012; 10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maglott D., Ostell J., Pruitt K.D., Tatusova T.. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007; 35:D26–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Deun J., Mestdagh P., Agostinis P., Akay O., Anand S., Anckaert J., Martinez Z.A., Baetens T., Beghein E., Bertier L. et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods. 2017; 14:228–232. [DOI] [PubMed] [Google Scholar]
- 15. Pathan M., Keerthikumar S., Chisanga D., Alessandro R., Ang C.S., Askenase P., Batagov A.O., Benito-Martin A., Camussi G., Clayton A. et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles. 2017; 6:1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A.. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015; 15:2597–2601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Vesiclepedia is a free to use web based database that is available at http://www.microvesicles.org. The data present in Vesiclepedia is available as tab-delimited files through the website or through FunRich enrichment analysis tool (http://www.funrich.org).