Abstract
Numerous non-inherited somatic mutations, distinct from those of germ-line origin, occur in somatic cells during DNA replication per cell-division. The somatic mutations, recording the unique genetic cell-lineage ‘history’ of each proliferating normal cell, are important but remain to be investigated because of their ultra-low frequency hidden in the genetic background of heterogeneous cells. Luckily, the recent development of single-cell genomics biotechnologies enables the screening and collection of the somatic mutations, especial single nucleotide variations (SNVs), occurring in normal cells. Here, we established DSMNC: a database of somatic mutations in normal cells (http://dsmnc.big.ac.cn/), which provides most comprehensive catalogue of somatic SNVs in single cells from various normal tissues. In the current version, the database collected ∼0.8 million SNVs accumulated in ∼600 single normal cells (579 human cells and 39 mouse cells). The database interface supports the user-friendly capability of browsing and searching the SNVs and their annotation information. DSMNC, which serves as a timely and valuable collection of somatic mutations in individual normal cells, has made it possible to analyze the burdens and signatures of somatic mutations in various types of heterogeneous normal cells. Therefore, DSMNC will significantly improve our understanding of the characteristics of somatic mutations in normal cells.
INTRODUCTION
Numerous non-inherited somatic mutations are occurring and accumulating with cell divisions, which record the unique genetic feature of each proliferating somatic cell beginning from a zygote. For these somatic mutations, the better understanding of their characteristics and potential roles in cell-lineage determination, aging or disease occurrence is extremely important (1–3). The somatic mutations that contribute to the rapid proliferation of abnormal cells were observable due to tumor clonality and thus were especially concerned in previous tumor genomic studies (4,5). But, the somatic mutations in heterogeneous cells remain largely unexplored and their signatures in most healthy cells are not well known.
The current development of advanced biotechnologies of single-cell genomics enables the screening of the somatic mutations hidden in genetic background of heterogeneous normal cells. Single-cell genomics biotechnologies have advanced rapidly with the two most common trace-DNA amplification strategies. The first strategy, for example linear amplification via transposon insertion (LIANTI), comprises a straightforward way to extract and amplify pg-level nucleic acids of a single cell using reaction reagents (6–8). The latter is a strategy of single-cell-derived clonal cultured, for example organoid formation, thereby enabling the ancestor cell genome to undergo high-fidelity expansion with the benefit of mitotic cell divisions (9,10). In recent studies, investigators have successfully surveyed somatic mutations in various types of single cells utilizing these single-cell genomics technologies. The number of somatic mutations, especial single nucleotide variations (SNVs), explored in normal cells has been growing gradually in past two years. However, to date, no database has been designed to capture these data for subsequent analysis.
We gathered single-cell DNA amplification and sequencing data, and developed the unique DSMNC database (a Database of Somatic Mutations in Normal Cells). DSMNC collects somatic SNVs occurring in single normal cells into a high quality, comprehensive resource. All of the SNVs were supported by advanced single-cell DNA amplification strategies and reliable deep sequencing data. We expect that this elaborate database will serve as an important catalyst for biologists in broad research areas to understand the signatures of somatic mutations occurring in normal cells.
DATA COLLECTION AND DATABASE CONTENT
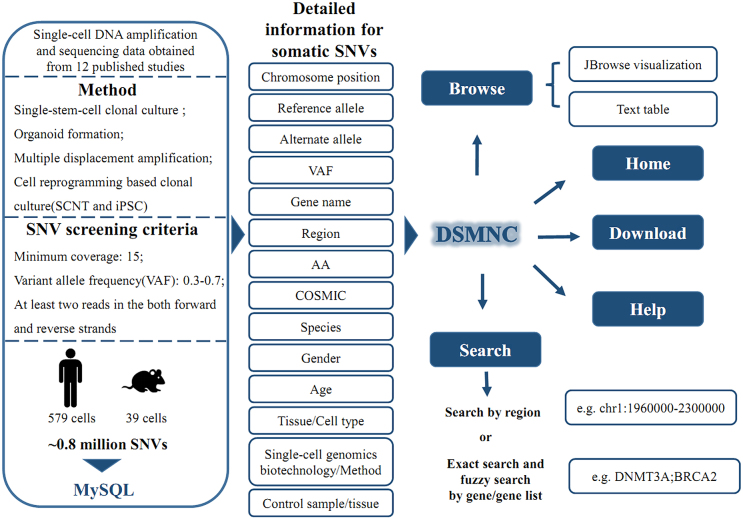
We gathered single-cell DNA amplification and sequencing data from resources of 12 published studies and our in-house studies (1,2,9,11–19) (Figure 1, Supplementary Table S1). The single-cell DNA amplification strategies include pg-level nucleic acids amplification strategy using reaction reagents and single-cell-derived clonal culture strategy. The detailed single-cell DNA amplification methods such as multiple displacement amplification (MDA), multiple annealing and looping-based amplification cycles (MALBAC), linear amplification via transposon insertion (LIANTI), single-stem-cell organoid formation, and cell reprogramming based single-cell clonal culture, have been proved to be effective in published literatures (6–10). The raw sequencing reads were aligned to the human and mouse genome assemblies (hg19 and mm9). To exclude germ-line variants from the sequencing data of each single cell (or cell clone), we used the bulk DNA sequencing data of tissue samples of the same human (or mouse) body where single cells (or cell clones) were obtained as controls. Reliable somatic SNVs in each cell were then screened and filtered from the single-cell genomic sequencing data via the MuTect algorithm with the following strict criteria (20), e.g. (a) variant sites had a minimum coverage of 15 and Phred-scaled base quality above 15; (b) the mutant allele SNV frequency was in the range of 0.3–0.7, whereas it was 0 or 1 in the corresponding bulk DNA sample; (c) the mutant allele was supported by at least two reads in the both forward and reverse strands (Figure 1).
Figure 1.
Schematically illustrates the general workflow and features of the database.
In summary, DSMNC currently contains a catalogue of ∼0.77 million somatic SNVs occurring in over 579 human cells and ∼0.014 million somatic SNVs in 39 mouse cells from ∼300 individuals (Figure 1; A detailed description please refer to Table 1). These single cells cover various cell types of blood, brain, colon, liver, skin, stomach, bowel and others (Table 1) (1,2,9,11–19). All the somatic SNVs and the related information were loaded into database server MySQL 5.7.19. Each row of the main database table contains the key item of somatic SNV ID and its annotation information with chromosome loci, nucleotide type in reference or mutation allele, supported read depths in genomic sequencing data, associated gene symbol, cell type, single-cell DNA amplification and sequencing method, and control bulk DNA samples (Figure 1). We will continue to collect more somatic SNVs in normal cells and update the database in the future. Besides, other genomic mutation data including germ-line SNPs from dbSNP137/dbSNP128 and COSMIC tumor SNVs were gathered and built into our database for further comparison study of mutational signatures (21,22).
Table 1.
Statistics of DSMNC database content
| Species | Categories | Single-Cell/ Individual numbers | SNVs numbers |
|---|---|---|---|
| HUMAN | Cell type | ||
| blood | 24/10 | 9171 | |
| brain | 186/21 | 86 838 | |
| colon | 21/6 | 44 404 | |
| liver | 10/5 | 13 915 | |
| skin | 324/204 | 594 790 | |
| small intestine | 14/9 | 21 261 | |
| Single-cell DNA amplification method | |||
| Multiple displacement amplification | 155/18 | 79 750 | |
| Single-stem-cell clonal culture | 51/12 | 10 402 | |
| Organoid formation | 45/19 | 79 580 | |
| Cell reprogramming based single-cell clonal culture | 328/205 | 600 647 | |
| Total | 579/254 | ∼770 000 | |
| MOUSE | cell type | ||
| brain | 7/5 | 1646 | |
| large bowel | 7/2 | 1625 | |
| prostate | 4/1 | 590 | |
| small bowel | 8/2 | 3477 | |
| stomach | 6/2 | 1022 | |
| mouse embryonic fibroblasts | 2/2 | 1950 | |
| adipocyte progenitor cells | 5/3 | 3893 | |
| Single-cell DNA amplification method | |||
| Organoid formation | 25/2 | 6714 | |
| Cell reprogramming based single-cell clonal culture | 14/10 | 7489 | |
| Total | 39/12 | ∼14 000 |
WEB INTERFACE
The webserver of the DSMNC database was built using Apache 2.4.27. The web interface was implemented in PHP and JavaScript. And the Search and Browse webpages were produced by Jbrowse 1.13.1. A fully functional database of DSMNC is freely available on the website at the link of http://dsmnc.big.ac.cn/. We recommend Google Chrome for visiting the database. The database interface supports the user-friendly capability of browsing, searching and downloading all the DSMNC data without login or registration (Figure 2).
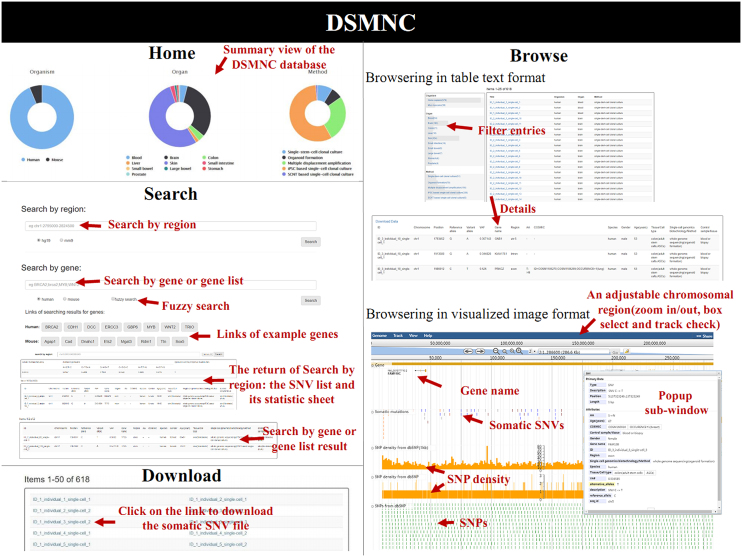
Figure 2.
Screenshots of the web interfaces in DSMNC. The web interface of DSMNC comprises four main functional components: Home webpage, Browse webpage, Search webpage and Download webpage. The view of DSMNC database summary is given in the Home webpage. Users can browse the detailed genomic information on selected group of somatic SNVs in text format or visualized image format. The Search webpage allows users to retrieve the list of somatic SNVs indexed by gene symbols or chromosome regions. And somatic SNVs indexed by the accession ID for each single cell can be downloaded in the Download webpage.
Five main webpages are been included in the database: Home, Browse, Search, Download and Help (Figure 2). Users can browse the detailed genomic information on somatic mutations in text format or visualized image format according to their own unique needs. In the table text format of Browse webpage, the list of somatic SNVs in a single cell and their annotation information is returned when clicking on the accession ID for each single cell (e.g. ID_7_individual_1_single-cell_1). Additionally, users can select the keywords below ‘organism’, ‘organ’ or ‘single-cell amplification method’ in the left toolbar menu to view the subset of the somatic SNVs in normal cells. By clicking on the menu ‘Browse → Browse by chromosome’ in the Browse webpage, users can change the table text format into the visualized image format. Somatic SNVs as well as comparable germ-line SNPs within an adjustable chromosomal region are visualized on JBrowse. User-friendly control elements such as zoom in/out, box select and track check are available for the creation of landscape maps of somatic SNVs in a genomic region. More detailed description on the SNV can be found in a popup sub-window when the users click the track of each SNV in JBrowse. In this genomic region selectable tracks of SNP density, calculated with two sliding window of 1 bp and 1 kb respectively, can be browsed. DSMNC also provides an option in the Search webpage that allows users to retrieve the list of somatic SNVs by gene symbols or chromosome regions. Besides the SNV list, the result of ‘Search by region’ also contains an additional statistic sheet including information of synonymous/non-synonymous ratios, mutation-type signatures and mutation density for the SNVs within a user-specified genomic region. ‘Search by gene’ has been designed to be capable of exact search and fuzzy search by a gene symbol. Also the item supports the searching input of multiple gene symbols, which should be separated by semicolon. All data in the database indexed by the accession ID for each single cell can be downloaded in the Download webpage. And finally, a detailed tutorial how to use DSMNC is available on the Help webpage.
PROSPECTIVE ANALYSIS ON SNVs IN DSMNC
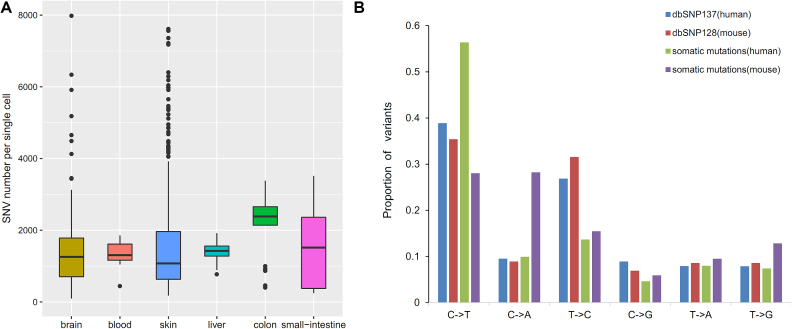
As the purpose of collecting somatic mutations in individual normal cells, the database DSMNC has made it possible to analyze the burdens and signatures of somatic mutations in various types of heterogeneous normal cells. We did some preliminary analysis on somatic SNVs collected in DSMNC database. In Figure 3A, we summarized the mutation loads of normal cells among different tissues. The data suggest that hundreds of somatic SNVs per cell accumulated beginning from a zygote, but cells from different cell types have distinct mutation loads. We further described the SNV spectra of normal cells compared with the germ-line SNP spectra (Figure 3B). The mutation signatures of somatic mutations are quite different from the ones of germ-line mutations. Beyond the above prospective analysis on SNVs in DSMNC, we believe that the DSMNC database is important and generally interesting to the biologists for further investigations on the characteristics of somatic mutations occurring in normal cells.
Figure 3.
Prospective analysis on SNV signatures in DSMNC. (A) Mutation loads in normal single cells from different types of human tissues. (B) The somatic SNV spectra in normal cells comparing with the SNP spectra. The SNP information of Human and Mouse was retrieved from the dbSNP137 and dbSNP128, respectively.
CONCLUSION AND DISCUSSION
Recent advances in single-cell DNA amplification technologies enable the construction of our database DSMNC, a collection of somatic mutations in heterogeneous normal cells. We implemented the user-friendly database interface with capability of browsing, searching and downloading the detailed SNVs information at the website link of http://dsmnc.big.ac.cn/.
To our knowledge, DSMNC is unique for collection somatic mutations data occurring in single normal cells. The data in DSMNC would facilitate the understanding of the mutation signatures in normal cells. DSMNC is of broad appeal and utility for the biologists in the research areas such as genetics, evolution, development and cancer. For example, numbers of somatic SNVs in this database would enable the comparisons of mutation loads and hotspot regions during mitosis among various normal cell types, as concerned about by the genetic biologists; Moreover, the single-cell profiles of somatic mutation from age-spanning donors collected in our DSMNC database are necessary for the studies of ageing. The natural selection with somatic mutations, enabling selection for or against somatic normal cells, would attract the evolutionary biologists’ attentions; Lastly, the observable somatic mutations in a clonal tumor mass are composed of the ones accumulated in normal cells and the tumor-driver ones occurring during tumorigenesis. The database DSMNC is providing a chance for cancer biologists to re-define the true driver mutations in cancer which did not occur before tumorigenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the contributors for providing the single-cell genomics data. We are also grateful to our institute colleague Mingyuan Sun for his assistance with database implementation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFC0910400, 2017YFC0908402, 2018YFC1003102]; National Natural Science Foundation of China [31571307, 31501020]; Open Project of Key Laboratory of Genomic and Precision Medicine, Chinese Academy of Sciences. Funding for open access charge: National Natural Science Foundation of China [31571307 and 31501020].
Conflict of interest statement. None declared.
REFERENCES
- 1. Behjati S., Huch M., van Boxtel R., Karthaus W., Wedge D.C., Tamuri A.U., Martincorena I., Petljak M., Alexandrov L.B., Gundem G. et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014; 513:422–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lodato M.A., Rodin R.E., Bohrson C.L., Coulter M.E., Barton A.R., Kwon M., Sherman M.A., Vitzthum C.M., Luquette L.J., Yandava C.. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2017; 359:eaao4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poduri A., Evrony G.D., Cai X., Walsh C.A.. Somatic mutation, genomic variation, and neurological disease. Science. 2013; 341:1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013; 499:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tao Y., Ruan J., Yeh S.H., Lu X., Wang Y., Zhai W., Cai J., Ling S., Gong Q., Chong Z. et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12042–12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C., Xing D., Tan L., Li H., Zhou G., Huang L., Xie X.S.. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science. 2017; 356:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dean F.B., Nelson J.R., Giesler T.L., Lasken R.S.. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001; 11:1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zong C., Lu S., Chapman A.R., Xie X.S.. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012; 338:1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hazen J.L., Faust G.G., Rodriguez A.R., Ferguson W.C., Shumilina S., Clark R.A., Boland M.J., Martin G., Chubukov P., Tsunemoto R.K. et al. The complete genome sequences, unique mutational spectra, and developmental potency of adult neurons revealed by cloning. Neuron. 2016; 89:1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwon E.M., Connelly J.P., Hansen N.F., Donovan F.X., Winkler T., Davis B.W., Alkadi H., Chandrasekharappa S.C., Dunbar C.E., Mullikin J.C. et al. iPSCs and fibroblast subclones from the same fibroblast population contain comparable levels of sequence variations. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:1964–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bae T., Tomasini L., Mariani J., Zhou B., Roychowdhury T., Franjic D., Pletikos M., Pattni R., Chen B.J., Venturini E.. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2017; 359:eaan8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blokzijl F., de Ligt J., Jager M., Sasselli V., Roerink S., Sasaki N., Huch M., Boymans S., Kuijk E., Prins P. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016; 538:260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai J., Miao X., Li Y., Smith C., Tsang K., Cheng L., Wang Q.F.. Whole-genome sequencing identifies genetic variances in culture-expanded human mesenchymal stem cells. Stem Cell Rep. 2014; 3:227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng L., Hansen N.F., Zhao L., Du Y., Zou C., Donovan F.X., Chou B.K., Zhou G., Li S., Dowey S.N. et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012; 10:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao S., Zheng C., Chang G., Liu W., Kou X., Tan K., Tao L., Xu K., Wang H., Cai J. et al. Unique features of mutations revealed by sequentially reprogrammed induced pluripotent stem cells. Nat. Commun. 2015; 6:6318. [DOI] [PubMed] [Google Scholar]
- 16. Jager N., Schlesner M., Jones D.T., Raffel S., Mallm J.P., Junge K.M., Weichenhan D., Bauer T., Ishaque N., Kool M. et al. Hypermutation of the inactive X chromosome is a frequent event in cancer. Cell. 2013; 155:567–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J. et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017; 546:370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lodato M.A., Woodworth M.B., Lee S., Evrony G.D., Mehta B.K., Karger A., Lee S., Chittenden T.W., D’Gama A.M., Cai X. et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015; 350:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Welch J.S., Ley T.J., Link D.C., Miller C.A., Larson D.E., Koboldt D.C., Wartman L.D., Lamprecht T.L., Liu F., Xia J. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012; 150:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G.. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013; 31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smigielski E.M., Sirotkin K., Ward M., Sherry S.T.. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000; 28:352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.