Abstract
Sharing of research data in public repositories has become best practice in academia. With the accumulation of massive data, network bandwidth and storage requirements are rapidly increasing. The ProteomeXchange (PX) consortium implements a mode of centralized metadata and distributed raw data management, which promotes effective data sharing. To facilitate open access of proteome data worldwide, we have developed the integrated proteome resource iProX (http://www.iprox.org) as a public platform for collecting and sharing raw data, analysis results and metadata obtained from proteomics experiments. The iProX repository employs a web-based proteome data submission process and open sharing of mass spectrometry-based proteomics datasets. Also, it deploys extensive controlled vocabularies and ontologies to annotate proteomics datasets. Users can use a GUI to provide and access data through a fast Aspera-based transfer tool. iProX is a full member of the PX consortium; all released datasets are freely accessible to the public. iProX is based on a high availability architecture and has been deployed as part of the proteomics infrastructure of China, ensuring long-term and stable resource support. iProX will facilitate worldwide data analysis and sharing of proteomics experiments.
INTRODUCTION
The rapid development of genome research has substantially benefited from the open data sharing of the Human Genome Project, and over the last decade open data sharing has become best practice in proteomics too. The ProteomeXchange (PX) consortium (1,2) coordinates a stable, distributed infrastructure for proteomics data sharing.
The challenges to rapid proteomic data release, addressed at the 2008 International Summit on Proteomics Data Release and Sharing Policy, were divided into three categories: technical, infrastructural and policy (3). Considerable progress has been made in all three aspects in the past decade. Promoted by the Amsterdam principles (3) and its follow-up data quality metrics (4,5), data policies and guidelines supporting rapid and open sharing of proteomic data are being implemented within the proteomics community; several leading journals, including Molecular and Cellular Proteomics, Journal of Proteome Research and Proteomics, develop and maintain data submission guidelines. Over the past 10 years, significant advances have been made in data exchange formats, controlled vocabularies (i.e. PSI-MS CV) (6,7) and reporting guidelines (i.e. MIAPE (8)) for many aspects of mass spectrometry (MS)-based proteomics. The Human Proteome Organization-Proteomics Standards Initiative (HUPO-PSI) (9) has developed and published coordinated standards for proteomics data representation, including the mzML (10) standard representing MS raw data, the mzIdentML (11) standard designed to report peptide and protein identification data, the mzQuantML (12) format designed to report quantification results and a text-based format, mzTab (13), which can present both identification and quantification results in a simplified overview format. All of these standard formats and their conversion tools (14–17) have certainly helped overcome the technical challenges of data sharing in the proteomics community.
Infrastructure provision for the deposition of large experimental data and analyses is the most fundamental and essential requirement for public sharing of proteomic data. Members of the PX consortium have addressed this requirement in the past decade. At present, PRIDE archive (18) (http://www.ebi.ac.uk/pride/archive/), MassIVE (http://massive.ucsd.edu/) and jPOST (19) (https://jpostdb.org/) are primarily supporting tandem MS data submission, whereas targeted proteomics (SRM/MRM) data can be submitted to PASSEL (20) (http://www.peptideatlas.org/passel/) and Panorama Public (21) (https://panoramaweb.org/). Data from all proteomics workflows can be submitted using the ‘partial’ submission type. The ProteomeCentral (http://proteomecentral.proteomexchange.org) site provides central, metadata-based search capability for all datasets, whereas the actual data are stored in the original repository to which they are submitted. Multiple repositories are highly necessary to enable researchers to deposit data and to ensure the long-term security of public data.
In step with global developments, proteomics research in China has evolved rapidly. Recently, the Chinese Human Proteome Project (CNHPP) was launched to conduct human proteome research focusing on the ten most prevalent cancers in China. It has achieved fast proteome sequencing and produced large-scale datasets of several human tissues and body fluids. To facilitate open access to proteome data worldwide and support proteomics research projects in China and beyond, the integrated proteome resource iProX (http://www.iprox.org) was built as a public platform for collecting and sharing raw data, analysis results and standardized metadata obtained in proteomics experiments. Users can submit their proteomics datasets to iProX as public or private datasets. Public datasets are immediately openly accessible, while private datasets can only be accessed by authorized users until an associated manuscript has been published.
OVERVIEW OF IPROX
iProX is a public platform for collecting and sharing raw data, identifications and standardized metadata obtained from proteomics experiments. The data flow diagram of iProX is shown in Figure 1. iProX implements the data sharing guidelines formulated by the PX consortium, wherein registered users can submit their proteomics datasets to iProX in public or private mode. In private mode, datasets can be shared with specific users, usually for collaboration or for the purpose of peer review of unpublished data. At a time point specified by the data owner, and latest with the publication of an associated manuscript, a dataset becomes fully publicly accessible. As proposed by PX, two different submission workflows (‘Complete submission’ and ‘Partial submission’) are supported by iProX. Complete submissions contain all processed data in community standard file formats, and can be browsed through the web interface, down to peptide and PSM level. Partial submissions provide the processed data in formats that are not yet fully supported by the web interface and can only be downloaded for local analysis.
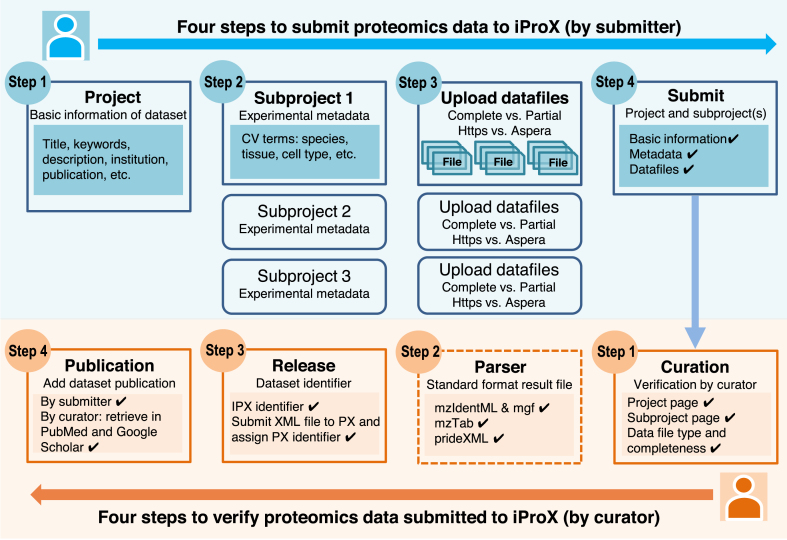
Figure 1.
Workflow of the proteomics data submission and curation process in iProX. The upper layer (blue rectangles) illustrates the data submission process for users, whereas the bottom layer (orange rectangles) represents the data curation process for iProX curators.
iProX became a full member of the PX Consortium in November 2017, both iProX identifier (IPX) and PX identifier (PXD) are assigned to any dataset submitted to iProX. An XML summary file with all necessary metadata is generated automatically on dataset release and submitted to PX for metadata exchange, enhancing visibility and citation probability of a dataset. All public iProX datasets are searchable through iprox.org itself, as well as through the central PX search interface at http://proteomecentral.proteomexchange.org/cgi/GetDataset and through the Omics Discovery Index (https://www.omicsdi.org/) (22).
DATASETS AVAILABLE IN IPROX
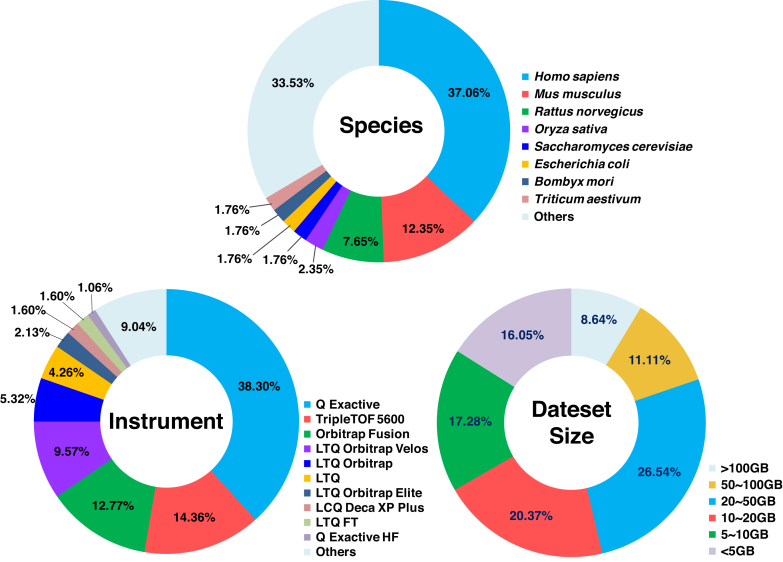
iProX initiated acceptance of regular data submissions in January 2017. By the end of July 2018, 417 datasets had been submitted, including 162 public datasets and 255 private datasets, with a total amount of 46 TB data. As shown in Figure 2, the six most frequent species in iProX are Homo sapiens, Mus musculus, Rattus norvegicus,Oryza sativa,Saccharomyces cerevisiae and Escherichia coli; MS instruments with high mass accuracy contribute most of the data deposited in iProX, 65% of the datasets were generated by Q Exactive, TripleTOF 5600 and Orbitrap Fusion. The size of datasets released in iProX ranges from several to hundreds of gigabytes, with 20–50 GB the most representative size. Moreover, large datasets with more than 100 GB of data in a single experiment are not uncommon.
Figure 2.
Summary of the datasets released in iProX. Distribution figures of the species, MS instrument and data size of datasets public available in iProX (by the end of July 2018).
Currently, significant amounts of data generated from the CNHPP have been made available to the public through iProX, including the proteome datasets generated in the first phase of the CNHPP (e.g. PXD008840 (23) and PXD010702) and from the Chromosome-Centric Human Proteome Project (e.g. PXD006654 (24), PXD004079 (25) and PXD001694 (26)).
PRIMARY FEATURES OF IPROX
To construct a high-quality public proteomics data repository, iProX implements several advanced features, including a web-based data submission process, structured management of datasets, standardized metadata collection, visualized data transmission, manual verification of deposited datasets and open sharing of MS-based proteomics datasets.
Web-based proteomics data submission
iProX provides a user-friendly web-based process for submitting proteomics data files and experimental metadata. As shown in Figure 1, it simplifies the data submission process in four steps: creating a project, presenting one or several subproject(s), uploading data files and submitting the dataset.
The iProX submission system employs a navigation bar to guide the processes of filling and uploading experiment information and data files. All metadata are provided in the form of project and subproject(s). Basic information including dataset title, institution, submitter name, etc. is filled in on the project page. Detailed sample and experimental information (e.g. the species, tissue, cell type, MS instrument type, and proteomics experimental and data analysis procedures) is included on the subproject page. Two different submission methods (‘Complete submission’ and ‘Partial submission’) are supported by iProX, and users can select the method in the data file upload step. In ‘Complete submission,’ raw and result files with supported formats (mzTab, mzIdenML files with peak files or prideXML files) are required, whereas in ‘Partial submission’, raw and search files can be in non-standard formats.
Structured management of proteomics experimental datasets
Users can manage their proteomics experimental datasets based on the project–subproject hierarchical structure of the iProX system. In iProX, each project is accepted as a complete dataset. One or several subprojects can be created within one project to represent different experimental designs, such as biological repeats, technical repeats and clinical samples. A project in iProX is equivalent to a dataset in PX, and an XML summary file is automatically generated once the project is released. The XML files are produced in the latest PX XML format (version 1.4.0 in August 2018) and contain all necessary metadata for each dataset. Take the dataset PXD008840 (the IPX-ID is IPX0001046000) as an example (http://www.iprox.org/page/project.html?id=IPX0001046000), which constructed a proteomic landscape of diffuse-type gastric cancer, contained a total of 168 samples (84 tumors and 84 paired nearby tissues). Six Liquid Chromatography-tandem Mass Spectrometry (LC-MS/MS) runs were generated for each sample and 1008 total raw files were produced. The raw and search files of 84 paired samples are included in 84 subprojects (IPX0001046001-IPX0001046084, only IPX-IDs are available for subprojects), with each subproject containing 24 files (12 raw files and 12 search files).
Use of controlled vocabularies for standardized metadata collection
Metadata collection, another important function of a proteome data repository, is useful in searching and reusing published proteomic datasets. Adequate information on experimental design and MS/MS data generation must be provided in a standardized manner to ensure that an answer to a query with particular terms includes all relevant data. Thus, iProX makes extensive use of controlled vocabularies and ontologies embedded in the Ontology Lookup Service (OLS) system (27) to annotate the entries of proteomics datasets in the subproject information page, e.g. NCBITAXON (NCBI organismal classification) for organismal taxonomy, DOID (Human Disease Ontology) (28) for human disease states, BTO (BRENDA tissue/enzyme source) (29) for tissues, CL (Cell Ontology) (30) for cell types, and PRIDE CV (PRIDE Controlled Vocabulary), PSI-MS CV (MS ontology) (6), MOD (Protein modification) (31) and SEP (Sample processing and separations controlled vocabulary) for MS experiments and data analysis procedure annotation. A CV term viewer, which has been developed in iProX (Supplementary Figure S1), enables data submitters to browse or search for relevant ontology terms directly on the website. Several common values are provided as defaults for each term in iProX, and users can also present their specific CV terms, which can be reused in different data submissions with similar experimental settings.
Visualized data transmission with high speed
iProX implements the rules proposed by PX in that two different submission workflows (‘Complete submission’ and ‘Partial submission’) are supported by iProX. In iProX, users are prompted to select one of these methods before uploading data files, following which the system automatically checks the file type rules.
Both web-based and fast Aspera (https://asperasoft.com/) based transfer protocols are offered by iProX for submitter data upload. The web interface is used to straightforwardly upload small files, whereas the fast Aspera-based transfer method is recommended for multiple large files, which is common in proteomic studies. By installing a free Aspera plug-in client, data file transfer with almost no restriction on file size and number can be achieved between users and iProX in a visualized manner. All file upload tasks can be monitored by clients. If the file upload is interrupted by network failure or other reasons, the task can be continued directly in the Aspera client without duplicating the file selection process. We have performed multi-person data transmission tests on the iProX system, with the limitation of bandwidth set to 45 Mbps for a single thread, the continuous uploading record was 96 h for ∼1.9 TB data and a single file of 142.1 GB requiring ∼7.5 h.
Manual verification of submitted data and metadata
The iProX repository implements an automatic submission process for the submitter and a manual verification process by curators. As shown in the bottom layer of Figure 1, a dataset is locked when it is handed to the iProX system by a submitter, followed by a curator checking both the data and metadata within 24 h and determining whether the deposited dataset meets the criteria of the PX consortium. A real-time and automatic notification mechanism in iProX uses emails and internal messages. If a dataset meets the standard requirements, the submission process is completed and a message is sent to submitters with the assigned IPX and PXD identifiers.
Open sharing of proteomics datasets in iProX
Generally, researchers who submit data can take complete control of their data before they are released. All released datasets in iProX are freely accessible to the public without login requirements, and the complete proteomic data files for a certain project are available for download via http or Aspera GUI directly from its web interface (details are shown in the ‘An example dataset at iProX’ section in the Supplementary Data). Meanwhile, users can create a sharing URL with a password for peer-review of their private datasets, with validity limited to 30, 90, 180 or 360 days (details are shown in the ‘Access to the shared dataset in iProX’ section in the Supplementary Data). iProX provides a RESTful Web Service API for bulk download. Users can query the web service to get a list of projects based on date, month or year. Then, users can download all the data files for each project with an Aspera command. iProX is completely compatible with the data sharing policies proposed by PX. For all datasets deposited after 1 July 2017, data are released by the data submitter or curator once the respective paper is published. iProX staff performs weekly checks for new publications with iProX datasets in PubMed and Google Scholar.
IPROX SYSTEM ARCHITECTURE AND INFRASTRUCTURE WITH HIGH AVAILABILITY
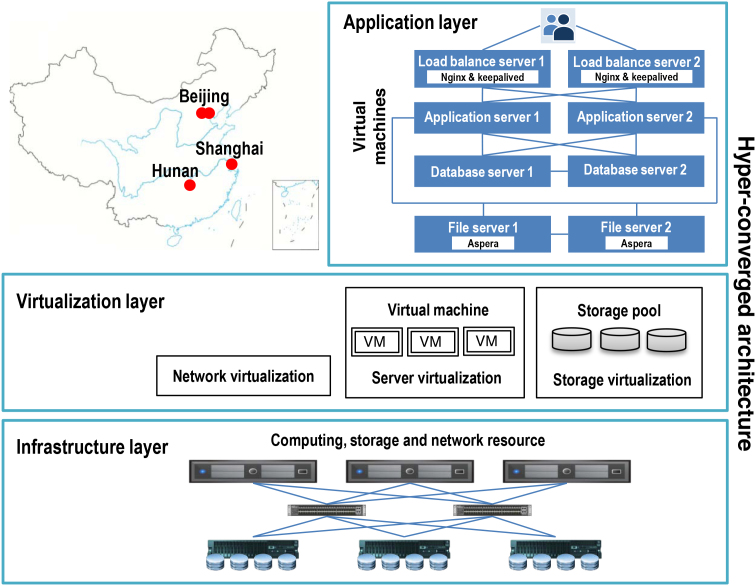
iProX is developed based on a hyper-converged architecture with high scalability, enabling the computing, storage and network environment requirements through a unified and fully virtualized resource pool. As shown in Figure 3, the hyper-converged platform integrates server, storage and network virtualization, and can achieve high scalability for iProX, ensuring continuous service using a distributed scale-out design for storage and computing resource expansion. This architecture also supports the localizable backup of datasets to ensure data security in iProX.
Figure 3.
System architecture and infrastructure of iProX. Based on a hyper-converged architecture, the application, virtualization and infrastructure layer are implemented for the iProX repository. The infrastructure layer includes hardware resources of computing, storage and network, while the virtualization layer enables a unified and virtualized resource pool to provide the real-time running resources required by iProX. The application layer uses virtual machines to achieve load balance and high availability, including load balance, application, database and datafile servers. The load balance is achieved by using the nginx and keepalived technologies on load balance and application servers, whereas two database servers are real-time synchronized to guarantee the high availability, and two file servers are used for high-speed data transfer and backup.
iProX is constructed for high availability via server load balancing technology, wherein the application-level load balancing is achieved using nginx and keepalived technologies. iProX has been deployed at the National Center for Protein Sciences (Beijing), as part of the proteomics infrastructure of China, which will ensure long-term and stable IT resource support. This site is the main location of deployment and the only submission site, whereas three other deployments are implemented for offsite data backups, including the Computer Network Information Center of Chinese Academy of Sciences (Beijing, Northern China), Shanghai Center for Bioinformation Technology (Shanghai, Eastern China) and the National Supercomputing Center in Changsha (Hunan, Southern China). Newly submitted data will be transformed to all three deployments within 24 h. Notably, all four sites provide simultaneous dataset downloading service using global load balance technology.
DISCUSSION AND CONCLUSION
With the advance of high-resolution MS, proteomics research has broken through the bottleneck of experimental technology and rapidly developed in the past few years to produce large amounts of proteomic data such that hundreds of MS raw data files can be generated in a single experiment. With the accumulation of massive datasets, requirements for network bandwidth and storage have rapidly developed. The PX consortium implements a mode of centralized metadata and distributed raw data management, promoting effective sharing of proteome data. We propose that such a distributed shared framework could be the future development trend for both independent and collaborating big-data repositories. The iProX repository is built in this way, based on a hyper-converged architecture with high scalability and high availability for proteome data collection and sharing. By solving the technical bottlenecks of network data transmission, the iProX repository implements a ‘physical centralized and distributed shared’ platform and efficiently integrates the proteome data deposited in different research institutions separately.
In this study, the integrated proteome resource center iProX was constructed. A user-friendly data submission system offering long-term and structured storage for large datasets of different types was built. iProX provides an accessible platform for raw MS data and standardized metadata. By facilitating the sharing and analysis of proteome data obtained from proteomics experiments, iProX will play a critical role in the implementation of proteomics projects in China and worldwide.
Supplementary Material
ACKNOWLEDGEMENTS
The iProX team would like to thank all the data submitters and collaborators for their contributions, and the members of the PX consortium for their help and support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research Program of China [2016YFC0901701, 2016YFB0201702, 2016YFC0901601]; International Scientific and Technological Cooperation project of China [2014DFB30010, 2014DFB30030]; National High Technology Research and Development Program of China [2015AA020108]; National Science Foundation of China [21475150 to Z. Y.]; BBSRC International Partnering Award [BB/N022432/1]. Funding for open access charge: National Science Foundation of China [21475150].
Conflict of interest statement. None declared.
REFERENCES
- 1. Vizcaino J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J.A., Sun Z., Farrah T., Bandeira N. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014; 32:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S. et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017; 45:D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodriguez H., Snyder M., Uhlen M., Andrews P., Beavis R., Borchers C., Chalkley R.J., Cho S.Y., Cottingham K., Dunn M. et al. Recommendations from the 2008 international summit on proteomics data release and sharing Policy: the Amsterdam principles. J. Proteome Res. 2009; 8:3689–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kinsinger C.R., Apffel J., Baker M., Bian X., Borchers C.H., Bradshaw R., Brusniak M.Y., Chan D.W., Deutsch E.W., Domon B. et al. Recommendations for mass spectrometry data quality metrics for open access data (corollary to the Amsterdam principles). Proteomics Clin. Appl. 2011; 5:580–589. [DOI] [PubMed] [Google Scholar]
- 5. Kinsinger C.R., Apffel J., Baker M., Bian X., Borchers C.H., Bradshaw R., Brusniak M.Y., Chan D.W., Deutsch E.W., Domon B. et al. Recommendations for mass spectrometry data quality metrics for open access data (corollary to the Amsterdam principles). Mol. Cell Proteomics. 2011; 10:O111.015446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mayer G., Montecchi-Palazzi L., Ovelleiro D., Jones A.R., Binz P.-A., Deutsch E.W., Chambers M., Kallhardt M., Levander F., Shofstahl J.. The HUPO proteomics standards initiative-mass spectrometry controlled vocabulary. Database. 2013; 2013:bat009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayer G., Jones A.R., Binz P.A., Deutsch E.W., Orchard S., Montecchi-Palazzi L., Vizcaino J.A., Hermjakob H., Oveillero D., Julian R. et al. Controlled vocabularies and ontologies in proteomics: overview, principles and practice. Biochim. Biophys. Acta. 2014; 1844:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez-Bartolome S., Binz P.A., Albar J.P.. The minimal information about a proteomics experiment (MIAPE) from the proteomics standards initiative. Methods Mol. Biol. 2014; 1072:765–780. [DOI] [PubMed] [Google Scholar]
- 9. Deutsch E.W., Orchard S., Binz P.A., Bittremieux W., Eisenacher M., Hermjakob H., Kawano S., Lam H., Mayer G., Menschaert G. et al. Proteomics Standards Initiative: Fifteen Years of Progress and Future Work. J. Proteome Res. 2017; 16:4288–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W.H., Römpp A., Neumann S., Pizarro A.D.. mzML–a community standard for mass spectrometry data. Mol. Cell. Proteomics. 2011; 10:R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vizcaino J.A., Mayer G., Perkins S., Barsnes H., Vaudel M., Perez-Riverol Y., Ternent T., Uszkoreit J., Eisenacher M., Fischer L. et al. The mzIdentML Data standard version 1.2, supporting advances in proteome informatics. Mol. Cell. Proteomics. 2017; 16:1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walzer M., Qi D., Mayer G., Uszkoreit J., Eisenacher M., Sachsenberg T., Gonzalez-Galarza F.F., Fan J., Bessant C., Deutsch E.W. et al. The mzQuantML data standard for mass spectrometry-based quantitative studies in proteomics. Mol. Cell. Proteomics. 2013; 12:2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griss J., Jones A.R., Sachsenberg T., Walzer M., Gatto L., Hartler J., Thallinger G.G., Salek R.M., Steinbeck C., Neuhauser N. et al. The mzTab data exchange format: communicating mass-spectrometry-based proteomics and metabolomics experimental results to a wider audience. Mol. Cell. Proteomics. 2014; 13:2765–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu Q.W., Griss J., Wang R., Jones A.R., Hermjakob H., Vizcaino J.A.. jmzTab: a java interface to the mzTab data standard. Proteomics. 2014; 14:1328–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perez-Riverol Y., Uszkoreit J., Sanchez A., Ternent T., Del Toro N., Hermjakob H., Vizcaino J.A., Wang R.. ms-data-core-api: an open-source, metadata-oriented library for computational proteomics. Bioinformatics. 2015; 31:2903–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qi D., Zhang H., Fan J., Perkins S., Pisconti A., Simpson D.M., Bessant C., Hubbard S., Jones A.R.. The mzqLibrary–An open source Java library supporting the HUPO-PSI quantitative proteomics standard. Proteomics. 2015; 15:3152–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cote R.G., Griss J., Dianes J.A., Wang R., Wright J.C., van den Toorn H.W., van Breukelen B., Heck A.J., Hulstaert N., Martens L. et al. The PRoteomics IDEntification (PRIDE) Converter 2 framework: an improved suite of tools to facilitate data submission to the PRIDE database and the ProteomeXchange consortium. Mol. Cell. Proteomics. 2012; 11:1682–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okuda S., Watanabe Y., Moriya Y., Kawano S., Yamamoto T., Matsumoto M., Takami T., Kobayashi D., Araki N., Yoshizawa A.C. et al. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res. 2017; 45:D1107–D1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farrah T., Deutsch E.W., Kreisberg R., Sun Z., Campbell D.S., Mendoza L., Kusebauch U., Brusniak M.Y., Huttenhain R., Schiess R. et al. PASSEL: the PeptideAtlas SRMexperiment library. Proteomics. 2012; 12:1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharma V., Eckels J., Schilling B., Ludwig C., Jaffe J.D., MacCoss M.J., MacLean B.. Panorama Public: a public repository for quantitative data sets processed in skyline. Mol. Cell. Proteomics. 2018; 17:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perez-Riverol Y., Bai M., da Veiga Leprevost F., Squizzato S., Park Y.M., Haug K., Carroll A.J., Spalding D., Paschall J., Wang M. et al. Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 2017; 35:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ge S., Xia X., Ding C., Zhen B., Zhou Q., Feng J., Yuan J., Chen R., Li Y., Ge Z. et al. A proteomic landscape of diffuse-type gastric cancer. Nat. Commun. 2018; 9:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang W., Chen X., Yan Z., Chen Y., Cui Y., Chen B., Huang C., Yin X., He Q.Y., He F. et al. Detergent-Insoluble proteome analysis revealed aberrantly aggregated proteins in human preeclampsia placentas. J. Proteome Res. 2017; 16:4468–4480. [DOI] [PubMed] [Google Scholar]
- 25. Guo J., Cui Y., Yan Z., Luo Y., Zhang W., Deng S., Tang S., Zhang G., He Q.Y., Wang T.. Phosphoproteome characterization of human colorectal cancer SW620 Cell-Derived exosomes and new phosphosite discovery for C-HPP. J. Proteome Res. 2016; 15:4060–4072. [DOI] [PubMed] [Google Scholar]
- 26. Chen Y., Li Y., Zhong J., Zhang J., Chen Z., Yang L., Cao X., He Q.Y., Zhang G., Wang T.. Identification of missing proteins defined by Chromosome-Centric proteome project in the cytoplasmic Detergent-Insoluble proteins. J. Proteome Res. 2015; 14:3693–3709. [DOI] [PubMed] [Google Scholar]
- 27. Perez-Riverol Y., Ternent T., Koch M., Barsnes H., Vrousgou O., Jupp S., Vizcaino J.A.. OLS Client and OLS Dialog: Open source tools to annotate public omics datasets. Proteomics. 2017; 17:1700244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bello S.M., Shimoyama M., Mitraka E., Laulederkind S.J.F., Smith C.L., Eppig J.T., Schriml L.M.. Disease Ontology: improving and unifying disease annotations across species. Dis. Model Mech. 2018; 11:dmm032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Placzek S., Schomburg I., Chang A., Jeske L., Ulbrich M., Tillack J., Schomburg D.. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 2017; 45:D380–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diehl A.D., Meehan T.F., Bradford Y.M., Brush M.H., Dahdul W.M., Dougall D.S., He Y., Osumi-Sutherland D., Ruttenberg A., Sarntivijai S. et al. The cell ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semantics. 2016; 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montecchi-Palazzi L., Beavis R., Binz P.A., Chalkley R.J., Cottrell J., Creasy D., Shofstahl J., Seymour S.L., Garavelli J.S.. The PSI-MOD community standard for representation of protein modification data. Nat. Biotechnol. 2008; 26:864–866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.