Abstract
Since its 2015 update, MaizeGDB, the Maize Genetics and Genomics database, has expanded to support the sequenced genomes of many maize inbred lines in addition to the B73 reference genome assembly. Curation and development efforts have targeted high quality datasets and tools to support maize trait analysis, germplasm analysis, genetic studies, and breeding. MaizeGDB hosts a wide range of data including recent support of new data types including genome metadata, RNA-seq, proteomics, synteny, and large-scale diversity. To improve access and visualization of data types several new tools have been implemented to: access large-scale maize diversity data (SNPversity), download and compare gene expression data (qTeller), visualize pedigree data (Pedigree Viewer), link genes with phenotype images (MaizeDIG), and enable flexible user-specified queries to the MaizeGDB database (MaizeMine). MaizeGDB also continues to be the community hub for maize research, coordinating activities and providing technical support to the maize research community. Here we report the changes MaizeGDB has made within the last three years to keep pace with recent software and research advances, as well as the pan-genomic landscape that cheaper and better sequencing technologies have made possible. MaizeGDB is accessible online at https://www.maizegdb.org.
INTRODUCTION
Zea mays ssp. mays (maize, corn) has been the top production grain crop in the world for over a decade (http://faostat.fao.org/). This success is largely due to high productivity and commercial versatility. However, not only is maize an excellent source of food, feed, and fuel, it more recently has become a leading model for the development of sustainable feedstock grasses for biofuel production (1–4). Maize is also an organism of historical and current importance to all biologists in its role as a model for complex plants and animals, for evolutionary processes, domestication, development, and cell destinies (5). Researchers including George Beadle, Rollins Emerson, Barbara McClintock, Lewis Stadler and Marcus Rhoades made seminal genetic discoveries in maize (6–9) that hold true for all living organisms (10).
The Maize Genetics and Genomics Database (MaizeGDB – https://www.maizegdb.org) (11) is the community database and global web resource for Zea mays, used each year by tens of thousands of researchers to access millions of pages, images, tools, documents, and datasets. MaizeGDB’s overall aim is to provide long-term storage, support, and stability to the maize research community's data and to provide informatics services for access, integration, and knowledge discovery. The maize research community uses data at MaizeGDB to facilitate their research, and in return, their published data gets curated at MaizeGDB. Over the years, MaizeGDB has been cited thousands of times on a wide range of maize-related publications. These publications have cited data (e.g. genetic maps, markers, SSRs, sequence data, annotations etc.), tools (e.g. genome browser, sequence identity tools), and services (e.g. searching and ordering stocks and germplasm). In addition to providing a data resource, the MaizeGDB team coordinates community activities such as Maize Genetics Executive Committee elections, provides technical support to the maize research community, and helps coordinate the annual maize genetics conference. The information and data provided at MaizeGDB and facilitated through outreach has directly been used in research that has had broad commercial, social, and academic impacts.
MaizeGDB has evolved from its first incarnation in the early 1990’s as an integrated gene and genetic map database (12), to a sequence-centric integration of data (11,13–17). Since then, the genomes of several maize inbred lines have been sequenced, including W22 (18), PH207 (19), CML247 (20), two assemblies of Mo17 (21,22), the European Flint lines EP1 and F7 (publication submitted), and the Zea mays ssp. mexicana line PI566673 (22), all of which are currently hosted at MaizeGDB. In the near future we expect to receive dozens more sequenced maize genomes, including all 25 of the maize Nested Association Mapping (NAM) population founder lines (23). MaizeGDB has expanded its genome browser, BLAST targets, and gene model pages to incorporate this influx of sequenced genomes, and identified syntenic orthologs between these various lines. We have also developed new tools to study maize diversity, link genes to phenotype images, and visualize pedigree data. MaizeGDB has begun incorporating RNA-seq data for B73 (24,25) and other maize inbred lines within our genome browsers, as well as developing or integrating new tools such as qTeller to analyze RNA expression, and MaizeMine to integrate complex biological datasets. The transition to a multi-genome resource along with the development of related tools, resources, and curated datasets will allow MaizeGDB to meet the needs of plant research communities as new technologies and approaches reveal a pangenomic model for maize.
MULTI-GENOME RESOURCE
Decreasing sequencing costs along with advances in technology have allowed many maize genomes, each with their myriad of duplicated genomic regions and transposable elements, to be sequenced at a reasonable cost and within a reasonable timeframe. There are now multiple sequenced maize genomes publicly available. Furthermore, we expect to see dozens, if not hundreds, of de novo sequenced and assembled maize genomes in the near future. To facilitate the influx of sequenced maize genomes, a number of changes have been made to the MaizeGDB database and user interface to host and cross-reference these genomes.
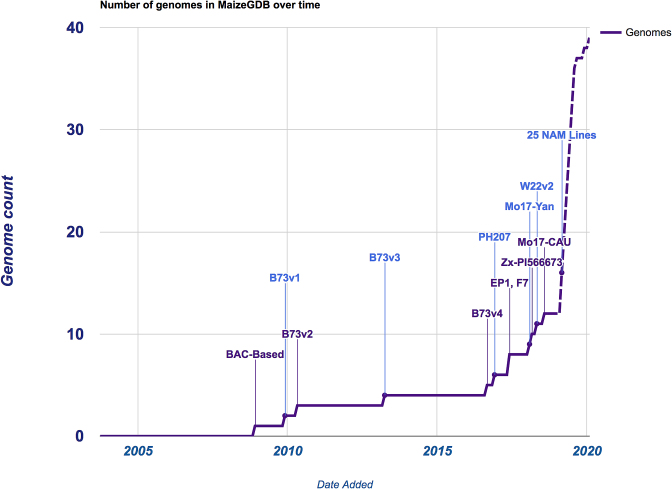
MaizeGDB currently hosts twelve fully sequenced and assembled maize genomes, including the inbred lines B73, W22, Mo17, PH207, EP1, F7, and by 2020 we expect to be hosting over 40 genomes (see Figure 2). MaizeGDB and the Maize Nomenclature Committee have established nomenclature guidelines for the consistent naming of genomes and gene model sets across all maize genomes (https://www.maizegdb.org/nomenclature). As this is an emerging issue in many species, we are spearheading an effort to standardize genome nomenclature among agriculturally important species. We have also expanded our genome browsers to include individual instances with many tracks for each of the above genomes (see below), generated gene model pages for each genome, and used CoGe's (https://genomevolution.org/coge/ (26)) comparative genomics tools to report syntenic orthologs between and among the sequenced genomes. We have created a genomes page, https://maizegdb.org/genome/assemblies_overview that gives an overview of all the genome assemblies currently hosted on MaizeGDB. From this page, users can navigate to separate pages for each genome that include metadata and download links. We require that a genome must be hosted in Genbank before we accept it into MaizeGDB. This ensures that all genomes that we host achieve a minimum standard of quality and metadata. Our genome metadata pages include information required by Genbank, plus our own expanded maize-specific requirements.
Figure 2.
This chart tracks the number of genome assemblies in MaizeGDB’s database since 2003. The first genome integrated into MaizeGDB was the B73 BAC-based assembly in November 2008, and until late 2016 additional genomes only included improved versions of B73. Following the release of B73 RefGen_v4, several assemblies of different maize lines have been sequenced de novo and incorporated into MaizeGDB. We anticipate this number to increase substantially as the cost of sequencing continues to fall, especially with the upcoming release of the 25 NAM founder lines. By 2020 we expect to be hosting ∼40 genomes.
NEW TOOLS AND FEATURES
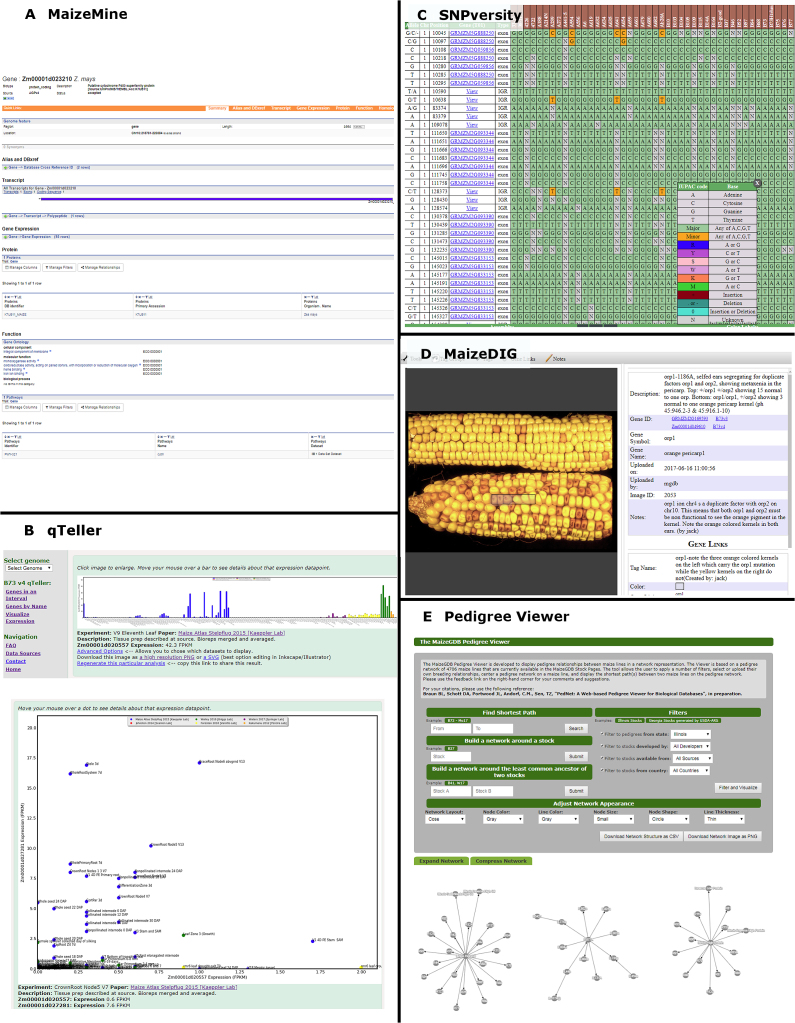
Since our 2015 update, MaizeGDB has developed and implemented multiple new tools for improved access and visualization of data. These tools provide the ability to view and query large-scale maize diversity data (SNPversity) (27), visualize pedigree data in graphical formats (Pedigree Viewer), view, query, and compare gene expression data (qTeller), link genes with phenotype images (MaizeDIG), and access flexible user specified queries to the MaizeGDB database (MaizeMine). Details of each tool are listed below and a composite image of each tool is shown in Figure 3.
Figure 3.
This image is a composite of the five new tools at MaizeGDB. Section (A) shows a MaizeMine results page for a particular gene model, which includes information on transcripts/proteins, expression, function, pathways, homology, and publications. Section (B) shows two images from qTeller. The top image show the expression values of a particular gene model across all of the tissues in the database. The bottom image shows a scatterplot of FPKM values between two gene models. Section (C) shows the results of a SNPversity query, which indicate the major/minor allele for a given inbred line at the top of the table and whether each SNP is located within a gene model's exon, intron or intergenic region. Section (D) shows the MaizeDIG interface, which allows curators to tag phenotypes in the image and link them to particular genes. The box outline in the middle of the image shows an example of what a tagged phenotype looks like. Section (E) shows the results of a query in the Pedigree Viewer of all stocks that were developed in Illinois.
SNPversity
SNPversity (27) allows maize researchers to select a customized set of maize lines and a genomic region, and visualize DNA changes for that genomic region. The tool is loaded with datasets from Panzea (20,28) that contain ∼1 million regions of DNA variation for over 17,000 public maize lines. The SNPversity tool, along with the PedNet tool described below, has been built according to a survey conducted among maize researchers (29). The tool was upgraded to improve the flexibility of searches, produce more detailed results, and provide estimates on query runtimes. This tool has utility for maize geneticists trying to identify and clone genes of interest, relate genomic regions to phenotype, and understand the diversity in maize.
Pedigree viewer (PedNet)
PedNet (publication in preparation) is a web-based pedigree viewer for maize lines. It allows users to build a network around a line, find the shortest path between two lines, build a network around the least common ancestor of two stocks, and filter pedigree networks by state, developer, source, and country. The tool has also been integrated into MaizeGDB’s stock pages to show interactive images of pedigree trees. The Pedigree Viewer is customizable and allows users to upload their own data. This tool will help maize breeders identify appropriate maize lines to use in breeding projects, which will improve available germplasm for researchers and farmers (manuscript in preparation).
qTeller
qTeller (http://www.qteller.com) is an RNA-seq processing pipeline and modular web interface that has been used to study quantitative expression variation in maize kernel row number (30), chitinase transcription during maize development (31), and other comparative expression analyses in maize, Arabidopsis thaliana, and Brassica rapa. qTeller draws RNA expression graphs for multiple RNA-seq datasets in real time for any gene of interest, and can be used to compare two genes at once by drawing a dot plot of relative expression for each RNA-seq dataset. MaizeGDB now hosts a version of qTeller (https://qteller.maizegdb.org) that currently has RNA-seq datasets from six different publications for B73 (covering over 150 different tissues, conditions and/or time points) and will soon host RNA-seq datasets for the inbred lines W22, Mo17 and others.
MaizeDIG
MaizeDIG is an easy to use and easily extensible web-based resource to link genes, alleles and genomes to phenotypes in images. MaizeDIG is based on the GMOD tool BioDIG (32), but has been enhanced to support multiple genomes and integrated with genome browsers to make tracks showing mutant phenotypes images within their genomic context. The ability to view images of mutant phenotypes at the genes’ position on our genome browser is a new and unique feature at MaizeGDB. MaizeDIG allows for custom tagging of images to highlight regions related to the phenotypes and to curate and search by gene model, gene symbol, gene name, and allele. MaizeDIG has over 2300 preloaded images available for 10 different genome browsers at MaizeGDB. Mutant phenotype images of 85 classical maize genes (one image per gene) have been manually tagged to clarify and highlight phenotypes.
MaizeMine
MaizeMine is a new data mining warehouse at MaizeGDB that accelerates genomic analysis by enabling researchers without scripting skills to create and export customized annotation datasets in a variety of formats that can be merged with their own research data for use in downstream analyses. MaizeMine uses the InterMine data warehousing system (33) to fully integrate genomic sequences from both the B73 RefGen_v3 and RefGen_v4 genome assemblies with the datasets described in Table. MaizeMine also provides simple and sophisticated search tools to MaizeGDB stakeholders, including a keyword search, built-in template queries with intuitive search menus, a list tool for creation of custom lists and a QueryBuilder tool for creating custom queries.
Table 1.
This table depicts all of the datasets loaded into MaizeMine as of this publication.
| Data Type | Description | Species | PubMed | Link |
|---|---|---|---|---|
| Genes | Maize community gene set for B73v4 | Z. mays | Lawrence et al - PubMed 14681441 Jiao et al - PubMed 28605751 | MaizeGDB Genomes |
| Maize community gene set for B73v3 | Z. mays | Law et al - PubMed 25384563 Lawrence et al - PubMed 14681441 | MaizeGDB Genomes | |
| NCBI annotation (RefSeq and Gene) | Z. mays | O'Leary et al - PubMed 26553804 | NCBI FTP | |
| Homology | Orthologue and paralogue relationships | A. thaliana B. distachyon ... O. sativa S. italica S. bicolor Z. mays | Monaco et al - PubMed 24217918 | EnsemblCompara Plant |
| Proteins | Protein annotations from UniProt | Z. mays | UniProt Consortium - PubMed 25348405 | UniProt FTP |
| Protein family and domain assignments to proteins from Interpro | Z. mays | Mitchell et al - PubMed 25428371 | InterPro FTP | |
| Gene Ontology | GO annotations | Z. mays | Huntley et al - PubMed 25378336 Gene Ontology Consortium - PubMed 25428369 | GO Consortium Annotation FTP |
| Pathways | Pathway information from Plant Reactome | A. thaliana Z. mays | Tello-Ruiz MK et al - PubMed 26553803 | Reactome Gramene FTP |
| Pathway information from KEGG | Z. mays | Kanehisa M et al - PubMed 22080510 | KEGG | |
| Pathway information from CornCyc 8.0 | Z. mays | Walsh JR et al - PubMed 27899149 | CornCyc FTP | |
| Publications | A mapping from genes to publications | Z. mays | UniProt Knowledgebase - PubMed 28150232 | |
| Gene Expression | Gene expression computed on reference gene set RefSeq, AGPv3, and AGPv4 | Z. mays | Stelpflug et al - PubMed 27898762 Sekhon et al - PubMed 23637782 | NCBI BioProject 171684 |
| Genome Assembly | Chromosome assembly for B73v4 | Z. mays | Jiao et al - PubMed 28605751 | NCBI FTP |
| Chromosome assembly for B73v3 | Z. mays | Schnable et al - PubMed 19965430 | Ensembl Genomes FTP |
CURATION
MaizeGDB continues to provide high quality curation of published literature. Priority for curation is given to (i) datasets associated with publications recommended by the MaizeGDB Editorial Board, an external group of maize researchers nominated annually that present a paper every month; (ii) datasets that describe mutant phenotypes, especially for germplasm curated by the Maize Genetics Cooperation Stock Center and germplasm of maize diversity panels; and (iii) datasets that increase the understanding of each gene's function and of orthologous or paralogous identities. We continue to provide high quality datasets as genome browser tracks, and include expression analysis in our new qTeller instance. MaizeGDB provides interfaces that allow community curation, but since June of 2015 there have only been 11 community annotations and so this feature remains poorly utilized.
Stock center
MaizeGDB provides information about stocks available from the Maize Genetics Cooperation Stock Center and the ability for a user to request stocks of interest directly. Stock Center staff curate information about the stocks, the mutant allele(s) carried by particular stocks, as well as information about the gene(s). Information about the stocks can be queried at: https://www.maizegdb.org/data_center/stock.
Reverse-genetics tools are also available. From sequence information, a user can examine a gene model of interest in the MaizeGDB Genome Browser and observe transposable element insert sites (e.g. Mutator, Ac/Ds). By holding the cursor over a particular insert one can see which stock carries it and go directly to that stock page and request the stock. These tools would help one determine the biological function of the sequence of interest.
Gene/gene model associations
In maize, making the connections between computationally predicted gene models and genetically mapped genes and markers is an issue requiring ongoing curation of recent literature. These connections link known or putative functions with gene models and chromosomal coordinates. Currently, MaizeGDB has 39,475 gene models for B73v3 and 39,498 gene models for B73v4, of which 6,564 are genetically mapped to genes/markers.
To better enable comparative research across maize lines, associations are calculated between annotations for the multiple genome assemblies and the annotation for the reference assembly of B73. Associations with gene models generated in earlier B73 annotations are also calculated along with the history of name changes, and split, merged and deleted gene models.
INTERFACE AND DATA UPDATE
Data centers
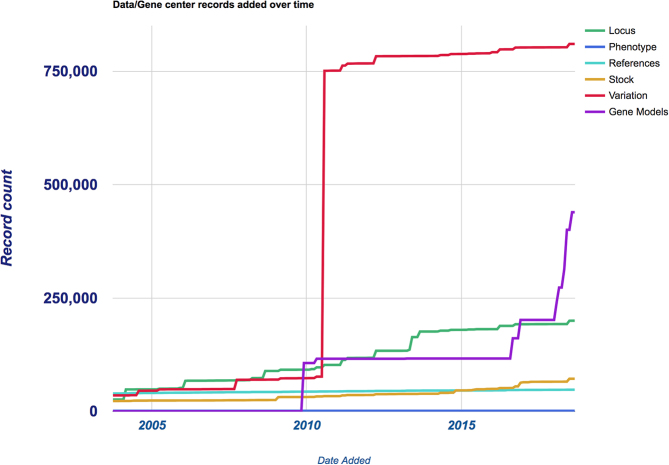
MaizeGDB users can quickly access a range of data types through the data center links on the top menu bar. We currently serve fifteen data centers that each contain anywhere from a few hundred records to hundreds of thousands of records (see Table 2). Most data centers have both simple search tools and advanced search tools that allow users to create filtered data searches. While the number of data centers has remained constant since the last MaizeGDB update, several categories have increased significantly in size and scope. The image data center now includes a guide to M.G. Neuffer's ∼8,600 mutant phenotype images (https://mutants.maizegdb.org), which are provided in a searchable wiki format with extensive image annotation. The gene model data center has expanded from containing only gene models for B73 RefGen_v3 nearly six fold to include gene models from seven additional reference assemblies (PH207, W22, Mo17 (two versions), EP1, F7 and PI566673), B73 RefGen_v4 gene models, and gene models in all previous B73 assemblies. Figure 1 shows how the number of records have changed since 2003 for these types of data. Organization of the many data types at MaizeGDB into distinct data centers has been a useful approach towards making data quickly accessible and MaizeGDB will continue to modify or add new data centers to meet the ever-evolving needs of our stakeholders.
Table 2.
This table shows the data centers and their content in MaizeGDB. Each data center groups similar data types for ease of custom querying, which also helps our users find what they need faster
| Data Center | Description | Record Count |
|---|---|---|
| Alleles/polymorphisms | Queryable data on alleles of known genes. | 810,229 alleles |
| BACs | Queryable BACs from the Maize Mapping Project isolated from B73 | 439,464 BACs |
| Cytogenetics | Cytogenetic data including: maps, knobs, centromeres, telomeres, karyotypes, stocks, and other resources | N/A |
| Diversity | Various tools and resources for downloading and querying diversity, SNP, and trait data | 419,061 traits |
| Expression | Lists of atlas type expression data | N/A |
| Genes/Gene Models | Queryable genes and gene models from all genomes hosted at MaizeGDB. Additional information about the reference annotation for B73 can also be found here. | 13,468 genes / 598,794 gene models |
| Gene Products | Queryable gene products. | 1,992 gene products |
| Images | Queryable images of traits, phenotypes, pests, gel patterns, and mutants | 10,115 images |
| Loci/QTLs | Queryable loci of all locus types. | 214,464 loci |
| Maps | Queryable genetic maps. | 2,117 maps |
| Metabolic Pathways | Metabolic pathway data and CornCyc 9.0 | 485 pathways (B73v3) / 505 pathways (B73v4) |
| Molecular Markers | Queryable markers of all probe types (BACs, ESTs, SSRs, etc.). | 771,136 markers |
| Phenotypes | Queryable phenotype and mutant data. | 1,120 phenotypes |
| References | Queryable references of all publication types (articles, reports, abstracts, books, etc). | 47,584 references |
| Stocks | Queryable stocks with links to order them from their respective repositories where applicable. | 66,825 stocks |
Figure 1.
This chart tracks the number of records in MaizeGDB’s database for the listed data types since 2003. Initially there were a comparable number of records for Loci, References, Stock, and Variation data types until July of 2010, when hundreds of thousands of Variation records from the first maize HapMap (43) were added to the database. Gene model records were not introduced until the release of the first maize reference assembly, B73 RefGen_v1 in late 2009, and then began to rise sharply in late 2016 with the release of each new maize genome assembly (see Figure 2). The number of Phenotype records has not changed significantly from 1,016 in 2003 to 1,136 in August of 2018.
Genome browsers
MaizeGDB genome browsers (34) are GBrowse (35) instances for exploring sets of genomic and genetic features on assembled genomes, including gene models, markers, gene expression, transposable elements, and many more. As visualization of genotypic and phenotypic datasets has always been a core part of MaizeGDB, there are tracks for viewing the gene and mobile element annotations of several maize genomes. Since 2015 there have been 208 new tracks created on the genome browsers for B73 and nine additional maize lines that show the visualization of eQTL, expression, gene, miRNA, proteomics, RNA-seq reads, SNP, synteny and other datasets. The RNA-seq reads in particular are a new type of data we visualize on the browser to show expression levels of each base pair in a specified maize tissue across the genome. Out of the 208 new tracks created since 2015, 140 of them are RNA-seq read tracks. The MaizeGDB genome browsers also have several custom tracks including histogram views of gene expression atlas data, BLAST hit alignments from the MaizeGDB BLAST tool, and tagged phenotype images from the MaizeDIG tool.
BLAST
The MaizeGDB custom BLAST tool (36,37) allows researchers to perform sequence similarity searches on genome assemblies and annotations. MaizeGDB BLAST targets currently include 21 genome assembly targets (different genomes, versions or masking options) and 46 annotations (including transcripts and protein translations). The MaizeGDB BLAST tool has several custom features: built in support for searching multiple target databases in parallel, links to results that are stored for one week, whole genome views using CViT (38), semantic alignment views, and custom BLAST hit tracks that are dynamically rendered on the MaizeGDB genome browsers.
CornCyc
CornCyc (39,40) is a pathway-genome database developed by the Plant Metabolic Network (PMN) in collaboration with MaizeGDB. The current version 9.0 was built using the B73 RefGen_v4 assembly and Pathway Tools v22.0 (41). MaizeGDB hosts the newest version of CornCyc featuring B73 RefGen_v4, which boasts not only the newest assembly, but also an increase in the number of pathways/reaction/genes. It has reliably-annotated maize metabolic resource for over 3,305 enzymatic reactions on 565 pathways for 7,399 enzymes. The Metabolic Pathway Data Center contains data as well as Pathway Tools installation files, video tutorials and training materials. MaizeGDB has also collected manually curated GO annotations that are included in the pathway databases.
COMMUNITY SUPPORT AND OUTREACH
The maize research community's success in maintaining a collaborative and cooperative spirit can be largely attributed to the coordinated activities of the Maize Genetics Executive Committee (MGEC), the Maize Genetics Conference Steering Committee (MGCSC), and the outstanding cooperative spirit of individual maize researchers. Emblematic of this spirit are years of individual contributions to the Maize Genetics Cooperation Newsletter (MNL), which was initiated in 1929; cover-to-cover PDF copies of all issues have been incorporated into MaizeGDB recently. To further foster this collaborative spirit, MaizeGDB provides a wide-range of community support services and outreach activities. MaizeGDB coordinates elections, collects award nominations, and conducts surveys of the maize community as directed by the MGEC/MGCSC. MaizeGDB also plays a significant support role in the Maize Genetics Conference, that includes handling submission of abstracts, student financial aid applications, community contests, hosting the conference website maintaining mailing lists, creating the conference program, and providing in-person technical support at the conference. In the past three years, MaizeGDB has hosted 13 pre-conference workshops on topics pertinent to the maize community, including maize genome assemblies, epigenetics, transformation and gene editing, and how to use MaizeGDB and other maize related tools and resources. MaizeGDB is also a founding member of the AgBioData Consortium (42) which focuses on creating and implementing common standards, tools and curation pipelines which can be used by multiple genome databases across different agriculturally important plants and animals.
ACKNOWLEDGEMENTS
MaizeGDB is part of the Corn Insects and Crop Genetics Research Unit in the Midwest Area office of USDA-ARS. MaizeGDB efforts are guided by the USDA-ARS National Program staff and with suggestions from the MaizeGDB Working Group as well as feedback and comments from the maize research community. Support, assistance, and guidance from the above groups along with the MaizeGDB Editorial Board, Maize Nomenclature Committee, Maize Genetics Cooperation Stock Center, Plant Introduction Station, Maize Genetics Conference Steering Committee, and Maize Genetics Executive Committee are deeply appreciated. We would like to give a special thank you to Ed Coe as not only the founder of the original Maize Genetics Database and continued contributor to the database, but also for his contribution to the generation of searchable PDFs of the Maize Newsletters.
FUNDING
United States Department of Agriculture-Agricultural Research Service [5030-21000-068-00-D] by funding salaries and open access charges. The funding agency played no role in in the design of the study and collection, analysis and interpretation of data or in writing the manuscript. Funding for open access charge: United States Department of Agriculture–Agricultural Research Service.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bosch M., Mayer C.D., Cookson A., Donnison I.S.. Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J. Exp. Bot. 2011; 62:3545–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carpita N.C., McCann M.C.. Maize and sorghum: genetic resources for bioenergy grasses. Trends Plant Sci. 2008; 13:415–420. [DOI] [PubMed] [Google Scholar]
- 3. Lawrence C.J., Walbot V.. Translational genomics for bioenergy production from fuelstock grasses: maize as the model species. Plant Cell. 2007; 19:2091–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Penning B.W., Hunter C.T. 3rd, Tayengwa R., Eveland A.L., Dugard C.K., Olek A.T., Vermerris W., Koch K.E., McCarty D.R., Davis M.F. et al. Genetic resources for maize cell wall biology. Plant Physiol. 2009; 151:1703–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Strable J., Scanlon M.J.. Maize (Zea mays): a model organism for basic and applied research in plant biology. Cold Spring Harb. Protoc. 2009; 2009:doi:10.1101/pdb.emo132. [DOI] [PubMed] [Google Scholar]
- 6. Creighton H.B., McClintock B.. A correlation of cytological and genetical crossing-over in Zea mays. Proc. Natl. Acad. Sci. U.S.A. 1931; 17:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beadle G.W. The relation of crossing over to chromosome association in Zea-Euchlaena hybrids. Genetics. 1932; 17:481–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stadler L.J. Genetic effects of X-Rays in maize. Proc. Natl. Acad. Sci. U.S.A. 1928; 14:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rhoades M.M. The early years of maize genetics. Annu. Rev. Genet. 1984; 18:1–29. [DOI] [PubMed] [Google Scholar]
- 10. Shull G. The composition of a field of maize. Am. Breeders' Assoc. Rpt. 1908; 4:296–301. [Google Scholar]
- 11. Andorf C.M., Cannon E.K., Portwood J.L., Gardiner J.M., Harper L.C., Schaeffer M.L., Braun B.L., Campbell D.A., Vinnakota A.G., Sribalusu V.V. et al. MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res. 2016; 44:D1195–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polacco M., Coe E., Fang Z., Hancock D., Sanchez-Villeda H., Schroeder S.. MaizeDB - a functional genomics perspective. Comp. Funct. Genomics. 2002; 3:128–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harper L., Gardiner J., Andorf C., Lawrence C.J.. MaizeGDB: The maize genetics and genomics database. Methods Mol. Biol. 2016; 1374:187–202. [DOI] [PubMed] [Google Scholar]
- 14. Lawrence C.J., Dong Q., Polacco M.L., Seigfried T.E., Brendel V.. MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res. 2004; 32:D393–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lawrence C.J., Harper L.C., Schaeffer M.L., Sen T.Z., Seigfried T.E., Campbell D.A.. MaizeGDB: The maize model organism database for basic, translational, and applied research. Int. J. Plant Genomics. 2008; 2008:496957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lawrence C.J., Schaeffer M.L., Seigfried T.E., Campbell D.A., Harper L.C.. MaizeGDB's new data types, resources and activities. Nucleic Acids Res. 2007; 35:D895–D900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sen T.Z., Andorf C.M., Schaeffer M.L., Harper L.C., Sparks M.E., Duvick J., Brendel V.P., Cannon E., Campbell D.A., Lawrence C.J.. MaizeGDB becomes 'sequence-centric'. Database. 2009; 2009:bap020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Springer N.M., Anderson S.N., Andorf C.M., Ahern K.R., Bai F., Barad O., Barbazuk W.B., Bass H.W., Baruch K., Ben-Zvi G. et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat. Genet. 2018; 50:1282–1288. [DOI] [PubMed] [Google Scholar]
- 19. Hirsch C.N., Hirsch C.D., Brohammer A.B., Bowman M.J., Soifer I., Barad O., Shem-Tov D., Baruch K., Lu F., Hernandez A.G. et al. Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell. 2016; 28:2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu F., Romay M.C., Glaubitz J.C., Bradbury P.J., Elshire R.J., Wang T., Li Y., Li Y., Semagn K., Zhang X. et al. High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 2015; 6:6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun S., Zhou Y., Chen J., Shi J., Zhao H., Zhao H., Song W., Zhang M., Cui Y., Dong X. et al. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 2018; 50:1289–1295. [DOI] [PubMed] [Google Scholar]
- 22. Yang N., Xu X.W., Wang R.R., Peng W.L., Cai L., Song J.M., Li W., Luo X., Niu L., Wang Y. et al. Contributions of Zea mays subspecies mexicana haplotypes to modern maize. Nat. Commun. 2017; 8:1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu J., Holland J.B., McMullen M.D., Buckler E.S.. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008; 178:539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stelpflug S.C., Sekhon R.S., Vaillancourt B., Hirsch C.N., Buell C.R., de Leon N., Kaeppler S.M.. An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome. 2016; 9:doi:10.3835/plantgenome2015.04.0025. [DOI] [PubMed] [Google Scholar]
- 25. Walley J.W., Sartor R.C., Shen Z., Schmitz R.J., Wu K.J., Urich M.A., Nery J.R., Smith L.G., Schnable J.C., Ecker J.R. et al. Integration of omic networks in a developmental atlas of maize. Science. 2016; 353:814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lyons E., Freeling M.. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008; 53:661–673. [DOI] [PubMed] [Google Scholar]
- 27. Schott D.A., Vinnakota A.G., Portwood J.L. 2nd, Andorf C.M., Sen T.Z.. SNPversity: a web-based tool for visualizing diversity. Database. 2018; 2018:doi:10.1093/database/bay037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Romay M.C., Millard M.J., Glaubitz J.C., Peiffer J.A., Swarts K.L., Casstevens T.M., Elshire R.J., Acharya C.B., Mitchell S.E., Flint-Garcia S.A. et al. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 2013; 14:R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sen T.Z., Braun B.L., Schott D.A., Portwood Ii J.L., Schaeffer M.L., Harper L.C., Gardiner J.M., Cannon E.K., Andorf C.M.. Surveying the Maize community for their diversity and pedigree visualization needs to prioritize tool development and curation. Database. 2017; 2017:doi:10.1093/database/bax031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu L., Du Y., Shen X., Li M., Sun W., Huang J., Liu Z., Tao Y., Zheng Y., Yan J. et al. KRN4 controls quantitative variation in maize kernel row number. PLoS Genet. 2015; 11:e1005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hawkins L.K., Mylroie J.E., Oliveira D.A., Smith J.S., Ozkan S., Windham G.L., Williams W.P., Warburton M.L.. Characterization of the maize chitinase genes and their effect on aspergillus flavus and aflatoxin accumulation resistance. PLoS One. 2015; 10:e0126185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oberlin A.T., Jurkovic D.A., Balish M.F., Friedberg I.. Biological database of images and genomes: tools for community annotations linking image and genomic information. Database. 2013; 2013:bat016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith R.N., Aleksic J., Butano D., Carr A., Contrino S., Hu F., Lyne M., Lyne R., Kalderimis A., Rutherford K. et al. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012; 28:3163–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sen T.Z., Harper L.C., Schaeffer M.L., Andorf C.M., Seigfried T.E., Campbell D.A., Lawrence C.J.. Choosing a genome browser for a Model Organism Database: surveying the maize community. Database. 2010; 2010:baq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stein L.D. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief. Bioinform. 2013; 14:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 37. Cannon E.K., Birkett S.M., Braun B.L., Kodavali S., Jennewein D.M., Yilmaz A., Antonescu V., Antonescu C., Harper L.C., Gardiner J.M. et al. POPcorn: An online resource providing access to distributed and diverse maize project data. Int. J. Plant Genomics. 2011; 2011:923035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cannon E.K., Cannon S.B.. Chromosome visualization tool: a whole genome viewer. Int. J. Plant Genomics. 2011; 2011:373875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chae L., Kim T., Nilo-Poyanco R., Rhee S.Y.. Genomic signatures of specialized metabolism in plants. Science. 2014; 344:510–513. [DOI] [PubMed] [Google Scholar]
- 40. Walsh J.R., Schaeffer M.L., Zhang P., Rhee S.Y., Dickerson J.A., Sen T.Z.. The quality of metabolic pathway resources depends on initial enzymatic function assignments: a case for maize. BMC Syst. Biol. 2016; 10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karp P.D., Latendresse M., Paley S.M., Krummenacker M., Ong Q.D., Billington R., Kothari A., Weaver D., Lee T., Subhraveti P. et al. Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief. Bioinform. 2016; 17:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harper L., Campbell J., Cannon E.K., Jung S., Main D., Poelchau M., Walls R.L., Andorf C.M., Arnaud E., Berardini T.Z. et al. AgBioData consortium recommendations for sustainable genomics and genetics databases for agriculture. Database. 2018; 2018:doi:10.1093/database/bay088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gore M.A., Chia J.M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer J.A., McMullen M.D., Grills G.S., Ross-Ibarra J. et al. A first-generation haplotype map of maize. Science. 2009; 326:1115–1117. [DOI] [PubMed] [Google Scholar]