Abstract
RNA editing, as an essential co-/post-transcriptional RNA modification type, plays critical roles in many biological processes and involves with a variety of human diseases. Although several databases have been developed to collect RNA editing data in both model and non-model animals, there still lacks a resource integrating associations between editome and human disease. In this study, we present Editome-Disease Knowledgebase (EDK; http://bigd.big.ac.cn/edk), an integrated knowledgebase of RNA editome-disease associations manually curated from published literatures. In the current version, EDK incorporates 61 diseases associated with 248 experimentally validated abnormal editing events located in 32 mRNAs, 16 miRNAs, 1 lncRNA and 11 viruses, and 44 aberrant activities involved with 6 editing enzymes, which together are curated from more than 200 publications. In addition, to facilitate standardization of editome-disease knowledge integration, we propose a data curation model in EDK, factoring an abundance of relevant information to fully capture the context of editome-disease associations. Taken together, EDK is a comprehensive collection of editome-disease associations and bears the great utility in aid of better understanding the RNA editing machinery and complex molecular mechanisms associated with human diseases.
INTRODUCTION
RNA editing, as an essential co-/post-transcriptional modification, alters the base pairing properties of RNA molecules by means of substitutions, insertions and/or deletions (1–3). The exponential growth of reported RNA editing events has revealed the pervasive nature of RNA editing in human transcriptome (4–6), indicating the functional implications in gene expression and regulation (7). Evidence has accumulated that RNA editing exerts critical roles in affecting mRNA stability (8), splicing (9), recoding protein sequence (10), regulating the biogenesis and base-pairing interaction of ncRNAs (11), as well as nuclear export and localization (12). More importantly, a growing body of research has shown that RNA editing is of broad physiologic significance in mammals (13–17). Aberrant editing enzyme activity and dysfunction of editing in either coding or noncoding region have been revealed to be correlated with a wide range of human diseases, including cardiovascular diseases (8), neurological and behavioral abnormalities (18), cancer (19), autoimmune disorders (20), viral infections (21), aging-related diseases (22), and metabolic diseases (23). Collectively, RNA editing associates closely with a variety of human diseases, leading to a demand for a comprehensive collection of the associations between RNA editing and human diseases.
With the advancement of next-generation sequencing technologies, millions of high-confidence RNA editing sites (5,6,24) have been identified across a variety of tissues in human. Accordingly, efforts have been made to develop different databases that integrate various RNA editing data, including REDIportal (24), dbRES (25), DARNED (26), RADAR (27) and LNCediting (28). However, none of them is devoted to the comprehensive collection of editome-disease associations in human. Specifically, dbRES focuses on collection of experimentally validated editing sites for different organisms, DARNED and RADAR collect A-to-I editing events based on either EST data or a limited number of RNA-Seq data, REDIportal houses a large number of A-to-I editing sites across 30 human tissues identified from thousands of RNA-Seq experiments, and LNCediting characterizes and predicts the effects of all editing sites in lncRNAs across model animals. Clearly, all these resources provide valuable annotations for RNA editing events but ignore the associations between RNA editing and diseases. Considering the rapid accumulation of knowledge on RNA editome-disease associations, there is a great need to build a specialized database that integrates disease-associated RNA editing data.
Toward this end, here we present Editome-Disease Knowledgebase (EDK; http://bigd.big.ac.cn/edk), a curated knowledgebase of RNA editome-disease associations. Unlike extant relevant data-oriented databases, EDK features comprehensive integration of knowledge on abnormal RNA editing events and aberrant RNA editing enzyme activities, which are associated with human diseases. Therefore, EDK would be of great help to provide a landscape of editome-disease associations and to explore the RNA editing machineries underlying different human diseases.
MATERIALS AND METHODS
Literatures were initially retrieved from NCBI PubMed by specifying keywords ‘RNA editing’ and ‘disease’ and then manually filtered and double-checked by expert curators to remove irrelevant publications. Considering that the reported diseases are mostly associated with either aberrant activities of editing enzymes or abnormal editing events occurring on transcribed coding/noncoding genes and viruses, a curation model was proposed to standardize the curation process, through literature curation of over 200 relevant publications. Based on this model, editome-disease associations were curated from four aspects, namely, diseases, editing enzymes, edited genes, and viruses. For each aspect, a variety of curated information were further collected, such as disease name, gene symbol, miRNA ID, virus name, editing enzyme, editing type, molecular effects, phenotype, etc. Then, control vocabularies and standardized descriptive terms were abstracted and categorized to depict each item. More detailed information can be found at http://bigd.big.ac.cn/edk/faq.
To unify disease name and definition, terms or identifiers of Disease Ontology (DO), Experimental Factor Ontology (EFO), Online Mendelian Inheritance in Man (OMIM) and National Cancer Institute (NCI) were retrieved and mapped to the corresponding diseases. Consequently, all RNA editing associated diseases were grouped into predefined categories based on the modified primary and secondary classification systems of DO. In addition, external links to several related resources, such as REDIportal (24), RADAR (27), UniProt (29), GenBank (30), GeneCard (31), miRBase (32), GTEx (33), etc., were also provided for convenience. Furthermore, enrichment analysis was done to deduce the pathways affected by abnormal RNA editing using IPA version 2.0. The ‘Core Analysis’ function in IPA was used to interpret the edited genes in the context of biological processes, pathways and networks.
EDK was implemented using Spring Boot as back-end framework. JSP, Bootstrap, SemanticUI and JQuery, which provide a series of templates for designing web pages with consistent interface components, were adopted as front-end framework. Curated data were stored in MySQL database and data visualization was rendered by HighCharts and DataTables. EDK organizes all curated editome-disease associations at different levels; it provides a whole picture of editome-disease associations with conclusion items, presents detailed association information in terms of edited mRNA/miRNA/lncRNA, editing enzyme and virus, and summarizes different editome-disease associations for a specific disease, edited gene, editing enzyme and virus.
DATABASE CONTENTS AND USAGE
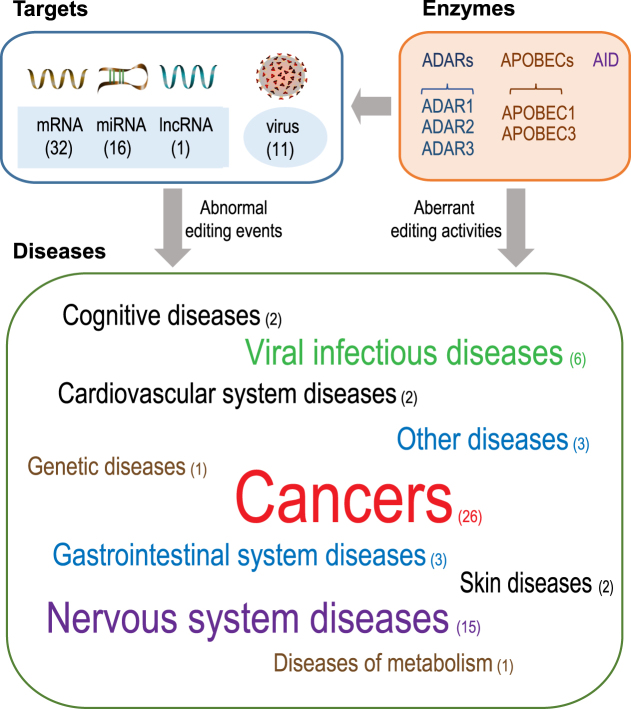
EDK features comprehensive integration of editome-disease associations as well as their relevant data manually curated from published literatures. Collectively, the current version of EDK incorporates 61 diseases (grouped into 10 categories) associated with 248 experimentally validated abnormal editing events that occur in 32 mRNAs, 16 miRNAs, 1 lncRNA and 11 viruses, as well as 44 aberrant activities involved with 6 editing enzymes, which are all manually curated from over 200 publications (Figure 1). Therefore, EDK serves as a curated knowledgebase of editome-disease associations, providing a comprehensive landscape of RNA editing in human diseases.
Figure 1.
Statistics of curated editome-disease associations in EDK (as of August 2018).
In the current implementation, EDK integrates 61 editome-associated diseases, including 26 cancers (43%), 15 nervous system diseases (24%), 6 viral infectious diseases (10%), 3 gastrointestinal system diseases (5%), 2 cognitive disorders (3%), 1 diseases of metabolism (2%), 1 genetic diseases (2%), 2 cardiovascular system disease (3%), 2 skin disease (3%) and 3 other types (5%) (http://bigd.big.ac.cn/edk/browse/disease) (Figure 2A). Among these diseases, cancers and nervous system diseases are extensively studied, which together cover nearly 70% of all collected diseases in EDK. Interestingly, one disease may be associated with aberrant RNA editing mechanisms on multiple levels. Taking ‘Glioblastoma’ as an example, it is associated with the abnormal editing events that occur on the mRNA of CDC14B and the miRNA of hsa-miR-589-3p/hsa-miR-221/hsa-miR-222 as well as the aberrant activity of the editing enzyme ADAR2 (http://bigd.big.ac.cn/edk/disease/Glioblastoma).
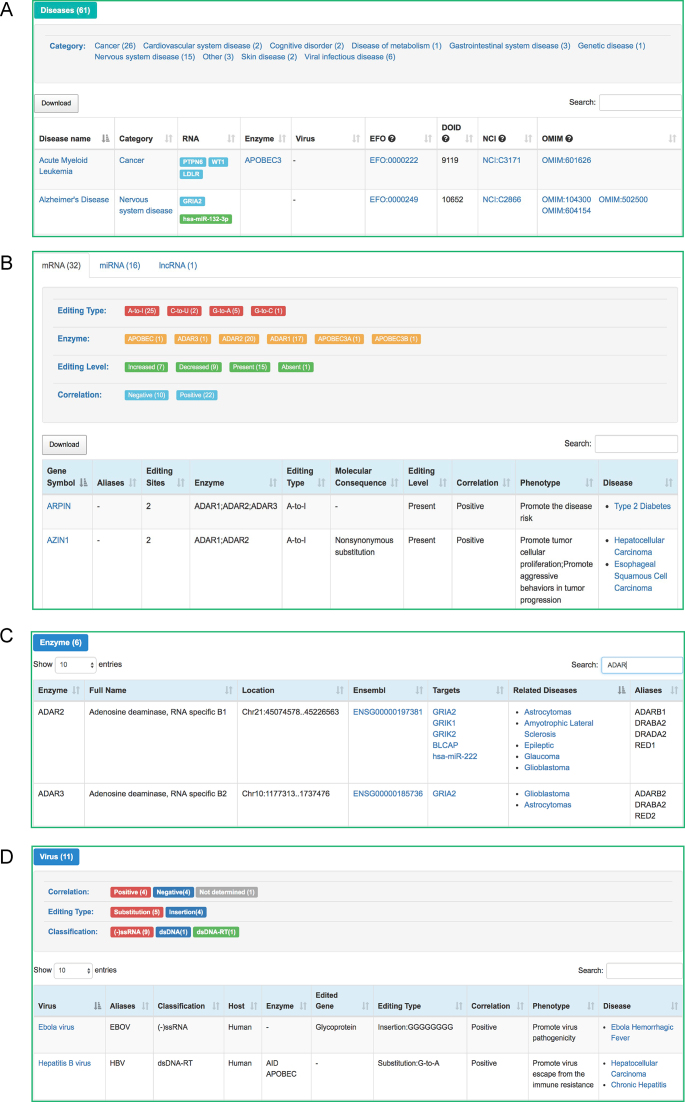
Figure 2.
Screenshots of EDK webpages, including browse pages of (A) editome-associated human diseases, (B) endogenous edited genes, (C) editing enzymes and (D) exogenous viruses.
To clarify the associations between aberrant edited genes and diseases, EDK categorizes disease-associated edited genes into 3 types, namely, mRNA, miRNA and lncRNA (http://bigd.big.ac.cn/edk/browse/RNA) (Figure 2B). To date, 153 aberrant editing sites in 32 mRNAs (AZIN1, CTSS, GRIA1, FLNB, BLCAP, etc.), 38 sites in 16 miRNAs (mir-214, miR-200b, miR-455-5p, etc.) and 1 site in 1 lncRNA (lncPCA3), are annotated to be associated with 46 human diseases. IPA analysis shows that the curated edited coding/noncoding genes and their target genes are significantly enriched in a number of networks and categories, which are relevant to cell-to-cell signalling and interaction, nervous system development, cell cycle and embryo development (http://bigd.big.ac.cn/edk/ipa). Regarding the mRNA type, enzyme families of ADARs (adenosine deaminase acting on RNA), APOBECs (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) and AID (activation-induced cytidine deaminase) are responsible for the characterized abnormal editing events (mainly A-to-I and C-to-U types). Most abnormal editing events, locating in the regions of CDS and 3′-UTR, induce several molecular consequences, such as nonsynonymous substitution, alternative splicing, mRNA stability change and mRNA expression change. One typical example is that the clustered RNA editing events within 3′-UTR region affect the stability of CTSS mRNA (a proinflammatory gene), which leads to cardiovascular disease (http://bigd.big.ac.cn/edk/mRNA?name=CTSS). Of note, both increased and decreased editing patterns can contribute to human diseases. For instance, decreased editing at 11 sites of CDC14B transcripts promotes malignant glioblastoma (http://bigd.big.ac.cn/edk/mRNA?name=CDC14B), and increased editing of AZIN1 (encoding antizyme inhibitor 1) promotes hepatocellular carcinoma (http://bigd.big.ac.cn/edk/mRNA?name=AZIN1). As to the miRNA type, both ADAR1 and ADAR2 are responsible for abnormal editing events, which may further affect diseases by retargeting miRNA or regulating the expression of miRNA itself or tumour-related target genes (34). One representative example is that hypo-editing within miR-455-5p promotes melanoma growth and metastasis by inhibiting the tumour suppressor gene CPEB1 (http://bigd.big.ac.cn/edk/miRNA/hsa-miR-455-5p). Moreover, it is noted that abnormal editing in long non-coding RNA has been identified to be associated with disease; there is only one example to date collected in EDK that the editing of the duplex of intronic long noncoding RNA lncPCA3 and pre-mRNA PRUNE2 augments the malignance of prostate cancer (35) (http://bigd.big.ac.cn/edk/lncRNA/PCA3).
To delineate the associations between aberrant editing enzymes and diseases, EDK collects 6 types of editing enzymes, including ADAR1, ADAR2 and ADAR3 in the ADAR family, APOBEC1 and APOBEC3 in the APOBEC family, and AID, which together associate with 27 diseases based on 44 independent experimental evidences (http://bigd.big.ac.cn/edk/browse/enzyme) (Figure 2C). As depicted on the enzyme category page, genetic mutations occurring in editing enzymes as well as aberrant expression of editing enzymes are both shown to be associated with human diseases. Notably, the enzyme-mediated mutagenesis is highly active in human cancers because abnormal editing may induce activation and/or inactivation of various tumour-related genes. For instance, over-expression of ADAR1 promotes ESCC tumour migration and invasion by increasing the editing level of AZIN1 and FLNB (36) (http://bigd.big.ac.cn/edk/enzyme/ADAR1). In addition to cancer, individual cases of other diseases related to aberrant editing enzyme are also curated and listed in EDK. One example is that mutation in ADAR1 is associated with the hereditary autoimmune diseases Aicardi–Goutieres syndrome (AGS) (20). However, it should be noted that not all editing enzyme aberrances affect diseases dependant on RNA editing mechanism, which means that editing enzymes may also participate in other cellular pathways to regulate physiological and pathological processes (37). For example, mutation in ADAR1 promotes HTLV-1 infection by the inhibition of PKR phosphorylation, which eventually causes anxiety disorder (38).
Additionally, RNA editing events occurring on exogenous viral dsRNA are also associated with some human diseases, especially the infectious diseases. In the current version, EDK also incorporates 13 editing events, residing in 11 edited viruses and associating with 9 diseases (among which 5 are infectious diseases) (http://bigd.big.ac.cn/edk/browse/virus) (Figure 2D). Based on the Baltimore classification, all collected viruses are classified into three types, namely, (–)ssRNA, dsDNA-RT and dsDNA virus. It is also found that ADARs, APOBECs and AID are responsible for the characterized editing events in these viruses. Different from the edited mRNA/miRNA/lncRNA, RNA editing events in virus includes not only substitution but also insertion of single or multiple nucleotides. Strikingly, most RNA editing events occur in viral structural genes, such as phosphoprotein gene and glycoproteins, which may change virus structure and affect virus function by recoding virus genes, inducing the generation of new transcripts or a new virus subclass, promoting or inhibiting virus replication, helping virus to escape from the immune resistance or inhibiting antiviral activity (37). For instance, editing in the phosphoprotein gene of measles virus may affect its pathogenicity (http://bigd.big.ac.cn/edk/virus/Measles%20virus).
DISCUSSION AND FUTURE DEVELOPMENTS
EDK is a comprehensive knowledgebase that provides the landscape of RNA editing events in a wide range of human diseases. Based on manual curation from more than 200 publications, the current implementation of EDK integrates 61 diseases that are associated with 248 experimentally validated abnormal editing events in 32 mRNAs, 16 miRNAs, 1 lncRNA and 11 viruses, as well as 44 aberrant enzyme activities involving with six editing enzymes. As a core resource of the BIG Data Center (http://bigd.big.ac.cn) (39,40), EDK is aimed at serving as an open access resource for dissecting the complex RNA editing mechanisms underlying human diseases. Considering the increasing volume of RNA editing studies in human diseases, we will frequently integrate experimentally validated RNA editome-disease associations based on literature curation from newly published literatures. Future directions of EDK also include development of community-curation interfaces to enable the scientific community in contributing their research data and/or expertise.
ACKNOWLEDGEMENTS
We thank a number of users for reporting bugs and providing suggestions as well as the anonymous reviewers for their valuable comments on this work.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDA19050302 and XDB13040500]; National Key Research & Development Program of China [2017YFC0907502]; National Programs for High Technology Research and Development [2015AA020108 and 2012AA020409]; International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008]; The Youth Innovation Promotion Association of Chinese Academy of Science [2018134]; 13th Five-year Informatization Plan of Chinese Academy of Sciences [XXH13505-05]; The 100-Talent Program of Chinese Academy of Sciences. Funding for open access charge: The Youth Innovation Promotion Association of Chinese Academy of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1. Schaefer M., Kapoor U., Jantsch M.F.. Understanding RNA modifications: the promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017; 7:170077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Farajollahi S., Maas S.. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010; 26:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xiong X., Yi C., Peng J.. Epitranscriptomics: toward a better understanding of RNA modifications. Genomics Proteomics Bioinformatics. 2017; 15:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramaswami G., Li J.B.. Identification of human RNA editing sites: a historical perspective. Methods. 2016; 107:42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M.J., Eterovic A.K., Yuan Y. et al. . The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015; 28:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Picardi E., Manzari C., Mastropasqua F., Aiello I., D’Erchia A.M., Pesole G.. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci. Rep. 2015; 5:14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zipeto M.A., Jiang Q., Melese E., Jamieson C.H.. RNA rewriting, recoding, and rewiring in human disease. Trends Mol. Med. 2015; 21:549–559. [DOI] [PubMed] [Google Scholar]
- 8. Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F.F., Jae N., Rossbach O., Amrhein C., Sigala F. et al. . Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016; 22:1140–1150. [DOI] [PubMed] [Google Scholar]
- 9. Deffit S.N., Hundley H.A.. To edit or not to edit: regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA. 2016; 7:113–127. [DOI] [PubMed] [Google Scholar]
- 10. Sommer B., Kohler M., Sprengel R., Seeburg P.H.. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991; 67:11–19. [DOI] [PubMed] [Google Scholar]
- 11. Shoshan E., Mobley A.K., Braeuer R.R., Kamiya T., Huang L., Vasquez M.E., Salameh A., Lee H.J., Kim S.J., Ivan C. et al. . Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015; 17:311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gommans W.M., Mullen S.P., Maas S.. RNA editing: a driving force for adaptive evolution. Bioessays. 2009; 31:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganem N.S., Ben-Asher N., Lamm A.T.. In cancer, A-to-I RNA editing can be the driver, the passenger, or the mechanic. Drug Resist. Update. 2017; 32:16–22. [DOI] [PubMed] [Google Scholar]
- 14. Gatsiou A., Vlachogiannis N., Lunella F.F., Sachse M., Stellos K.. Adenosine-to-Inosine RNA editing in health and disease. Antioxid. Redox Signal. 2017; 29:846–863. [DOI] [PubMed] [Google Scholar]
- 15. Dominissini D., Moshitch-Moshkovitz S., Amariglio N., Rechavi G.. Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis. 2011; 32:1569–1577. [DOI] [PubMed] [Google Scholar]
- 16. Maas S., Kawahara Y., Tamburro K.M., Nishikura K.. A-to-I RNA editing and human disease. RNA Biol. 2006; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Slotkin W., Nishikura K.. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013; 5:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamashita T., Akamatsu M., Kwak S.. Altered intracellular milieu of ADAR2-Deficient motor neurons in amyotrophic lateral sclerosis. Genes. 2017; 8:E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rayon-Estrada V., Papavasiliou F.N., Harjanto D.. RNA editing dynamically rewrites the cancer code. Trends Cancer. 2015; 1:211–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rice G.I., Kasher P.R., Forte G.M., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A. et al. . Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 2012; 44:1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bankamp B., Lopareva E.N., Kremer J.R., Tian Y., Clemens M.S., Patel R., Fowlkes A.L., Kessler J.R., Muller C.P., Bellini W.J. et al. . Genetic variability and mRNA editing frequencies of the phosphoprotein genes of wild-type measles viruses. Virus Res. 2008; 135:298–306. [DOI] [PubMed] [Google Scholar]
- 22. Yeo J., Goodman R.A., Schirle N.T., David S.S., Beal P.A.. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. NatI. Acad. Sci. U.S.A. 2010; 107:20715–20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanoue A., Koshimizu T.A., Tsuchiya M., Ishii K., Osawa M., Saeki M., Tsujimoto G.. Two novel transcripts for human endothelin B receptor produced by RNA editing/alternative splicing from a single gene. J. Biol. Chem. 2002; 277:33205–33212. [DOI] [PubMed] [Google Scholar]
- 24. Picardi E., D’Erchia A.M., Lo Giudice C., Pesole G.. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017; 45:D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He T., Du P., Li Y.. dbRES: a web-oriented database for annotated RNA editing sites. Nucleic Acids Res. 2007; 35:D141–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiran A.M., O’Mahony J.J., Sanjeev K., Baranov P.V.. Darned in 2013: inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 2013; 41:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramaswami G., Li J.B.. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gong J., Liu C., Liu W., Xiang Y., Diao L., Guo A.Y., Han L.. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res. 2017; 45:D79–D84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018; 46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J., Pruitt K.D., Sayers E.W.. GenBank. Nucleic Acids Res. 2018; 46:D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jomini V., Oppliger-Pasquali S., Wietlisbach V., Rodondi N., Jotterand V., Paccaud F., Darioli R., Nicod P., Mooser V.. Contribution of major cardiovascular risk factors to familial premature coronary artery disease: the GENECARD project. J. Am. Coll. Cardiol. 2002; 40:676–684. [DOI] [PubMed] [Google Scholar]
- 32. Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J.. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006; 34:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keen J.C., Moore H.M.. The Genotype-Tissue expression (GTEx) Project: Linking clinical data with molecular analysis to advance personalized medicine. J. Pers. Med. 2015; 5:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y.M., Liang H.. When MicroRNAs meet RNA editing in cancer: a nucleotide change can make a difference. Bioessays. 2018; 40:doi:10.1002/bies.201700188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salameh A., Lee A.K., Cardo-Vila M., Nunes D.N., Efstathiou E., Staquicini F.I., Dobroff A.S., Marchio S., Navone N.M., Hosoya H. et al. . PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. NatI. Acad. Sci. U.S.A. 2015; 112:8403–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan T.H., Lin C.H., Qi L., Fei J., Li Y., Yong K.J., Liu M., Song Y., Chow R.K., Ng V.H. et al. . A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014; 63:832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song C., Sakurai M., Shiromoto Y., Nishikura K.. Functions of the RNA editing enzyme ADAR1 and their relevance to human diseases. Genes. 2016; 7:E129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cachat A., Alais S., Chevalier S.A., Journo C., Fusil F., Dutartre H., Boniface A., Ko N.L., Gessain A., Cosset F.L. et al. . ADAR1 enhances HTLV-1 and HTLV-2 replication through inhibition of PKR activity. Retrovirology. 2014; 11:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. BIG Data Center Members The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017; 45:D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. BIG Data Center Members Database Resources of the BIG Data Center in 2018. Nucleic Acids Res. 2018; 46:D14–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]