Abstract
The Plant Promoter Analysis Navigator (PlantPAN; http://PlantPAN.itps.ncku.edu.tw/) is an effective resource for predicting regulatory elements and reconstructing transcriptional regulatory networks for plant genes. In this release (PlantPAN 3.0), 17 230 TFs were collected from 78 plant species. To explore regulatory landscapes, genomic locations of TFBSs have been captured from 662 public ChIP-seq samples using standard data processing. A total of 1 233 999 regulatory linkages were identified from 99 regulatory factors (TFs, histones and other DNA-binding proteins) and their target genes across seven species. Additionally, this new version added 2449 matrices extracted from ChIP-seq peaks for cis-regulatory element prediction. In addition to integrated ChIP-seq data, four major improvements were provided for more comprehensive information of TF binding events, including (i) 1107 experimentally verified TF matrices from the literature, (ii) gene regulation network comparison between two species, (iii) 3D structures of TFs and TF-DNA complexes and (iv) condition-specific co-expression networks of TFs and their target genes extended to four species. The PlantPAN 3.0 can not only be efficiently used to investigate critical cis- and trans-regulatory elements in plant promoters, but also to reconstruct high-confidence relationships among TF–targets under specific conditions.
INTRODUCTION
Regulatory factors contain transcription factors (TFs), modified histones, and other DNA-binding proteins that affect gene transcriptional activity and chromatin remodeling due to interactions with their target DNA sequences. Recognizing transcription factor binding sites (TFBSs), and actual regulatory regions of modified histones has been one of the most important problems in functional genomics. In the last few decades, high-throughput techniques, such as chromatin immunoprecipitation sequencing (ChIP-seq), DNase sequencing (DNase-seq) and DNA affinity purification sequencing (DAP-seq), have been carried out to reveal the genomic binding landscapes of regulatory factors (1–3). Specifically, these techniques can help scientists reveal the regulations occurring in a specific biological process or under condition of interest. Although high-throughput sequencing data have grown exponentially in the public domain, the diverse data processed methods and file formats cause low utility of these big data. Thus, there is a strong need to set up an integrative resource to interpret complicated transcriptional regulatory networks from various datasets. To date, several databases have been developed to explore the occupancy landscapes of regulatory factors, such as, Cistrome DB (4), ReMap (5), Factorbook (6), GTRD (7), PlantTFDB (8), ChIPBase (9) and Expresso (10). However, most of such resources only support mammalian and Drosophila research, and a limited number were designed for plants. Among them, PlantTFDB, ChIPBase and Expresso focus on a restricted number of ChIP-seq data, and only support Arabidopsis thaliana (8–10). Furthermore, these systems do not provide customized sequence analysis capabilities, depending on their experimental DNA binding matrices. In this update version (PlantPAN 3.0), the genomic regulatory sites of each genes will be supplied from experimental sequencing data. Additionally, the experimental matrices will be utilized in plant promoter sequence analysis.
TFs regulate cell processes by binding a specific DNA motif on promoter regions and affecting downstream gene expression. However, the recognized mechanisms between TF and DNA remain unclear. To address this question, high-resolution crystal structures of DNA-binding domains have been generated to interpret protein–DNA recognition (11,12). In addition, the tertiary structures refine the functional features of proteins, such as dimerization, protein-protein interaction sites, and transcriptional activation sites in an effector domain (13). The Protein Data Bank (PDB) serves as a searchable platform for the 3D structures of proteins, nucleic acids, and multiprotein complexes (14,15). Besides tertiary structures, several primary and secondary structural features have also been used to infer protein functions. For example, variants located in pivotal amino acids or domains might confer deleterious effects on protein functions (16), and functional domains might suggest evolutionary relationships in protein families and DNA-binding preferences (13,17). The ePlant provides useful function to identify DNA binding domains and non-synonymous SNPs in 3D structures (18). Therefore, it is worth integrating these features in public TF repositories to interpret the DNA-binding structure and biological function of TFs.
PlantPAN is aimed toward reconstructing transcriptional regulatory networks and providing actual cis-regulatory elements for plant genes. This study reports the first introduction of genomic binding events from 421 Chip-seq datasets for 99 regulatory factors across seven plant species into the PlantPAN system. In addition to TFBSs, ChIP-seq occupancy of histone modifications and DNA-binding proteins, such as the chromatin remodeling complex protein, have been added to illustrate the transcriptional activity of genomic landscapes. In newly constructed PCBase resource, users can not only access the ChIP-seq results of interest via four functions: Gene Search, Protein Search, Genome Browse, and Promoter Analysis, but also download the processed result files for further analysis. With the incorporation of gene annotation and the promoter sequence in seven plant species, PlantPAN 3.0 offers an efficient platform on which to identify important regulatory elements (binding sites of regulatory factors, CpG islands, tandem repeats, and conserved regions) of user queries. In addition, by adding protein sequence annotation and structural basis features, PlantPAN 3.0 is expected to help users explore the critical interaction motif among TFs and binding DNA molecules. An overview and several handpicked results of PlantPAN 3.0 are shown in Supplementary Figure S1.
DATA CONTENT AND WEB INTERFACE
ChIP-seq data collection
The plant ChIP-seq data were collected systematically from Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) (19,20). Only the dataset containing a pair of a ChIP-seq experimental sample and an input sample for a regulatory factor were collected. The datasets were manually checked using the following three criteria: (i) methods for the ChIP-seq experiments, (ii) available raw data (FASTQ or SRA formats) and (iii) comprehensive description of the experimental purpose for each ChIP-seq sample. Datasets lacking any of the above information were filtered out. In total, 421 datasets (662 samples) were used to reveal regulatory relationships for 99 regulatory factors across seven species including A. thaliana, Oryza sativa, Zea mays, Glycine max, Solanum lycopersicum, Gossypium hirsutum and A. lyrata (Table 1).
Table 1.
The ChIP-seq data statistics from seven species
| Species | Regulatory factors | Datasets | samples | Binding sites | Binding relationships |
|---|---|---|---|---|---|
| Arabidopsis thaliana | 82 | 355 | 535 | 3 456 486 | 966 251 |
| Oryza sativa | 1 | 1 | 3 | 2019 | 1480 |
| Zea mays | 6 | 35 | 71 | 31 436 | 13 623 |
| Glycine max | 4 | 20 | 38 | 798 340 | 163 698 |
| Solanum lycopersicum | 1 | 1 | 2 | 1274 | 66 |
| Gossypium hirsutum | 1 | 2 | 4 | 117 768 | 44 830 |
| Arabidopsis lyrata | 4 | 7 | 9 | 167 014 | 44 051 |
| Total | 99 | 421 | 662 | 4 574 337 | 1 233 999 |
ChIP-seq data processing and motif discovery
All datasets were systematically processed according to the following procedures: Raw data in SRA (.sra) format were converted to FASTQ (.fastq) by using SRA toolkit (version 2.8.2-1, http://www.ncbi.nlm.nih.gov/Traces/sra). The FASTX-Toolkit (version 0.0.13, http://hannonlab.cshl.edu/fastx_toolkit/) and cutadapt (version 1.16) (21) were used to remove low quality reads (reads ≧Q30 and reads up to 30 bp) and adapters from single-end and paired-end datasets, respectively. Bowtie (version 1.2.2) was used to align reads to the reference genome (22). The reference genomes used in this study are shown in Supplementary Table S1. SAMtools (version 1.4) was used to sort and cut the reads labelled ‘reads unmapped’, ‘not primary alignment’ and ‘reads are PCR or optical duplicates’ as FLAGs in Bowtie results (23). The duplicate reads were discarded using Picard (version 2.18.5, http://broadinstitute.github.io/picard/). For peak calling, SPP (version 1.14) in AQUAS pipeline and MACS2 (version 2.1.0) programs were applied to identify the protein binding sites for single-end and paired-end datasets, respectively (24–26). Consequently, a total of 4 574 337 protein binding sites were obtained (Table 1). De novo motif discovery of each protein was performed using MEME-ChIP in MEME SUITE (version 4.12.0) (27). The top three position-specific probability matrices from two motif discovery algorithms, MEME and DREME, were then used to scan the binding sites on any input promoter sequences. Totally, 2449 PWMs (position weight matrices) and 2459 motif logos were obtained.
Regulatory linkage construction
The genome locations of regulatory factor binding sites were required to construct the regulatory relationship between a regulatory factor and a target gene. For target gene recognition, all genomic binding sites from the ChIP-seq datasets were overlapped with the potential regulatory regions of all genes on their chromosome coordinates. The potential regulatory protein-coding gene regions were defined as promoter (upstream 2000 bp from transcription start sites (TSS)), 5’UTR, exon, intron, 3’UTR and downstream regions (downstream 350 bp from transcription stop sites for genes without 3’UTR). In three species (A. thaliana, Z. mays and A. lyrata), the regulatory relationships among regulatory factor and non-coding RNAs (ncRNAs) were also considered. Finally, 1,233,999 regulatory linkages were identified among the target genes (including protein coding genes and ncRNAs) and 99 regulatory factors.
Integrative information of regulatory factors
The regulatory factors collected from ChIP-seq datasets can be divided into TFs, histones, and other DNA-binding proteins. Proteins not found in PlantPAN 2.0, PlantTFDB and HistoneDB 2.0 were classified into other DNA-binding proteins (8,28,29). The detailed information about the regulatory factors was retrieved from the UniProt database (30). The additional histone modification descriptions were extracted from HistoneDB 2.0 (28).
Structure-based and protein sequence-based annotation of TFs
The secondary and tertiary structures of protein functions are significant features. Specifically, the local sequence-specific binding structures may contribute to the binding specificity of TFs. Thus, in this release, the PlantPAN provides protein–DNA complex tertiary structures and protein sequence-based annotation. The protein tertiary structures were retrieved from the PDB through the ID mapping files from UniProt (14,15,30). Due to the limited amount of structural data in PDB, the tertiary structures of homologous proteins were also collected to facilitate making a homology model and to compare structural models. The homologs of TFs in other species were congregated from the HomoloGene Database (https://www.ncbi.nlm.nih.gov/homologene) and InParanoid, (31). The JSmol applet was applied to visualize the tertiary structures. ProtVista was used to provide a graphical representation of protein sequence annotation (such as functional domains, secondary structures, post-translational modification and variants, etc.) (32). In PlantPAN 3.0, users can access these advanced functional features via the TF/TFBS Search function.
Prediction of TFBSs in promoter sequences
Since genomic binding sites are good clues to capture the appearance of conserved TF binding variations, PWMs created from high-throughput experiments have become vital for TFBS prediction. By incorporating 1100 PWMs from three studies and 2449 PWMs from ChIP-seq datasets, a total of 4703 PWMs for 17 230 regulatory factors were collected in PlantPAN 3.0 (1,3,33). The putative TFBS predictions in a given promoter sequence was implemented using the Match™ program (34). The procedure for creating the cut-off profiles is described in our previous paper (29).
New PCBase function for identifying protein–DNA regulatory relationships derived from ChIP-seq experiments
To facilitate user access to ChIP-seq data, a new portal ChIP-seq search, called PCBase was developed in PlantPAN 3.0. Two main functions, ‘Gene Search’ and ‘Protein Search’, were designed for the most frequent queries regarding transcriptional regulation.
To investigate the transcriptional regulatory networks and histone modification of target genes, PlantPAN 3.0 provides six types of potential regulatory regions (i.e. upstream, 5’UTR, exon/ncRNA, intron, 3’UTR, and downstream). In the ‘Gene Search’ mode of PCBase, users can input their genes of interest to explore which regulatory factors are located on the potential regulatory regions. Moreover, users can choose a regulatory factor and a serious of datasets to obtain graphical diagrams of genomic binding locations by using the D3.js JavaScript library (https://d3js.org).
In the ‘Protein Search’, users can search their datasets of interest by browsing proteins for a specific species. The result page provides (i) detailed information from each dataset, including name and type of regulatory factor, tissue, and experimental treatment, (ii) motif logos generated from MEME or DREME, (iii) Target Browse (iv) Binding Proportion, (v) Peak Browse and (vi) a downloadable table of the processed results. The ‘Protein Search’ output interfaces are summarized in Figure 1. The Target Browse function allows users to retrieve all target genes through filter options such as dataset ID, replicate, chromosome, and potential regulatory region (Figure 1B). To characterize the preferred potential regulatory region of regulatory factors, the Binding Proportion function shows the distribution of all binding events relying on six types of potential regulatory regions (Figure 1C). Furthermore, Peak Browse helps users restrict their genomic regions of interest, and each binding sites with gene structures can be visualized in the JBrowse genome browser (Figure 1E) (35). To assist users with conducting further analysis, PlantPAN 3.0 compiles the locations and sequences of peak-calling results into BED and FASTA formats, respectively (Figure 1F).
Figure 1.
The output interfaces of ‘Protein Search’ in PCBase in PlantPAN 3.0. (A) After users select a species and a regulatory factor (marked in red boxes), detailed information for the selected dataset (right) is displayed. The result page also provides (B) a searchable table for Target Browse, where user can click ‘Visualize’ to identify the location of a binding site, (C) binding proportion, (D) motif logos, (E) Peak Browse for a dataset and (F) tables to download processed files and link with external databases.
Enhancement of the existing functions
For a cis-regulatory element search in PlantPAN, the input can be either a transcript locus or a group of genes. Four new plant species, G. max, S. lycopersicum, G. hirsutum and A. lyrata, were added (Supplementary Table S1) (36–39). Gene annotations for A. thaliana and Z. mays were updated to Araport11 and AGV.4 version (37,40). For Z. mays, the gene IDs of AGV.3 were converted to AGV.4 by using geneIDhistory files obtained from Gramene (41). Furthermore, the expression profiles of 58 samples were imported from SoyBase to illustrate co-expression networks among TFs and target genes in G. max (Supplementary Table S2) (42).
APPLICATION AND DISCUSSION
Extended usage of ChIP-data in TFBS predictions
Determining how to reduce false positive in TFBS predictions remains a difficult task in bioinformatic methods. Additionally, inferring promoter activity under different conditions or for different plant tissues is a major challenge to the study of the transcriptional regulation of genes. To handle these problems, CpG islands, tandem repeats, conserved regions, and co-expression profiles were used to identify the actual TFs/TFBSs in PlantPAN 2.0 (29). In this release, PlantPAN 3.0 provides a significant improvement in terms of elucidating the transcriptional regulation of genes by integrating experimental binding sites for TFs and other DNA-binding proteins, as well as histone modification marks from ChIP-seq data. In the proposed PlantPAN 3.0, the predicted and experimental TFBSs were shown in Jbrowse, which enables a straightforward comparison of different regulatory tracks on any genomic regions.
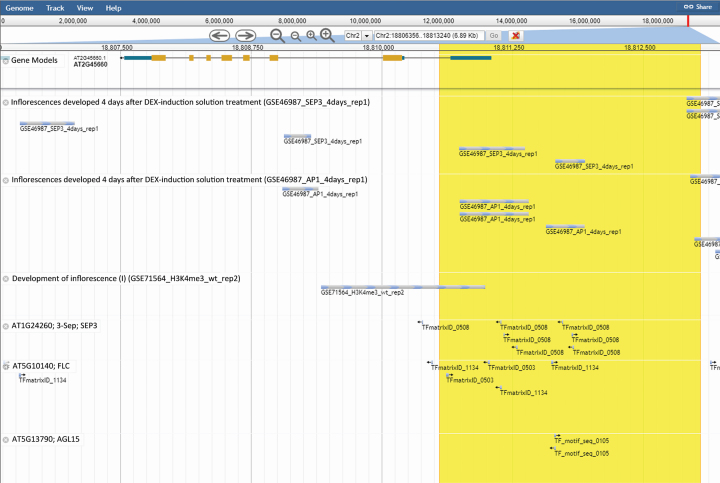
Here, a case study of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) is given below to demonstrate applications of PlantPAN 3.0. SOC1 is known to regulate flowering time in A. thaliana. A previous study suggests that APETALA1 (AP1) and SEPALLATA 3 (SEP3) may play an important role as regulators to modify the gene expression of SOC1 and change chromatin accessibility (43). Based on the analysis in PCBase function, the results show that AP1 and SEP3 occupied the promoter and intron of SOC1 in inflorescences (Figure 2), which is consistent with pervious findings. Interestingly, the H3K4me3 was found to locate in adjacent TFBSs and TSS regions of SOC1. This may imply that the activation of SOC1 gene expression is related to co-occupancy of TFs and H3K4me3 in the early developmental stage of the flower. In addition, based on the results of the TFBS prediction via PWM similarities, six binding sites of 11bps core motifs (CCAAAAA[AT]GGA) for SEP3 were found to overlap with the peak signals in the ChIP-seq experiments. On the other hand, several TFs that have been studied to regulate SOC1, such as Flowering Locus C (FLC) and AGAMOUS‐like 15 (AGL15), also can be observed in the upstream regions of SOC1 (44,45). This case reveals the comparability of the ChIP-seq dataset results and the predicted TFBSs in PlantPAN 3.0. By integrating multiple features of promoters, PlantPAN 3.0 is expected to help users reconstruct complex transcriptional regulatory genes networks and to decrease the false positives in TFBS predictions.
Figure 2.
The binding sites for SEP3, AP1, H3K4me3, FLC, and AGL15 across SOC1 gene (AT2G45660) in the Jbrowse viewer. The upstream region (+500 to -2000; Chr2:18813047–18810548) of SOC1 is highlighted with a yellow background. The experimental binding sites from ChIP-seq are shown in the first three tracks. The last three lines are the predicted TFBSs via PWM patterns.
Construction of transcription regulatory networks across different species
Isolation of homologs of an already-known gene has been widely used in tracking evolutionary conservation in gene regulations and characterizing necessary changes during genetic divergence (46,47). To assist users in comparing the gene regulation between homologous genes, PlantPAN 3.0 offers an effective ‘Cross Species’ function for detecting TFBSs in conserved regions of promoters. Furthermore, a transcriptional regulatory network in a model plant can be easily referred to understand the mechanism in several important crops, G. max, S. lycopersicum, G. hirsutum and Z. mays.
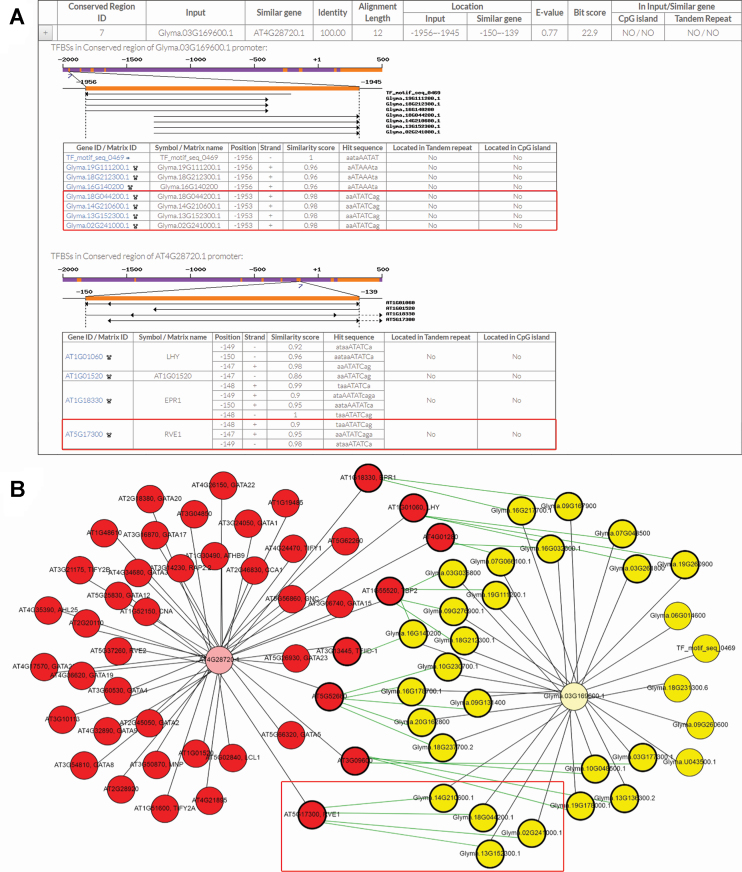
In several plant species, circadian rhythms have been reported to be essential in the regulation of plant growth (48,49). For example, circadian-regulated MYB-like transcription factor, REVEILLE 1 (RVE1, AT5G17300) is able to regulate hypocotyl growth by increasing IAA concentrations through activation of the auxin biosynthesis‐related genes YUCCA8 (YUC8, AT4G28720) (50). Based on these findings, the cross species analysis between A. thaliana and G. max was performed to illustrate the usage of PlantPAN 3.0. In ‘Cross Species’ function, eleven conserved regions were identified in promoters of YUC8 and its homolog (Glyma.03G169600) (Supplementary Figure S2). As expected, the TFBS prediction results show that YUC8 promoter harbors RVE1 binding sites within seventh conserved regions (Figure 3A). In G. max, the predicted binding sites of four homeodomain-like superfamily proteins (Glyma.18G044200, Glyma.14G210600, Glyma.13G15230 and Glyma.02G241000) were found in the corresponding seventh conserved regions of Glyma.03G169600 (Figure 3A). Expectedly, there are homologous relationships among RVE1 and four homeodomain-like superfamily proteins of G. max (Figure 3B). The transcriptional regulatory network of YUC8 and Glyma.03G169600 displays similar transcriptional regulations between A. thaliana and G. max (Figure 3B). These results might imply that the circadian regulation of auxin biosynthesis is functionally conserved in G. max through auxin biosynthesis‐related genes and homeodomain-like superfamily proteins. Accordingly, other A. thaliana homeodomain-like superfamily proteins, such as LATE ELONGATED HYPOCOTYL (LHY, AT1G01060) and EARLY PHYTOCHROME RESPONSIVE 1 (EPR1, AT1G18330) also show the homologous relationships with G. max TFs. These TFs might be the new candidates involved in the regulation of circadian rhythms in both A. thaliana and G. max. This case reveals that PlantPAN 3.0 can helps users compare both similarities and differences in transcriptional regulatory networks between homologous genes across different plant species.
Figure 3.
Conserved transcription regulation between YUC8 and its homolog (Glyma.03G169600). (A) The partial TFBS prediction results in the seventh conserved regions, where RVE1 and four homeodomain-like superfamily proteins were marked in red. (B) The transcriptional regulatory network of YUC8 (pink node) and Glyma.03G169600 (light yellow node). Red and yellow nodes represent predicted TFs from A. thaliana and G. max, respectively. Green line were used to link homologous TFs.
Significance and utility of PlantPAN3.0
The utility and comparisons of PlantPAN 3.0 with other similar resources are illustrated in Table 2.
Table 2.
A comparison of PlantPAN 3.0 with previous version and similar resources
| PlantPAN 3.0 | PlantPAN 2.0a | PlantTFDB 4.0b | ChIPBase v2.0c | Expressod | ReMap 2018e | PCSDf | |
|---|---|---|---|---|---|---|---|
| Number of species in this databases | 78 | 76 | 165 | 10 | 1 | 1 | 3 |
| Number of TFs | 17 230 | 16 960 | 320 370 | 26 | 20 | NAg | 46 |
| Number of TF matrices | 4 703 | 1143 | 674 | NAg (∼6 200)h | 0 | 0 | 0 |
| Number of plant species in ChIP-seq datasets | 7 | 0 | 2 | 1 | 1 | 0 | 3 |
| Number of regulatory factors in ChIP-seq datasets | 99 | 0 | 14 | 29 (1414)h | 20 | 0 (485)h | 110 |
| Number of ChIP-seq samples | 662 | 0 | NA | NA | NA | NA | NA |
| Number of ChIP-seq datasets | 421 | 0 | 26 | 54 i (10 216)h | 20 | 0 (2 829)h | 303 |
| Annotation of Target genes | Yes | Yes | No | Yes | Yes | No | Yes |
| Histon/Nuclesome Binding regions | Yes | No | Yes | Yes | No | Yes | Yes |
| Genome Browse for binding regions | Yes | No | Yes | Yes i | No | Yes | Yes |
| Uniform ChIP-seq data processing | Yes | No | Yes | No | No | Yes | Yes |
| Download whole genomic binding peaks (bed/bigwig files) | Yes | No | No | No | No | Yes | Yes |
| Comprehensive curation of TF information (i.e. functional domain, response conditions, target genes, activator or repressor, and sequence logos of binding motifs) | Yes (increase secondary and 3D structures, PTM, and variants) | Yes | Yes | No | No | No | No |
| Co-expression profiles of TFs and their target genes | Yes | Yes | No | Yes | Yes | No | No |
| Cis-regulatory element prediction | Yes | Yes | Yes | No | No | No | No |
aPlantPAN 2.0: http://PlantPAN2.itps.ncku.edu.tw/ (29).
bPlantTFDB 4.0: http://planttfdb.cbi.pku.edu.cn/ (8).
cChIPBase v2.0: http://rna.sysu.edu.cn/chipbase/ (9).
dExpresso: http://bioinformatics.cs.vt.edu/expresso/ (10).
eReMap 2018: http://remap.cisreg.eu (5).
gEach TF/matrices can be accessed from the database separately. However, the total number of TFs/matrices cannot be calculated via the resource.
hThe number of data for plant species is shown without brackets, whereas the total number of data for plant and non-plant species is indicated within brackets.
iRecently, this function/resource has not been available on the website (http://rna.sysu.edu.cn/chipbase/).
Based on the rapid accumulation of ChIP-seq data, several public web-based resources were developed. For example, ReMap currently increased their ChIP-seq collection and provided an exhaustive review of regulatory maps (5). Unfortunately, ReMap only supports humans. For plant species, there are several databases devoted to collecting plant ChIP-seq data and identifying their gene regulation mechanisms. For example, Expresso was created to identify the regulatory relationships among 20 A. thaliana TFs and their target genes from public ChIP-seq data (10). PlantTFDB 4.0 is a useful TF repository for green plants, providing various information leading to an understanding of the functions of TFs as well as genome-wide regulatory maps for 14 TFs (8). However, their collection in ChIP-seq experiments has only included for A. thaliana, and there is a lacks of annotation of target genes. Although ChIPBase v2.0 provided A. thaliana ChIP-seq data and co-expression profiles for TFs and target genes, the ChIP-seq data were not processed consistently (9). Moreover, recently, the function for many species has not been available on the website (http://rna.sysu.edu.cn/chipbase/). Furthermore, PCSD is dedicated to recognizing the chromatin states of A. thaliana, O. sativa and Z. mays based on both public and in-house epigenomic data (51). However, this resource did not provide the specific condition or the tissue where the regulation or chromatin state was observed.
Compared among these resources and PlantPAN 3.0, the distinctive advantages of PlantPAN 3.0 are listed as follows: (i) PlantPAN 3.0 collected comprehensive public ChIP-seq datasets, which cover 421 datasets for 99 regulatory factors across seven plant species. (ii) The collected ChIP-seq datasets were processed systematically, and all analysis results are downloadable. (iii) The detailed information for each dataset and functional annotation of both TFs and target genes are available. (iv) The most complete plant PWMs are provided for analyzing TFBSs in a promoter or a set of promoters. Additionally, users can graphically compare predicted TFBSs with the experimental binding sites of regulatory factors and other important regulatory elements (CpG islands, tandem repeats, and conserved regions). (v) PlantPAN 3.0 also can be used to elucidate similarities among the expression profiles of TFs and target genes under various environmental stresses, hormone treatments, or developmental stages across four species. In summary, PlantPAN 3.0 facilitates an understanding of complicated transcriptional regulatory networks in plants.
DATA AVAILABILITY
The PlantPAN 3.0 is available via a web interface and is freely to all interested user, at http://PlantPAN.itps.ncku.edu.tw/.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the Ministry of Science and Technology (MOST 105-2311-B-006-004-MY3) and Academia Sinica (Innovative Translational Agricultural Research Grant) of the Republic of China for financially supporting this research. Computational analyses, data mining and storage resources were performed using the system provided by the Bioinformatics Core at the National Cheng Kung University and National Center for High-performance Computing (NCHC) of National Applied Research Laboratories (NARLabs), supported by Ministry of Science and Technology, Taiwan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [MOST 105-2311-B-006-004-MY3]; Academia Sinica (Innovative Translational Agricultural Research Grant). Funding for open access charge: Ministry of Science and Technology [MOST 105-2311-B-006-004-MY3].
Conflict of interest statement. None declared.
REFERENCES
- 1. O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R.. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016; 165:1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K. et al. . Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sullivan A.M., Arsovski A.A., Lempe J., Bubb K.L., Weirauch M.T., Sabo P.J., Sandstrom R., Thurman R.E., Neph S., Reynolds A.P. et al. . Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014; 8:2015–2030. [DOI] [PubMed] [Google Scholar]
- 4. Mei S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L. et al. . Cistrome data browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017; 45:D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheneby J., Gheorghe M., Artufel M., Mathelier A., Ballester B.. ReMap 2018: An updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res. 2018; 46:D267–D275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang J., Zhuang J., Iyer S., Lin X.Y., Greven M.C., Kim B.H., Moore J., Pierce B.G., Dong X., Virgil D. et al. . Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013; 41:D171–D176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yevshin I., Sharipov R., Valeev T., Kel A., Kolpakov F.. GTRD: A database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2017; 45:D61–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin J., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J., Gao G.. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017; 45:D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou K.R., Liu S., Sun W.J., Zheng L.L., Zhou H., Yang J.H., Qu L.H.. ChIPBase v2.0: Decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017; 45:D43–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aghamirzaie D., Raja Velmurugan K., Wu S., Altarawy D., Heath L.S., Grene R.. Expresso: A database and web server for exploring the interaction of transcription factors and their target genes in Arabidopsis thaliana using ChIP-Seq peak data [version 1; referees: 2 approved, 1 approved with reservations]. F1000Research. 2017; 6:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boer D.R., Freire-Rios A., van den Berg W.A., Saaki T., Manfield I.W., Kepinski S., Lopez-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R. et al. . Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014; 156:577–589. [DOI] [PubMed] [Google Scholar]
- 12. Wilson K.A., Kellie J.L., Wetmore S.D.. DNA-protein pi-interactions in nature: abundance, structure, composition and strength of contacts between aromatic amino acids and DNA nucleobases or deoxyribose sugar. Nucleic Acids Res. 2014; 42:6726–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olsen A.N., Ernst H.A., Leggio L.L., Skriver K.. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005; 10:79–87. [DOI] [PubMed] [Google Scholar]
- 14. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rose P.W., Prlic A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z. et al. . The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taketa S., Amano S., Tsujino Y., Sato T., Saisho D., Kakeda K., Nomura M., Suzuki T., Matsumoto T., Sato K. et al. . Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:4062–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lehti-Shiu M.D., Panchy N., Wang P., Uygun S., Shiu S.H.. Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochim. Biophys. Acta. 2017; 1860:3–20. [DOI] [PubMed] [Google Scholar]
- 18. Waese J., Fan J., Pasha A., Yu H., Fucile G., Shi R., Cumming M., Kelley L.A., Sternberg M.J., Krishnakumar V. et al. . ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell. 2017; 29:1806–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edgar R., Domrachev M., Lash A.E.. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kodama Y., Shumway M., Leinonen R. International Nucleotide Sequence Database Collaboration . The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011; 17:10. [Google Scholar]
- 22. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing Subgroup . The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kharchenko P.V., Tolstorukov M.Y., Park P.J.. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008; 26:1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koh P.W., Pierson E., Kundaje A.. Denoising genome-wide histone ChIP-seq with convolutional neural networks. Bioinformatics. 2017; 33:i225–i233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Machanick P., Bailey T.L.. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Draizen E.J., Shaytan A.K., Marino-Ramirez L., Talbert P.B., Landsman D., Panchenko A.R.. HistoneDB 2.0: A histone database with variants–an integrated resource to explore histones and their variants. Database (Oxford). 2016; 2016:baw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chow C.N., Zheng H.Q., Wu N.Y., Chien C.H., Huang H.D., Lee T.Y., Chiang-Hsieh Y.F., Hou P.F., Yang T.Y., Chang W.C.. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016; 44:D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sonnhammer E.L., Ostlund G.. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015; 43:D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Watkins X., Garcia L.J., Pundir S., Martin M.J., UniProt C.. ProtVista: visualization of protein sequence annotations. Bioinformatics. 2017; 33:2040–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hume M.A., Barrera L.A., Gisselbrecht S.S., Bulyk M.L.. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein–DNA interactions. Nucleic Acids Res. 2015; 43:D117–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E.. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003; 31:3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu J., Jung S., Cheng C.H., Ficklin S.P., Lee T., Zheng P., Jones D., Percy R.G., Main D.. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014; 42:D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kersey P.J., Allen J.E., Allot A., Barba M., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Grabmueller C. et al. . Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018; 46:D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al. . Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandez-Pozo N., Menda N., Edwards J.D., Saha S., Tecle I.Y., Strickler S.R., Bombarely A., Fisher-York T., Pujar A., Foerster H. et al. . The Sol Genomics Network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res. 2015; 43:D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E.. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis. 2015; 53:474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tello-Ruiz M.K., Naithani S., Stein J.C., Gupta P., Campbell M., Olson A., Wei S., Preece J., Geniza M.J., Jiao Y. et al. . Gramene 2018: unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res. 2018; 46:D1181–D1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grant D., Nelson R.T., Cannon S.B., Shoemaker R.C.. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010; 38:D843–D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pajoro A., Madrigal P., Muino J.M., Matus J.T., Jin J., Mecchia M.A., Debernardi J.M., Palatnik J.F., Balazadeh S., Arif M. et al. . Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014; 15:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Immink R.G., Pose D., Ferrario S., Ott F., Kaufmann K., Valentim F.L., de Folter S., van der Wal F., van Dijk A.D., Schmid M. et al. . Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 2012; 160:433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Andres F., Coupland G.. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012; 13:627–639. [DOI] [PubMed] [Google Scholar]
- 46. Silva P.A., Silva J.C., Caetano H.D., Machado J.P., Mendes G.C., Reis P.A., Brustolini O.J., Dal-Bianco M., Fontes E.P.. Comprehensive analysis of the endoplasmic reticulum stress response in the soybean genome: Conserved and plant-specific features. BMC Genomics. 2015; 16:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou Q.Y., Tian A.G., Zou H.F., Xie Z.M., Lei G., Huang J., Wang C.M., Wang H.W., Zhang J.S., Chen S.Y.. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008; 6:486–503. [DOI] [PubMed] [Google Scholar]
- 48. Covington M.F., Harmer S.L.. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007; 5:e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Atamian H.S., Harmer S.L.. Circadian regulation of hormone signaling and plant physiology. Plant Mol. Biol. 2016; 91:691–702. [DOI] [PubMed] [Google Scholar]
- 50. Rawat R., Schwartz J., Jones M.A., Sairanen I., Cheng Y., Andersson C.R., Zhao Y., Ljung K., Harmer S.L.. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:16883–16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu Y., Tian T., Zhang K., You Q., Yan H., Zhao N., Yi X., Xu W., Su Z.. PCSD: A plant chromatin state database. Nucleic Acids Res. 2018; 46:D1157–D1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PlantPAN 3.0 is available via a web interface and is freely to all interested user, at http://PlantPAN.itps.ncku.edu.tw/.