Abstract
PIWI-interacting RNAs are a class of small RNAs that is most abundantly expressed in animal germline. Substantial research is going on to reveal the functions of piRNAs in the epigenetic and post-transcriptional regulation of transposons and genes. To collect and annotate these data, we developed piRBase, a database assisting piRNA functional study. Since its launch in 2014, piRBase has integrated 264 data sets from 21 organisms, and the number of collected piRNAs has reached 173 million. The latest piRBase release (v2.0, 2018) was more focused on the comprehensive annotation of piRNA sequences, as well as the increasing number of piRNAs. In addition, piRBase release v2.0 also contained the potential information of piRNA targets and disease related piRNA. All datasets in piRBase is free to access, and available for browse, search and bulk downloads at http://www.regulatoryrna.org/database/piRNA/.
INTRODUCTION
Up to now, various small RNAs can be classified into three types: microRNA (miRNA), endogenous small interfering RNA (siRNA), and PIWI-interacting RNA (piRNA), based on their biogenesis and associated proteins (1). piRNAs are mainly expressed in mammalian germline (2–5), and are generally 24–31 nucleotides in length with 2′-O-methylation at their 3′ ends in most animals (6–8). Compared with miRNAs and siRNAs, piRNAs are much more diverse in sequences. PIWI proteins, which are associated with piRNAs, are also enriched in germline (9–11).
piRNAs play various functions in germline and somatic tissues. PIWI/piRNA complex can repress the activity of transposon, which has a high risk of damaging the genome, through post-transcriptional silencing or heterochromatin formation (6,12–20). However, many piRNAs do not match transposon sequences, which implies additional functions of piRNAs (21–23). Recent studies have found that piRNAs can target mRNA by base pairing in mouse (24,25), Drosophila (26,27) and Caenorhabditis elegans (28–32). In silkworm (33) and C. elegans (31), piRNAs are reported to be involved in sex determination through down-regulation of target genes. Besides transposon and mRNA, a large number of lncRNAs are revealed to be mediated by retrotransposon derived piRNAs in mouse late spermatocytes (34).
As piRNAs are implicated in transposon and gene regulation, there has been a budding interest in defining the role of piRNAs in human disease (35,36). piRNAs are garnering more and more attention in a variety of cancers. For example, 106 piRNAs are up-regulated and 91 are down-regulated in bladder cancer (37). The abnormal expression of piRNAs are also demonstrated in other types of cancers such as breast cancer (38,39) and gastric cancer (36,40,41).
In light of the rapidly increasing studies on piRNA, several piRNA related databases, such as piRNABank (42), piRNAQuest (43), piRNA cluster database (44) and IsopiRBank (45), are generated. However, these databases only contain limited amounts of piRNAs from limited species, and the information of piRNA functions are rarely included. piRBase is the expert database about piRNA included by RNAcentral (46) and is the first database that systematically integrates various piRNA associated data to support piRNA functional analysis.
In the latest piRBase release, the number of unique piRNA sequences increased to 173 million, including 21 species. In addition, the piRNA target mRNA records were expanded and potential piRNA target lncRNAs were added in piRBase release v2.0. The information about eight types of cancers (breast, bladder, pancreas, gastric, liver, kidney, myeloma and colorectal cancer) related piRNAs was also added to the new version. We also provided new web tools and improved user interface in piRBase release v2.0.
DATA COLLECTION AND PROCESSING
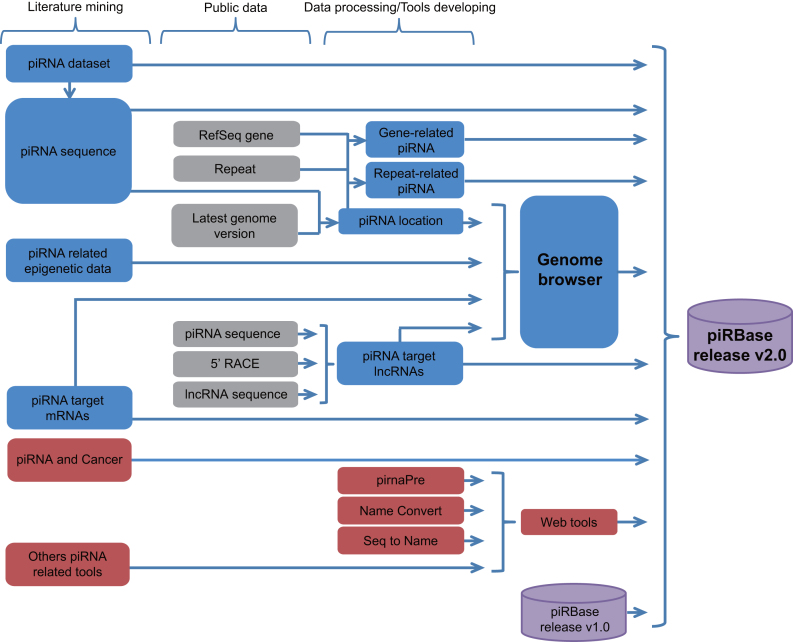
Based on the first release of piRBase database (47), new datasets and processed piRNA sequences from the literature, supplementary files, and NCBI GEO were collected. We only included sequences that were taken as piRNA or piRNA candidates in corresponding papers. Other piRNA related information was manually extracted from the relevant articles. The contents in piRBase release v2.0 are shown in Figure 1.
Figure 1.
The contents in piRBase release v2.0. piRNA loci, piRNA target sites (mRNAs and lncRNAs) and piRNA related epigenetic data are visualized by UCSC genome browser. Blue boxes: the main updated modules; Red boxes: new modules.
Each piRNA was shown with a piRBase name and other related information such as NCBI accession number, organism of origin, sequence, length, the number of papers reporting the piRNA and the number of different methods supporting the piRNA. In the detailed information page, we also provided additional information about the piRNA, such as aliases (NCBI and RNAdb piRNA name), related datasets with tissue and method, genome location, targets and associated cancer. In piRBase, if a piRNA sequence is a subsequence of another piRNA, both of them were considered as different sequences and were assigned distinct piRBase names.
As described in the first release of piRBase, all piRNA sequences were mapped to its latest genome using Bowtie (48) with parameter ‘-v 1 -a --best --strata’ in order to obtain the potential origin of every piRNA. piRNAs were referred to as gene- or repeat-related according to the overlapping of piRNA genome loci with RefSeq genes (49) or repeat elements.
DATABASE CONTENT
piRBase aims to provide comprehensive piRNA sequence data, annotation and targets assisting piRNA study. The first release in 2014 contains 77 million piRNA sequences from nine organisms. piRNAs in piRBase are marked as repeat- and gene-related according to their loci in genome, which imply involvement in the regulation of the corresponding elements. In additional, reported piRNA targets and piRNA related epigenetic data were collected. The information of piRNA loci, piRNA target sites, piRNA related epigenetic data, and some basic annotations like RepeatMasker annotations (50) and RefSeq genes are visualized in the UCSC genome browser (50,51) in piRBase release v1.0. Since piRBase release in 2014, we collected more piRNA related data and released in v2.0.
New piRNA records
In present release of piRBase, the number of unique piRNA sequences reaches 173 million, including 264 datasets from 21 species. The number is greatly increased in this version compared with the first release (47) and other piRNA related databases (Table 1). In order to assess credibility of piRNAs in piRBase, we also offered two evaluation score. One score is the number of papers which detected certain piRNA, and the other score is the number of different methods used to detect the sequence. These methods including protein IP, protein CLIP, chromatography, oxidization and small RNA-Seq.
Table 1.
The comparison of piRBase and other piRNA databases
| piRBase | |||||
|---|---|---|---|---|---|
| Species | piRNABank | piRNAQuest | IsopiRBankc | Release v1.0 | Release v2.0 |
| Human | 35 356 | 41 749 | 4 564 080 | 32 826 | 8 438 265 |
| Mouse | 55 359 | 890 078 | 38 093 003 | 51 664 769 | 68 054 594 |
| Rat | 46 444 | 66 758 | 63 182 | 4 081 625 | |
| D. melanogaster | 44 417a | 15 058 093 | 21 027 419 | 41 950 613 | |
| C. elegans | 28 219 | 28 219 | |||
| Zebrafish | 6 367 811 | 1 330 692 | 1 330 692 | ||
| Chicken | 508 437 | 1 716 795 | |||
| X. tropicalis | 6 142 904 | 6 142 904 | |||
| Silkworm | 1 174 963 | 1 253 987 | |||
| Starlet sea anemone | 11 811 | ||||
| Cow | 19 528 252 | ||||
| Crab-eating macaque | 5 819 946 | ||||
| Rhesus | 6 514 | ||||
| Marmoset | 8 311 210 | ||||
| Sea hare | 499 | ||||
| Tree shrew | 65 810 | ||||
| Pig | 1 301 866 | ||||
| D.erecta | 1 091 318 | ||||
| D.yakuba | 1 069 918 | ||||
| D.virilis | 907 289 | ||||
| Rabbit | 2 816 864 | ||||
| Platypus | 51b | ||||
aThe number of unique piRNAs may be less than shown.
bThe number of unique piRNAs comes from statistic of download file.
cThe number of canonical piRNAs is shown.
piRNA targets
Many studies have found that piRNAs can regulate the expression of mRNAs and lncRNAs. In the previous version, mRNAs regulated by piRNAs in fly and mouse were included. In this version, new mRNA targets (for silkworm and C. elegans) extracted from the literature were added. Moreover, we predicted piRNA target lncRNAs using the same way as predicting piRNA target mRNA cleavage (25) in mouse testis. In the web page of piRNA target table, we provided the piRBase name linking to detailed information of piRNA, target gene/transcript ID, target site if provided, and other related information.
piRNA and cancer
From piRNA and cancer related literature, we manually collected cancer related piRNAs. In the piRBase release v2.0, eight types of cancers (breast, bladder, pancreas, gastric, liver, kidney, myeloma and colorectal cancer) related piRNAs were included with the information of piRNA name, piRNA expression (up-regulated, down-regulated and fold change) and functional description in cancer tissues or cell lines.
New tools
piRBase release v2.0 added ‘Tools’ menu in navigation bar. We incorporated pirnaPre (52) for users to predict piRNA targets. In addition, some online tools were integrated under the ‘Tools’ menu. ‘Name Convert’ enabled users to convert a list of NCBI or RNAdb piRNA names and accession numbers to piRBase names. The users can also retrieve piRBase names by a list of piRNA sequences using ‘Seq To Name’. Other existing tools such as ‘Reverse Complement’ and ‘Bowtie Online’ tool are also incorporated in the ‘Tools’ menu. Beyond that, we also included the links of others tools for predicting piRNA-targeting sites or identifying piRNAs, such as pirScan (53) and 2L-piRNA (54).
Visualization using UCSC genome browser
In the current piRBase, the latest genome versions such as hg38 and mm10 were added in the UCSC genome browser. Many of the information in the present piRBase release v2.0, such as piRNA loci, piRNA target sites (mRNAs and lncRNAs) and piRNA related epigenetic data, have be visualized using UCSC genome browser. Moreover, we have added about 400 tracks of all related data from current version of piRBase in UCSC genome browser and users can choose to display content they are interested in.
Interface improvement
The web interface of the present piRBase database has been improved for better browse and search experience. The piRNA targets and related cancers are added in piRNA detailed information page. In the piRNA target page, users can browse and search the information by selecting organism, gene symbol and transcript ID. In piRNA search page, two more search options are added in piRNA sequence search. ‘SubStr’ can search all sub-sequences of input sequence. ‘Extend 3′ can search 3′ extended piRNAs.
On the basis of download page in the first release, new version data were included. We provided all piRBase data in both release v1.0 and v2.0 for downloading. In addition, users can also download the related data through the ‘Export’ button in corresponding pages.
CONCLUSION
piRBase is the first database that systematically integrates various types of data to support piRNA functional analysis. Compared with the first release in 2014, piRBase release v2.0 is more comprehensive. The total numbers of unique piRNAs and species have been increased. Besides piRNA target mRNAs, the current piRBase also included target lncRNAs. In light of piRNAs as potential biomarkers in cancers, the information of piRNA and cancer was added to the current release. More piRNA related epigenetic data were also added in piRBase UCSC genome browser. In addition, piRBase release v2.0 added new web tools and improved the user interface. As piRNA studies expanded rapidly, we will update piRBase with more related information supporting piRNA functional analysis.
ACKNOWLEDGEMENTS
Data analysis and computing resource was supported by Center for Big Data Research in Health (http://bigdata.ibp.ac.cn), Institute of Biophysics, Chinese Academy of Sciences.
FUNDING
National Key R&D Program of China [2016YFC0901702]; National Natural Science Foundation of China [31871294]. Funding for open access charge: National Key R&D Program of China [2016YFC0901702].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim V.N., Han J., Siomi M.C.. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009; 10:126–139. [DOI] [PubMed] [Google Scholar]
- 2. Girard A., Sachidanandam R., Hannon G.J., Carmell M.A.. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006; 442:199–202. [DOI] [PubMed] [Google Scholar]
- 3. Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T. et al. . A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006; 442:203–207. [DOI] [PubMed] [Google Scholar]
- 4. Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E.. Characterization of the piRNA complex from rat testes. Science. 2006; 313:363–367. [DOI] [PubMed] [Google Scholar]
- 5. Grivna S.T., Beyret E., Wang Z., Lin H.. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006; 20:1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D.. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006; 313:320–324. [DOI] [PubMed] [Google Scholar]
- 7. Kirino Y., Mourelatos Z.. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007; 14:347–348. [DOI] [PubMed] [Google Scholar]
- 8. Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T.. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007; 14:349–350. [DOI] [PubMed] [Google Scholar]
- 9. Deng W., Lin H.. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002; 2:819–830. [DOI] [PubMed] [Google Scholar]
- 10. Kuramochi-Miyagawa S., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W. et al. . Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004; 131:839–849. [DOI] [PubMed] [Google Scholar]
- 11. Carmell M.A., Girard A., van de Kant H.J., Bourc’his D., Bestor T.H., de Rooij D.G., Hannon G.J.. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007; 12:503–514. [DOI] [PubMed] [Google Scholar]
- 12. Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J.. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007; 128:1089–1103. [DOI] [PubMed] [Google Scholar]
- 13. Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C.. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007; 315:1587–1590. [DOI] [PubMed] [Google Scholar]
- 14. Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B. et al. . A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007; 129:69–82. [DOI] [PubMed] [Google Scholar]
- 15. Carmell M.A., Xuan Z., Zhang M.Q., Hannon G.J.. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002; 16:2733–2742. [DOI] [PubMed] [Google Scholar]
- 16. Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., Hannon G.J.. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009; 137:522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C.. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006; 20:2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yin H., Lin H.. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007; 450:304–308. [DOI] [PubMed] [Google Scholar]
- 19. Aravin A.A., Sachidanandam R., Bourc’his D., Schaefer C., Pezic D., Toth K.F., Bestor T., Hannon G.J.. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008; 31:785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim A.K., Tao L., Kai T.. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 2009; 186:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Batista P.J., Ruby J.G., Claycomb J.M., Chiang R., Fahlgren N., Kasschau K.D., Chaves D.A., Gu W., Vasale J.J., Duan S. et al. . PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 2008; 31:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das P.P., Bagijn M.P., Goldstein L.D., Woolford J.R., Lehrbach N.J., Sapetschnig A., Buhecha H.R., Gilchrist M.J., Howe K.L., Stark R. et al. . Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell. 2008; 31:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aravin A.A., Hannon G.J., Brennecke J.. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007; 318:761–764. [DOI] [PubMed] [Google Scholar]
- 24. Gou L.T., Dai P., Yang J.H., Xue Y., Hu Y.P., Zhou Y., Kang J.Y., Wang X., Li H., Hua M.M. et al. . Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014; 24:680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang P., Kang J.Y., Gou L.T., Wang J., Xue Y., Skogerboe G., Dai P., Huang D.W., Chen R., Fu X.D. et al. . MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015; 25:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rouget C., Papin C., Boureux A., Meunier A.C., Franco B., Robine N., Lai E.C., Pelisson A., Simonelig M.. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010; 467:1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vourekas A., Alexiou P., Vrettos N., Maragkakis M., Mourelatos Z.. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature. 2016; 531:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee H.C., Gu W., Shirayama M., Youngman E., Conte D. Jr., Mello C.C.. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012; 150:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bagijn M.P., Goldstein L.D., Sapetschnig A., Weick E.M., Bouasker S., Lehrbach N.J., Simard M.J., Miska E.A.. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012; 337:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen E.Z., Chen H., Ozturk A.R., Tu S., Shirayama M., Tang W., Ding Y.H., Dai S.Y., Weng Z., Mello C.C.. Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans Germline. Cell. 2018; 172:937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang W., Seth M., Tu S., Shen E.Z., Li Q., Shirayama M., Weng Z., Mello C.C.. A sex chromosome piRNA promotes robust dosage compensation and sex determination in C. elegans. Dev. Cell. 2018; 44:762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang D., Tu S., Stubna M., Wu W.S., Huang W.C., Weng Z., Lee H.C.. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 2018; 359:587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kiuchi T., Koga H., Kawamoto M., Shoji K., Sakai H., Arai Y., Ishihara G., Kawaoka S., Sugano S., Shimada T. et al. . A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature. 2014; 509:633–636. [DOI] [PubMed] [Google Scholar]
- 34. Watanabe T., Cheng E.C., Zhong M., Lin H.. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015; 25:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu Y., Li C., Zhang K., Sun H., Tao D., Liu Y., Zhang S., Ma Y.. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 2010; 43:635–641. [DOI] [PubMed] [Google Scholar]
- 36. Cheng J., Guo J.M., Xiao B.X., Miao Y., Jiang Z., Zhou H., Li Q.N.. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta. 2011; 412:1621–1625. [DOI] [PubMed] [Google Scholar]
- 37. Chu H., Hui G., Yuan L., Shi D., Wang Y., Du M., Zhong D., Ma L., Tong N., Qin C. et al. . Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015; 356:561–567. [DOI] [PubMed] [Google Scholar]
- 38. Huang G., Hu H., Xue X., Shen S., Gao E., Guo G., Shen X., Zhang X.. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin. Transl. Oncol. 2013; 15:563–568. [DOI] [PubMed] [Google Scholar]
- 39. Zhang H., Ren Y., Xu H., Pang D., Duan C., Liu C.. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg. Oncol. 2013; 22:217–223. [DOI] [PubMed] [Google Scholar]
- 40. Martinez V.D., Enfield K.S., Rowbotham D.A., Lam W.L.. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric Cancer. 2016; 19:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng J., Deng H., Xiao B., Zhou H., Zhou F., Shen Z., Guo J.. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012; 315:12–17. [DOI] [PubMed] [Google Scholar]
- 42. Sai Lakshmi S., Agrawal S.. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008; 36:D173–D177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarkar A., Maji R.K., Saha S., Ghosh Z.. piRNAQuest: searching the piRNAome for silencers. BMC Genomics. 2014; 15:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosenkranz D. piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res. 2016; 44:D223–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang H., Ali A., Gao J., Ban R., Jiang X., Zhang Y., Shi Q.. IsopiRBank: a research resource for tracking piRNA isoforms. Database (Oxford). 2018; 2018:doi:10.1093/database/bay059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. The, R.C. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017; 45:D128–D134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang P., Si X., Skogerbo G., Wang J., Cui D., Li Y., Sun X., Liu L., Sun B., Chen R. et al. . piRBase: a web resource assisting piRNA functional study. Database (Oxford). 2014; 2014:bau110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. . Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D. et al. . The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018; 46:D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yuan J., Zhang P., Cui Y., Wang J., Skogerbo G., Huang D.W., Chen R., He S.. Computational identification of piRNA targets on mouse mRNAs. Bioinformatics. 2015; 32:1170–1177. [DOI] [PubMed] [Google Scholar]
- 53. Wu W.S., Huang W.C., Brown J.S., Zhang D., Song X., Chen H., Tu S., Weng Z., Lee H.C.. pirScan: a webserver to predict piRNA targeting sites and to avoid transgene silencing in C. elegans. Nucleic Acids Res. 2018; 46:W43–W48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu B., Yang F., Chou K.C.. 2L-piRNA: a two-layer ensemble classifier for identifying Piwi-Interacting RNAs and their function. Mol Ther Nucleic Acids. 2017; 7:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]