Abstract
Temporal and spatial protein phosphorylation dynamically orchestrates a broad spectrum of biological processes and plays various physiological and pathological roles in diseases and cancers. Recent advancements in high-throughput proteomics techniques greatly promoted the profiling and quantification of phosphoproteome. However, although several comprehensive databases have reserved the phosphorylated proteins and sites, a resource for phosphorylation quantification still remains to be constructed. In this study, we developed the qPhos (http://qphos.cancerbio.info) database to integrate and host the data on phosphorylation dynamics. A total of 3 537 533 quantification events for 199 071 non-redundant phosphorylation sites on 18 402 proteins under 484 conditions were collected through exhaustive curation of published literature. The experimental details, including sample materials, conditions and methods, were recorded. Various annotations, such as protein sequence and structure properties, potential upstream kinases and their inhibitors, were systematically integrated and carefully organized to present details about the quantified phosphorylation sites. Various browse and search functions were implemented for the user-defined filtering of samples, conditions and proteins. Furthermore, the qKinAct service was developed to dissect the kinase activity profile from user-submitted quantitative phosphoproteome data through annotating the kinase activity-related phosphorylation sites. Taken together, the qPhos database provides a comprehensive resource for protein phosphorylation dynamics to facilitate related investigations.
INTRODUCTION
As one of the most important post-translational modifications (PTMs), protein phosphorylation is involved in almost all biological processes and plays physiological and pathological roles in diseases and cancers (1–3). Phosphorylation is reversibly catalysed by kinases and phosphatases, which dynamically control the phosphorylation dynamics based on a temporal and spatial context (4,5). In 1992, Edmond H. Fischer and Edwin G. Krebs shared the Nobel Prize in Physiology or Medicine for their discovery of reversible protein phosphorylation as a biological regulatory mechanism (6), while the prize in 2001 was awarded to Leland H. Hartwell, Tim Hunt and Paul M. Nurse for identifying key regulators, including cyclin-dependent kinases (CDKs) and cyclins, which accurately orchestrate the cell cycle through phosphorylation (7). In recent decades, many studies have dissected the molecular mechanisms and biological functions of phosphorylation dynamics, and kinases and phosphatases are popular research areas for the development of target therapies (5,8).
Recently, the advancement of high-throughput proteomics techniques greatly promoted the profiling and quantification of the phosphoproteome in various cells and tissues under different conditions (9,10). For example, based on phosphoproteome quantification, Wojcechowskyj et al. dissected the molecular mechanism of cellular reprogramming during HIV-1 entry (11), and van den Biggelaar et al. unveiled the dynamic phosphorylation of thrombin signalling in human primary endothelial cells (12). Since dynamic phosphorylation events could provide helpful clues for kinase-mediated signalling, Ho et al. identified the activation of PI3K/AKT/mTORC1 signalling for cell survival by the ELABELA peptide in human embryonic stem cells during heart development (13), Bai et al. profiled the driver tyrosine kinases in sarcoma (14) and Casado et al. inferred the aberrant kinase activation in leukaemia cells through the quantitative phosphoproteome-based computational analyses (15). Thus, the quantitative phosphoproteomics data could provide great help in understanding phosphorylation-controlled biological processes.
Previously, several pioneering studies have contributed to constructing resources to host phosphorylation-related data. The currently available databases contain many human protein phosphorylation data. UniProt (16) is the most important infrastructure for protein annotations, which contain massive experimentally identified phosphorylation sites and annotations in the Human Protein Reference Database (HPRD) (17) also contain protein phosphorylation information. Databases including dbPTM (18), PhosphoSitePlus (19), SysPTM (20), PHOSIDA (21), dbPAF (22), Phospho.ELM (23) and PhosphoPep (24) curate and host experimentally identified phosphorylation sites from the published literature, and iPTMnet (25) contains information on the phosphorylation regulatory network and conservation. Furthermore, the PhosSNP (27) and ActiveDriverDB (28) databases analyse the genetic variations that influence phosphorylation, and the PTMcode (26) database provides the functional associations of phosphorylation sites. With the continuous improvement of high-throughput phosphoproteome techniques and the rapid increase of phosphoproteome datasets, ProteomeScout (29) and ProteomicsDB (30) were developed to store proteome and PTM proteome datasets and provide online analysis tools. However, although the ProteomeScout database contains protein quantification information, the quantification of phosphorylation events is still missing. Taken together, although databases for various aspects of protein phosphorylation are available, the database for the quantification of phosphorylation is still absent. Since the quantification/stoichiometry/dynamics of phosphorylation are critical for the molecular mechanisms under different temporal and spatial contexts, a comprehensive resource for protein phosphorylation quantification could facilitate the reuse of published quantitative phosphoproteome datasets and provide great help for phosphorylation-related studies.
In this study, we developed the qPhos database to host the quantitative phosphoproteome data generated in Homo sapiens. In total, 3 537 533 quantification events for 199 071 non-redundant phosphorylation sites on 18 402 proteins under 484 conditions were curated and integrated into the database, coupled with various annotations such as sequence and structure properties, potential upstream kinases and their inhibitors. For convenient usage, browse and search functions were implemented and the information in the database was carefully organized. Furthermore, based on curated activity-related phosphorylation sites, the qKinAct service was developed to dissect the kinase activity profile from phosphoproteome data. Thus, qPhos could serve as a one-stop database to investigate phosphorylation dynamics in H. sapiens.
CONSTRUCTION AND CONTENT
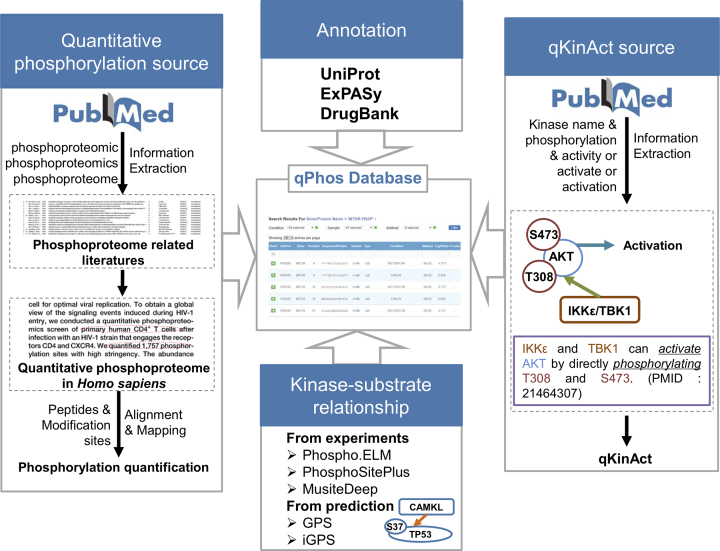
To establish the resource for protein phosphorylation dynamics, the published quantitative phosphoproteomics datasets were collected from the literature and annotations from various resource were integrated (Figure 1). Keywords including ‘phosphoproteomic’, ‘phosphoproteomics’ and ‘phosphoproteome’ were used to search PubMed to retrieve the phosphoproteome-related literature. Based on the criteria of high-throughput mass spectrometry-based site-level protein phosphorylation quantification, the published datasets of quantitative phosphoproteome datasets in H. sapiens were manually curated. Besides the quantified phosphorylation sites, details about the quantification including the experimental condition, phosphopeptide enrichment method, mass spectrometry and raw peptide were collected. All quantified phosphopeptides and modified residues were mapped to the H. sapiens reference proteome sequences downloaded from UniProt database (Release 2018_04) (Figure 1) (16). Due to the differences among the miscellaneous reference proteome datasets used in these literature, about 4.31% of raw phosphopeptides could not be mapped to the UniProt reference human proteome, and these limited unmapped data was dropped. In addition, the annotations from databases such as UniProt (16), ExPASy (31) and DrugBank (Release 2018_07) (32) were integrated into the qPhos database to provide comprehensive information for phosphorylation events (Figure 1). The experimental verified kinase–substrate relationships were integrated from Phospho.ELM (23), PhosphoSitePlus (19) and MusiteDeep (33), and potential relationship were predicted by sequence-based predictor GPS (34) and network-based predictor iGPS (35) with a high threshold (Figure 1). Furthermore, the activity-related phosphorylation sites in kinases were also curated from the literature. The human kinases and their names were retrieved from the EKPD database (36), and coupled with keywords including ‘phosphorylation’ and ‘activity or activate or activation’ to search PubMed for experimental evidence of activity-related phosphorylation (Figure 1).
Figure 1.
The schema for the construction of the qPhos database.
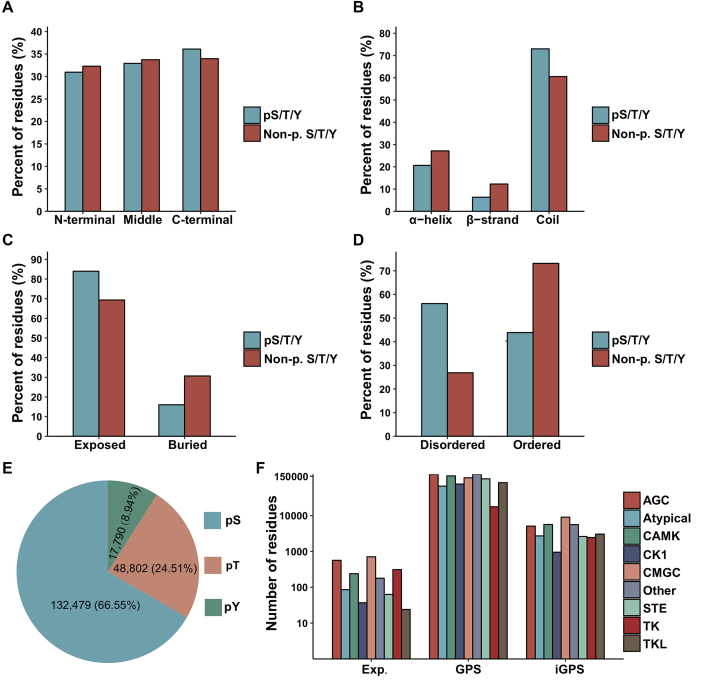
In total, 3 537 533 quantification events for 199 071 non-redundant phosphorylation sites on 18 402 proteins under 484 conditions were collected from 190 published studies. The primary references for the data were provided to ensure their quality and repeatability, and various annotations were integrated into the database. The sequence and structure preferences of the quantified phosphorylation sites were presented in Figure 2A–D. It seemed that the phosphorylation sites were generally equally distributed among the protein sequences but slightly enriched in C-terminal areas (E-ratio = 1.06, P-value < 10−17) (Figure 2A). Through computational annotation of secondary structures, surface accessibility and disorder region by NetSurfP (37) and IUPred (38), it was observed that the phosphorylation sites were enriched in the coil region (E-ratio = 1.21, P-value < 10−18) (Figure 2B), exposed region (E-ratio = 1.21, P-value < 10−18) (Figure 2C) and disordered region (E-ratio = 2.09, P-value < 10−18) (Figure 2D). As shown in Figure 2E, the distribution of phosphorylated serine, threonine and tyrosine residues was consistent with previous studies (19,22). Furthermore, the potential upstream kinases were annotated for these phosphorylation sites by various resources. The experimentally identified kinase-site regulatory relations were retrieved from previous studies, including Phospho.ELM (23), PhosphoSitePlus (19) and MusiteDeep (33). Additionally, the potential site-specific kinase-substrate relationships were predicted by sequence-based predictor GPS (34) and network-based predictor iGPS (35) with a high threshold. The annotations of kinases were summarized at the family level in Figure 2F.
Figure 2.
Summary of the sequence and structure preferences and kinase families of the quantified phosphorylation sites, including the summary of the position along the protein sequence (A), secondary structure (B), surface accessibility (C), disorder region (D), serine/threonine/tyrosine (E) and regulator kinase family (F) for the quantified phosphorylation sites.
USAGE
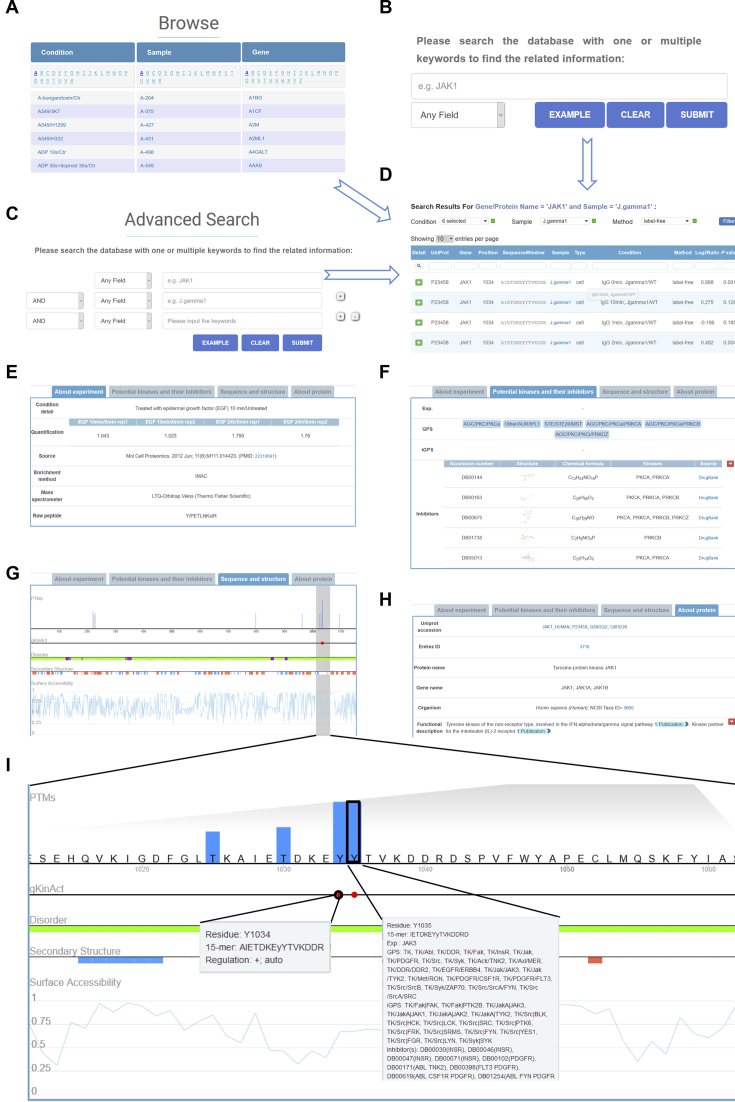
The qPhos database was developed for scientists to quickly access the quantitative phosphoproteome data in a user-friendly manner. For convenient usage, qPhos provided browse and search functions to query the database. Three browse options, including condition, sample and gene, were provided to browse the database by selecting the item from the list (Figure 3A). The experimental conditions, samples including cell lines and tissues, and gene symbols were sorted and organized in alphabetical order, which enabled the users to quickly choose the interesting data (Figure 3A). Simple and complex search functions were implemented at the home (Figure 3B) and search (Figure 3C) pages, respectively, which provided keyword-based queries in protein and gene names, protein functions and descriptions of conditions and samples. Furthermore, the retrieved phosphorylation sites from the browse or search functions could be further filtered by conditions, samples and methods in the results page (Figure 3D).
Figure 3.
The detailed information in qPhos. (A) Browse function. (B) Simple search function. (C) Advanced search function. (D) The returned search results. (E) The information about the protein. (F) The information about the quantification of the phosphorylation site. (G) The information on potential kinases and their inhibitors for the quantified phosphorylation site. (H) The sequence and structure properties of the phosphorylation site. (I) The enlarged view of the sequence and structure properties.
In the results page, the information was organized by the quantification events in a tabular format with UniProt accession, gene name, position, sequence window, sample information, sample type, experimental condition, quantification method, log2-transformed ratio and P-value (Figure 3D). Users could click the plus button to view the detailed information about the protein and phosphorylation sites. The detailed information had four sections, including ‘About experiment’, ‘Potential kinases and their inhibitors’, ‘Sequence and structure’ and ‘About protein’ (Figure 3E–I). The detailed description of the condition, raw quantifications, source reference, experimental method and instrument and raw peptide were shown in the ‘About Experiment’ section (Figure 3E). ‘Potential kinases and their inhibitors’ showed the experimental identified and predicted upstream kinases for the phosphorylation site (Figure 3F). Furthermore, the inhibitors annotated by DrugBank for the kinases were shown (Figure 3F). The sequence and structure properties of the protein were visualized in the ‘Sequence and Structure’ section, which included the quantified phosphorylation sites, activity-related phosphorylation sites if the protein was a kinase, disorder region, secondary structure and surface accessibility (Figure 3G). A magnifier was implemented to show the details by enlarging the selected region (Figure 3G). Furthermore, users could access descriptions about visualization by hovering over the content (Figure 3I). The ‘About Protein’ section presented the protein information including database accessions, protein/gene name/alias, functional descriptions, PTMs and sequences adopted from the UniProt database (Figure 3H).
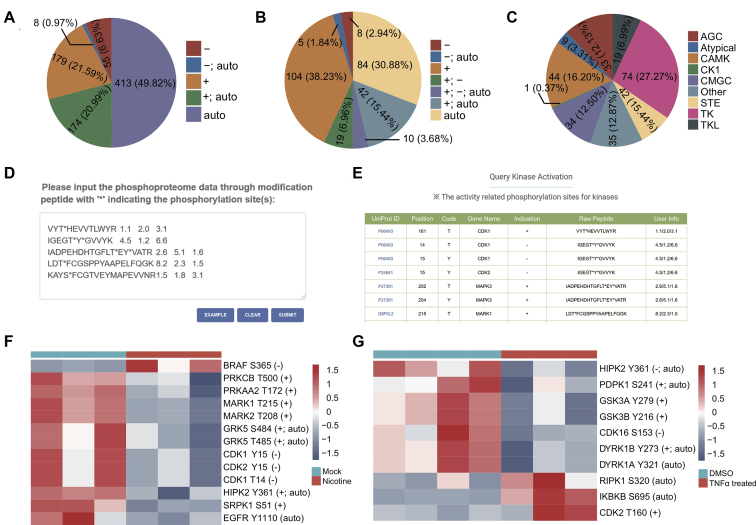
Since autophosphorylation or phosphorylation of the specified segment could activate or inactivate the kinases (39), the identification and quantification of these activity-related phosphorylation sites could indicate the activation status of these kinases. Through exhaustive curation of literature, we collected the experimentally identified activity-related phosphorylation sites for the human kinome. In total, 829 activity-related phosphorylation sites were curated in 272 kinases (Figure 4A), which covered over half of the kinome. Among the activity-related phosphorylation sites, nearly half were autophosphorylation sites. Most autophosphorylation sites could activate the kinases, while seven autophosphorylation sites could inactivate the kinases. Furthermore, most activity-related phosphorylation sites were positively related to the activities of the kinase, while only a small fraction were negatively related (Figure 4A). The distributions of activity-related phosphorylation sites among kinases and kinase families were summarized in Figure 4B and C, respectively. The qKinAct service was developed to query the quantification events of activity-related phosphorylation sites to annotate the kinase activity profile from quantitative phosphoproteome data (Figure 4D). The activity-related phosphorylation sites could be annotated through straightforward submission of the identified phosphorylated peptide and their quantifications (Figure 4E).
Figure 4.
The qKinAct service for the analysis of kinase activities. (A) The distribution of different types of activity-related phosphorylation sites in kinases. ‘+’, ‘−’ and ‘auto’ represent positively related, negatively related and autophosphorylation sites. (B) The distribution of kinases with activity-related phosphorylation sites in kinases. (C) The distribution of kinase families with activity-related phosphorylation sites in kinases. (D) The example for submission of quantitative phosphoproteome data. (E) The returned results for the query of kinase activity-related phosphorylation sites. The kinase activity profile for phosphorylation dynamics in nicotine-treated pancreatic stellate cells (F) and TNFα-stimulated phosphorylation dynamics (G).
Here, we provide two examples to show the application of the qKinAct service for kinase activity analysis. Paulo et al. globally analysed the phosphorylation dynamics in nicotine-treated pancreatic stellate cells and discovered the intensive dysregulation of protein phosphorylation in transcriptional events and nuclear function in response to nicotine (40). The kinase activity-related phosphorylation sites were analysed by qKinAct and shown in Figure 4F. It was observed that the phosphorylation level of activity-related phosphorylation sites was aberrant for kinases such as BRAF, PRKCB (PKCβ), PRKAA2 (AMPK) and EGFR. Interestingly, previous studies have identified the aberrant activation of EGFR (41) and PRKAA2 (42,43) induced by cigarette smoke. Moreover, the qKinAct analysis of TNFα-stimulated phosphorylation dynamics quantified by Mohideen et al. (44) identified ten aberrant activity-related phosphorylation sites for kinases such as PDPK1 (PDK1), GSK3, DYRK1 and RIPK1 (Figure 4G). Among the results, the observed inactivation of RIPK1 by S320 phosphorylation was also validated by their experiments (44). Thus, the qKinAct service could quickly provide helpful kinase activity profiles to elucidate the dynamics of phosphorylation signalling pathways regulated by kinases.
DISCUSSION
Protein phosphorylation occurs frequently over numerous biological processes, such as cell proliferation and differentiation, transcriptional activation and metabolic homeostasis (1–7). As one of the most important PTMs, phosphorylation greatly expands the complexity and diversity of proteome (1), while aberrant phosphorylation disorders the signalling pathways and is intensively correlated with diseases and cancers (2–5). The advancement of phosphoproteomic techniques greatly facilitated the high-throughput profiling and quantification of phosphorylation events and the accumulation of massive phosphoproteome data (9,10). However, although a number of systematic studies have contributed to recording the phosphorylation sites (16–30), their quantitative dynamics were still stored in the literature.
qPhos is the first repository to curate phosphorylation dynamics data. Various annotations, including protein information, potential upstream kinases and their inhibitors, and sequence and structure properties, were integrated to annotate the phosphorylation sites. Furthermore, the qKinAct service was developed to dissect the kinase activity profile from user-submitted quantitative phosphoproteome data through direct annotations of activity-related phosphorylation sites for kinases. Meanwhile, there were also various limitations in the database. The database was focused on human health-related phosphorylation dynamics, and currently only data from human tissues and cell lines were collected. However, several quantitative phosphoproteomics studies were based on a mouse model or other model organisms (45), which also provided helpful clues for medical investigations. Furthermore, previous studies showed that single nucleotide polymorphisms (SNPs) and somatic mutations at or around the phosphorylation sites could affect the phosphorylation dynamics (27), and currently qPhos has not linked the phosphorylation sites to the large quantity of SNP and mutation data. Taken together, although improvements remain to be achieved, qPhos could serve as a comprehensive resource to enable researchers to systematically and conveniently access the phosphorylation dynamics data under different experimental conditions, and its qKinAct service could help users to easily analyse the kinase activity profile from their own quantitative phosphoproteome data. The qPhos database will be regularly updated to keep pace with the progress of the quantitative dynamics of phosphorylation.
FUNDING
National Natural Science Foundation of China [31501069, 81802438]; National Key R&D Program of China [2018YFC1313300]; Program for Guangdong Introducing Innovative and Entrepreneurial Teams [2017ZT07S096]; Natural Science Foundation of Guangdong Province [2014A030312015]; Science and Technology Program of Guangdong [2015B020232008]; Science and Technology Program of Guangzhou [15570006, 201508020250, 201604020003]; Fundamental Research Funds for the Central Universities [SYSU: 16ykzd06]; Pearl River S&T Nova Program of Guangzhou [Ze-Xian Liu]; Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (the second phase). Funding for open access charge: Program for Guangdong Introducing Innovative and Entrepreneurial Teams [2017ZT07S096].
Conflict of interest statement. None declared.
REFERENCES
- 1. Cohen P. The origins of protein phosphorylation. Nat. Cell Biol. 2002; 4:E127–E130. [DOI] [PubMed] [Google Scholar]
- 2. Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 2001; 268:5001–5010. [DOI] [PubMed] [Google Scholar]
- 3. Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 2009; 21:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995; 80:225–236. [DOI] [PubMed] [Google Scholar]
- 5. Cohen P. Protein kinases–the major drug targets of the twenty-first century. Nat. Rev. Drug. Discov. 2002; 1:309–315. [DOI] [PubMed] [Google Scholar]
- 6. Anderson C. Nobel Prize given for work on protein phosphorylation. Nature. 1992; 359:570. [DOI] [PubMed] [Google Scholar]
- 7. Balter M., Vogel G.. Nobel prize in physiology or medicine. Cycling toward Stockholm. Science. 2001; 294:502–503. [DOI] [PubMed] [Google Scholar]
- 8. Krzyzosiak A., Sigurdardottir A., Luh L., Carrara M., Das I., Schneider K., Bertolotti A.. Target-based discovery of an inhibitor of the regulatory phosphatase PPP1R15B. Cell. 2018; 174:1216–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macek B., Mann M., Olsen J.V.. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 2009; 49:199–221. [DOI] [PubMed] [Google Scholar]
- 10. Mann M., Ong S.E., Gronborg M., Steen H., Jensen O.N., Pandey A.. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002; 20:261–268. [DOI] [PubMed] [Google Scholar]
- 11. Wojcechowskyj J.A., Didigu C.A., Lee J.Y., Parrish N.F., Sinha R., Hahn B.H., Bushman F.D., Jensen S.T., Seeholzer S.H., Doms R.W.. Quantitative phosphoproteomics reveals extensive cellular reprogramming during HIV-1 entry. Cell Host Microbe. 2013; 13:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van den Biggelaar M., Hernandez-Fernaud J.R., van den Eshof B.L, Neilson L.J., Meijer A.B., Mertens K., Zanivan S.. Quantitative phosphoproteomics unveils temporal dynamics of thrombin signaling in human endothelial cells. Blood. 2014; 123:e22–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho L., Tan S.Y., Wee S., Wu Y., Tan S.J., Ramakrishna N.B., Chng S.C., Nama S., Szczerbinska I., Chan Y.S. et al. ELABELA is an endogenous growth factor that sustains hESC Self-Renewal via the PI3K/AKT pathway. Cell Stem Cell. 2015; 17:435–447. [DOI] [PubMed] [Google Scholar]
- 14. Bai Y., Kim J.Y., Watters J.M., Fang B., Kinose F., Song L., Koomen J.M., Teer J.K., Fisher K., Chen Y.A. et al. Adaptive responses to dasatinib-treated lung squamous cell cancer cells harboring DDR2 mutations. Cancer Res. 2014; 74:7217–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casado P., Rodriguez-Prados J.C., Cosulich S.C., Guichard S., Vanhaesebroeck B., Joel S., Cutillas P.R.. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci. Signal. 2013; 6:rs6. [DOI] [PubMed] [Google Scholar]
- 16. Consortium U. UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. et al. Human protein reference Database–2009 update. Nucleic Acids Res. 2009; 37:D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang K.Y., Su M.G., Kao H.J., Hsieh Y.C., Jhong J.H., Cheng K.H., Huang H.D., Lee T.Y.. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 2016; 44:D435–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M.. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012; 40:D261–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J., Jia J., Li H., Yu J., Sun H., He Y., Lv D., Yang X., Glocker M.O., Ma L. et al. SysPTM 2.0: an updated systematic resource for post-translational modification. Database (Oxford). 2014; 2014:bau025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gnad F., Gunawardena J., Mann M.. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011; 39:D253–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ullah S., Lin S., Xu Y., Deng W., Ma L., Zhang Y., Liu Z., Xue Y.. dbPAF: an integrative database of protein phosphorylation in animals and fungi. Sci. Rep. 2016; 6:23534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinkel H., Chica C., Via A., Gould C.M., Jensen L.J., Gibson T.J., Diella F.. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 2011; 39:D261–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., Picotti P., Schlapbach R., Aebersold R.. PhosphoPep–a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 2008; 26:1339–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang H., Arighi C.N., Ross K.E., Ren J., Li G., Chen S.C., Wang Q., Cowart J., Vijay-Shanker K., Wu C.H.. iPTMnet: an integrated resource for protein post-translational modification network discovery. Nucleic Acids Res. 2018; 46:D542–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minguez P., Letunic I., Parca L., Bork P.. PTMcode: a database of known and predicted functional associations between post-translational modifications in proteins. Nucleic Acids Res. 2013; 41:D306–D311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y., Cheng H., Pan Z., Ren J., Liu Z., Xue Y.. Reconfiguring phosphorylation signaling by genetic polymorphisms affects cancer susceptibility. J. Mol. Cell Biol. 2015; 7:187–202. [DOI] [PubMed] [Google Scholar]
- 28. Krassowski M., Paczkowska M., Cullion K., Huang T., Dzneladze I., Ouellette B.F.F., Yamada J.T., Fradet-Turcotte A., Reimand J.. ActiveDriverDB: human disease mutations and genome variation in post-translational modification sites of proteins. Nucleic Acids Res. 2018; 46:D901–D910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matlock M.K., Holehouse A.S., Naegle K.M.. ProteomeScout: a repository and analysis resource for post-translational modifications and proteins. Nucleic Acids Res. 2015; 43:D521–D530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt T., Samaras P., Frejno M., Gessulat S., Barnert M., Kienegger H., Krcmar H., Schlegl J., Ehrlich H.C., Aiche S. et al. ProteomicsDB. Nucleic Acids Res. 2018; 46:D1271–D1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E. et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40:W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014; 42:D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang D., Zeng S., Xu C., Qiu W., Liang Y., Joshi T., Xu D.. MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction. Bioinformatics. 2017; 33:3909–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X.. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell Proteomics. 2008; 7:1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song C., Ye M., Liu Z., Cheng H., Jiang X., Han G., Songyang Z., Tan Y., Wang H., Ren J. et al. Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol. Cell Proteomics. 2012; 11:1070–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y., Liu Z., Cheng H., Gao T., Pan Z., Yang Q., Guo A., Xue Y.. EKPD: a hierarchical database of eukaryotic protein kinases and protein phosphatases. Nucleic Acids Res. 2014; 42:D496–D502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petersen B., Petersen T.N., Andersen P., Nielsen M., Lundegaard C.. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009; 9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dosztanyi Z., Csizmok V., Tompa P., Simon I.. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005; 21:3433–3434. [DOI] [PubMed] [Google Scholar]
- 39. Nolen B., Taylor S., Ghosh G.. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004; 15:661–675. [DOI] [PubMed] [Google Scholar]
- 40. Paulo J.A., Gaun A., Gygi S.P.. Global analysis of protein expression and phosphorylation levels in Nicotine-Treated pancreatic stellate cells. J. Proteome Res. 2015; 14:4246–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Filosto S., Becker C.R., Goldkorn T.. Cigarette smoke induces aberrant EGF receptor activation that mediates lung cancer development and resistance to tyrosine kinase inhibitors. Mol. Cancer Ther. 2012; 11:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang S., Zhang C., Zhang M., Liang B., Zhu H., Lee J., Viollet B., Xia L., Zhang Y., Zou M.H.. Activation of AMP-activated protein kinase alpha2 by nicotine instigates formation of abdominal aortic aneurysms in mice in vivo. Nat. Med. 2012; 18:902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Furlong H.C., Stampfli M.R., Gannon A.M., Foster W.G.. Cigarette smoke exposure triggers the autophagic cascade via activation of the AMPK pathway in mice. Biol. Reprod. 2015; 93:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mohideen F., Paulo J.A., Ordureau A., Gygi S.P., Harper J.W.. Quantitative phospho-proteomic analysis of TNFalpha/NFkappaB signaling reveals a role for RIPK1 phosphorylation in suppressing necrotic cell death. Mol. Cell Proteomics. 2017; 16:1200–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zanivan S., Meves A., Behrendt K., Schoof E.M., Neilson L.J., Cox J., Tang H.R., Kalna G., van Ree J.H., van Deursen J.M. et al. In vivo SILAC-based proteomics reveals phosphoproteome changes during mouse skin carcinogenesis. Cell Rep. 2013; 3:552–566. [DOI] [PubMed] [Google Scholar]