Abstract
The EncoMPASS online database (http://encompass.ninds.nih.gov) collects, organizes, and presents information about membrane proteins of known structure, emphasizing their structural similarities as well as their quaternary and internal symmetries. Unlike, e.g. SCOP, the EncoMPASS database does not aim for a strict classification of membrane proteins, but instead is organized as a protein chain-centric network of sequence and structural homologues. The online server for the EncoMPASS database provides tools for comparing the structural features of its entries, making it a useful resource for homology modeling and active site identification studies. The database can also be used for inferring functionality, which for membrane proteins often involves symmetry-related mechanisms. To this end, the online database also provides a comprehensive description of both the quaternary and internal symmetries in known membrane protein structures, with a particular focus on their orientation relative to the membrane.
INTRODUCTION
Protein structure determination and prediction, active site detection, and protein sequence alignment techniques all exploit information about relationships between protein structures. Hence, over the last few decades online projects for collecting and organizing structural data have proliferated, fuelled by the quasi-exponential growth of experimentally determined protein structures. However, membrane proteins, which constitute 20–30% of any given genome and >50% of all FDA-approved drug targets, are noticeably underrepresented in such databases. For example, membrane proteins account for only ∼2% of the Protein Databank (PDB) (1), of which ∼800 structures represent unique proteins as of September 2018 (http://blanco.biomol.uci.edu/mpstruc/). Membrane proteins also appear to pose a challenge to common strategies for relating protein structures such as those used by SCOP (2) and CATH (3), whose classifications for membrane proteins are inconsistent (4). This inconsistency may reflect the distinct features that membrane proteins exhibit. In particular, while the broad functional diversity of membrane proteins is reflected in the wide divergence of their amino acid sequences (5,6), the fold space available to these proteins is restricted by their anisotropic environment. Therefore, identifying differences and commonalities in their architectures might require the development of novel strategies. In addition, the membrane orientation is a unique feature that can potentially be leveraged to improve upon such methods.
Another striking feature of membrane protein architectures is that they are abundant in symmetries and pseudosymmetries, on both intramolecular and quaternary levels (7–9). These symmetries often reflect evolution and function and can be used to predict active sites, conformational changes and mechanisms (9–12). Despite their importance, the few online resources that address structural symmetries provide limited information. For example, the SymD webserver (13) can analyze a protein structure file for symmetries, but the reported results do not describe the repeating elements and the method can detect only one symmetry per structure. In the PDB database, the only information about symmetry refers to that found between chains in a complex (14), and not internal symmetries. Importantly, no available resource relates the symmetry axis to the membrane orientation, which can provide an important clue to protein function.
To address these issues, we created the Encyclopedia of Membrane Proteins Analyzed by Structure and Symmetry (EncoMPASS). Expanding on the offline, manually-curated membrane protein structure classification database HOMEP (15), the EncoMPASS database is automated and uses accurate structure and sequence alignments to relate membrane protein structures. Instead of building a hierarchical classification in the style of e.g. SCOP, we constructed EncoMPASS around the structural and sequence similarities of all protein chains with similar transmembrane topologies. This results in networks of sequence and structural homologues for each protein chain. Uniquely, EncoMPASS also provides detailed information about both quaternary and internal structural symmetries, including the type of symmetry, the symmetry axes and their orientation with respect to the membrane, as well as a sequence alignment of the symmetry-related residues. The database curates results of several symmetry detection approaches and allows the user to select the analysis that best fits their objectives.
On the online version of the EncoMPASS database, the user is guided through the data by graphical and interactive interfaces, such as: 3dmol representations (16) of each membrane protein complex, each chain, and all symmetry-related structural repeats; intuitive, interactive graphs that elucidate sequence and structure relationships among the database entries; and search bars that allow for either a straightforward or a more tailored exploration of the database. All data used to generate the displayed content are also available for download.
DATABASE CONTENT
To maximize the accuracy of our structural analysis, the EncoMPASS database collects crystallographic structures of membrane proteins with resolution ≤3.5 Å. EncoMPASS uses the manually-curated Orientations of Proteins in Membranes (OPM) database (17) as the primary source of structure coordinates that are reliably oriented relative to the predicted lipid bilayer. However, the information in the OPM coordinate files is occasionally incomplete or inconsistently formatted, or the biological assembly can be in disagreement with the one reported in the PDB. Indeed, predicting the correct biological assembly of membrane protein structures is a major challenge (bioRxiv: https://doi.org/10.1101/391961), leading to discrepancies between the assignments in the PDB (1), PDBTM (18) and OPM databases. To maximize the accuracy of the predicted biological assembly we follow the strategy implemented in PDBTM (18). First, each structure is classified as either potentially problematic or plausibly correct. Then, for each member of the former set, we check whether a structure in the latter set can serve as a template for the biological assembly. If no template is found, we assume the biological assembly described in the PDBTM web server. Thus, the coordinate file published on EncoMPASS is a revised version of the coordinate file in the PDB database, with its biological assembly optimized where necessary, and with membrane boundaries estimated by the algorithm underlying the OPM database, PPM (19). As of September 2018, EncoMPASS contains 2344 coordinate files, corresponding to 67% of all membrane protein entries present in the PDB; the majority of the excluded entries do not meet the resolution threshold.
All structural and sequence similarity networks presented in EncoMPASS are based on a large set of pairwise structure alignments between individual chains of proteins, carried out using Fr-TM-Align (20). However, aligning two structures with very different topologies can force one of the two structures to fragment excessively and thereby produce a fit with a biologically meaningless alignment. Therefore, we only compare structures with similar numbers of transmembrane regions. The quality of the structural alignment is measured using the root mean squared deviation (RMSD) of the aligned Cα-atoms, as well as the Template-Modeling score, or TM-score (21,22). The TM-score is the metric we use to establish a structural relationship between two proteins. Both the TM-score and RMSD are global scores, but we also wish to identify the most structurally conserved regions of a membrane protein chain. To this end, we also report the per-residue Cα–Cα distances between a given chain and all others aligned to it.
Protein sequence similarity provides complementary information to structural similarity and can highlight, for example, proteins undergoing conformational changes during their function. Thus, independent of the structural alignment, the sequences of all the same pairs of topologically-related proteins are aligned with MUSCLE (23).
For each protein complex and chain we also report the results from a comprehensive symmetry analysis. First, EncoMPASS curates the results of two standard algorithms for symmetry detection, CE-Symm 2.2 (bioRxiv: https://doi.org/10.1101/297960) and SymD 1.61 (24). In addition, the database includes results from a multi-step symmetry detection (MSSD) procedure (bioRxiv: https://doi.org/10.1101/391961). The MSSD method processes structures using CE-Symm and SymD with a range of customized parameters and then, among other considerations, filters the results based on the position of the detected structural repeats with respect to the membrane. MSSD can report multiple symmetries within either a complex or a chain; the corresponding symmetry axes and transformations; the residue ranges and multiple alignment of the structural repeats; and the orientation of the repeats and the symmetry axes relative to the membrane. Finally, taking advantage of the networks of structural and sequence similarities described above, the database reports so-called inferred symmetries. These are obtained by assuming the presence of repeated or symmetric elements in a given chain based on their detection with the MSSD method in related protein chains. The results of this final analysis allow for informative comparisons of the degree of symmetry in different conformations of the same protein or between different proteins in the same protein family.
A detailed description of the procedures used to create the dataset is provided in (bioRxiv: https://doi.org/10.1101/391961).
DATABASE ACCESS
The EncoMPASS web server is hosted at http://encompass.ninds.nih.gov and is equipped with a a simple PDB-code search as well as a more versatile search tool that allows the user to explore the many features of the database. The complete list of search criteria organized by category is reported in Supplementary Table S1.
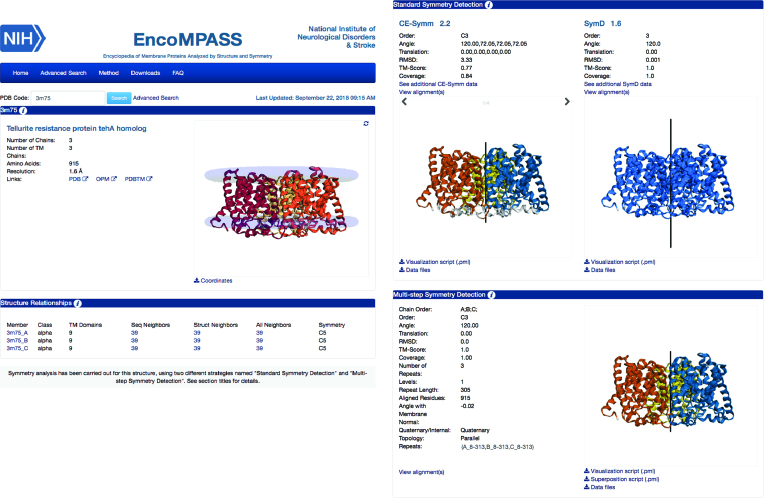
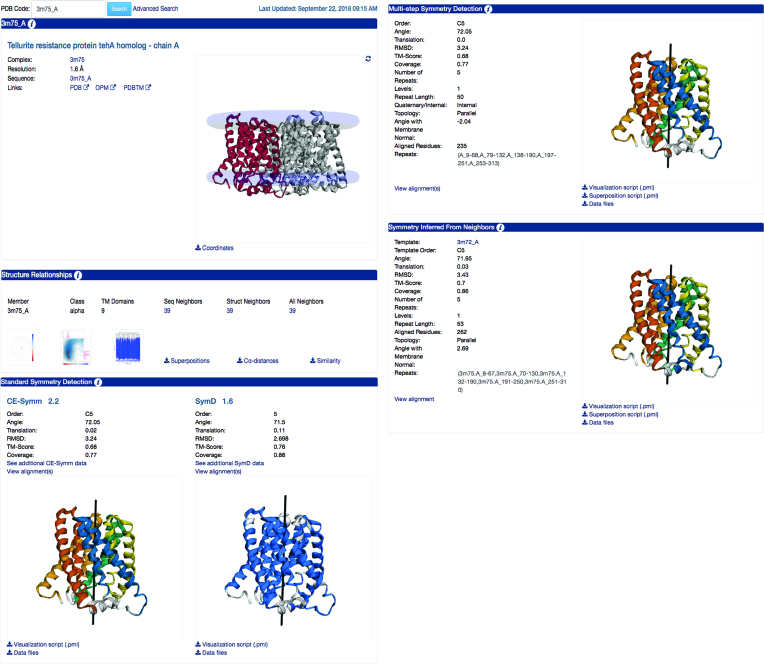
Information is provided for each PDB entry on two levels: entire protein complexes (Figure 1) and individual transmembrane chains (Figure 2). The pages for each protein complex are divided into four sections, containing a general description of the protein, a summary of its transmembrane chains and their structure relationships, and descriptions of the symmetries obtained by the standard symmetry detection algorithms and by the MSSD procedure.
Figure 1.
Example EncoMPASS online database complex page. Representative page for the whole structure of PDB entry 3M75 (a TehA homologue), including a brief summary of the characteristics of each of its transmembrane subunits (left) and analysis of the symmetries within the complex (right) using two standard symmetry detection algorithms as well as the multi-step symmetry detection (MSSD) method. Note that on these pages the MSSD section contains results only if quaternary symmetries (i.e., involving multiple chains) have been detected. According to all three methods (CE-Symm, SymD, and MSSD), the TehA homologue forms a three-fold circularly (C3) symmetric assembly. Note that in the molecular viewer in the SymD section, the repeats are not differentiated by color, since SymD does not report residue ranges for the symmetry-related repeats. According to the MSSD results, the axis of this C3 symmetry is perpendicular to the membrane plane, i.e., the protomers have parallel topologies.
Figure 2.
Example EncoMPASS online database chain page. Representative page for chain A of PDB entry 3M75. These pages follow the same outline as for the complex in Figure 1, but contain analysis specific to each chain, including sequence and structural relationships, and any detected internal symmetries. Chain A of 3M75 has 39 sequence and structure neighbors. (Visualization of the polar or scatter plots, shown here as clickable icons, reveals that these structures are all closely related, both structurally and by sequence.) In terms of the internal symmetry, all methods agree that chain A contains a five-fold circular pseudo-symmetry (C5), although there are some differences in the details. As in the pages for complexes, the MSSD section reports the relationship of the symmetry axis to the membrane plane. The last section (‘Symmetry Inferred from Neighbors’), only available for chains, reports results based on inference from putative structural homologues, in cases where MSSD detected a more comprehensive symmetry than in the chain of interest. By inferring the symmetry information from another TehA-homologue structure, 3M72 chain A, this approach detected a symmetry relationship for 3M75 chain A with greater coverage (86% of the chain, compared to 77% when using MSSD) and a higher similarity (TM-score 0.70 rather than 0.68).
The first section of each whole structure page reports general information about the complex and provides links to the corresponding entry in the PDB and OPM databases, as well as a graphical interface that allows real-time visualization of the structure from different perspectives. This section also provides a button for downloading the coordinates of the protein. Note that the displayed structure is not identical to that in the PDB: for example, the biological assembly might differ, and the membrane boundaries are highlighted by two planes of pseudo-atoms.
The second section contains a table summarizing the characteristics of each of the transmembrane chains in the complex. This includes: the number of transmembrane regions, the number of sequence and structural neighbors, and the symmetry order of any internal symmetry detected by either the MSSD, CE-Symm or inferred symmetry methods in that order of preference. By clicking on the transmembrane chain identifier, the user is redirected to the relevant page for that chain, while the sequence-, structure- and all-neighbors fields are linked to tables describing the corresponding structures.
The third and fourth sections of each complex page are dedicated to symmetry analyses. The third section summarizes the results of the two standard symmetry detection algorithms, CE-Symm and SymD. CE-Symm can detect multiple symmetries for the same structure, so the user can scroll through the different symmetries. Each symmetry is represented using different colors for each repeat and a black line for the symmetry axis. By comparison, SymD can detect only one symmetry axis in a structure and its output does not indicate the boundaries of each repeat. Hence, in the 3dmol visualization of the SymD results, all residues that are related by the detected symmetry axis are colored blue. The raw data and a PyMOL visualization script can be downloaded directly from this section. It is important to note that the output of the two symmetry recognition programs does not distinguish between quaternary and internal symmetries and, therefore, the results for complexes might include information on both. On the other hand, the subsequent section (the fourth section) contains results from our MSSD procedure focused exclusively on quaternary symmetries. The layout of the MSSD results section mirrors that of the CE-Symm results, but the reported data also includes classification of the topology of the repeats with respect to the membrane (antiparallel or parallel), as well as the angle of each reported symmetry axis to the membrane normal. Finally, the user can download a PyMOL script that allows visualization of the superposition of all repeats.
The web pages for individual chains are extended versions of the whole structure pages, with a total of five sections (Figure 2). The second section on each chain page is notable, in that it includes visual representations of the analysis of sequence and structure relationships. Three graphs are presented. The first graph illustrates the structural similarity between the chain of interest and its close structural neighbors (TM-score ≥ 0.6), where the distance between any two points is proportional to their similarity (i.e., inversely proportional to the TM-score). Since large structural differences could be explained simply by a difference in size, each point on this plot is colored in shades from blue to red to show the (greater or fewer, respectively) number of transmembrane regions relative to that of the chain of interest. Hovering over the points with the cursor brings up their PDB identifiers, while clicking on a point navigates to the EncoMPASS page describing the corresponding protein chain. All the information provided in this first plot, however, is focused on structural similarity. The user may also be interested in the sequence similarity between protein chains that have related structures, or conversely, the structural variability among entries with similar sequences. Thus, in the second graph, the structural similarity according to the TM-score is plotted against the sequence identity from the MUSCLE pair-wise sequence alignment, for every compared chain. The contour lines on the graph illustrate the abundance of pairs of compared structures across the entire database. Finally, to illustrate how different regions of the protein chain compare, the third graph provides the structural similarity (i.e. distance between Cα atoms) as a function of residue number for each structure alignment. All similarity measures and structure alignment outputs for the protein chain of interest are also downloadable.
Sections three and four of the individual chain pages follow the same layout as those on the complex page, but obviously only report symmetries within the given chain. In the fifth section, each chain page includes results from an additional procedure that relies on the structural and sequence neighbors to infer symmetry. That is, if the chain has a neighbor, for which the MSSD analysis detected a more extensive symmetric relationship, the symmetry of that neighbor is used as a template for mapping out the repeats in the current chain. The symmetry axis and structural similarity between these repeats are reported in a similar fashion as in the MSSD results section.
The EncoMPASS dataset is created and updated automatically using a set of python libraries available at https://www.github.com/EncoMPASS-code/EncoMPASS. All data entries and processed PDB files can be downloaded as a single file from http://encompass.ninds.nih.gov/downloads. The online database for EncomPASS is designed using the Oracle Relational Database System, and utilizes the Java 2 Enterprise Edition platform, the Spring Framework, and JavaScript libraries.
CASE STUDY: INVESTIGATING SODIUM-DEPENDENT TRANSPORT
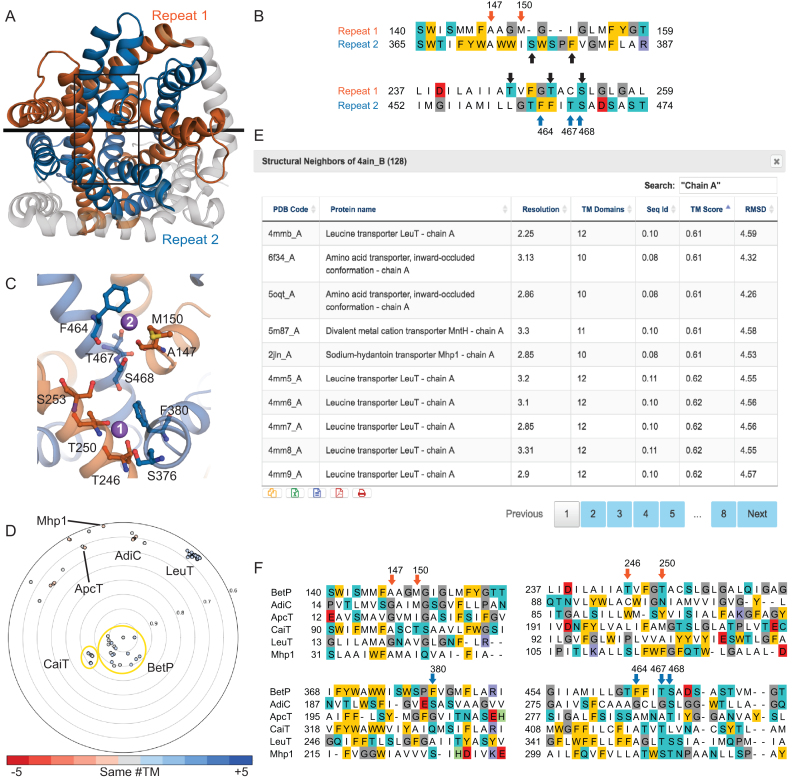
As an illustration of how EncoMPASS can be used to generate function-related hypotheses about membrane proteins, we consider the example of the sodium-coupled betaine symporter BetP. BetP uses the translocation of two sodium ions to energize the transport of each betaine molecule (25). The available structural data for BetP, however, only indicate the location of one of the two required sodium ions, at the so-called Na2 site, for example in PDB identifier 4AIN, chain B) (26). Unfortunately, no evidence has been found for the other sodium ion (Na1) in the six available structures of BetP. Interestingly, however, EncoMPASS reports the presence of two inverted-topology structural repeats in the BetP protomer (Figure 3A, 4AIN_B). Hence, examining the region of the protein that is structurally related to the Na2 binding site may reveal residues contributing to the Na1 site. This task is made straightforward by using the structural alignment of the repeats provided in the MSSD section of EncoMPASS (Figure 3B). Mapping the related residues onto the structure narrows the list of candidate residues that could interact with the ion to those within one or two helical turns and on the same face of the helix, which could contribute either side chain or backbone groups (Figure 3C). Residues S376, F380, T246, T250 and S253 satisfy these requirements, and hence, might contribute to the binding site. Khafizov et al. applied similar reasoning to arrive at this prediction, which they then tested using molecular dynamics simulations, as well as biochemical, biophysical and electrophysiological experiments, to conclude that F380, T246 and T250 are indeed important for sodium binding (12).
Figure 3.
Examining structure-function relationships in the sodium-coupled betaine transporter BetP and its homologues using EncoMPASS. (A) Structural symmetry reported by MSSD for BetP (PDB identifier 4AIN chain B), in which residues 135–326 (repeat 1, orange) are pseudo-C2-symmetric to residues 360–542 (repeat 2, blue), and related by an axis (black line) that lies in the plane of the membrane. The protein is viewed from the extracellular side of the membrane. The protein is shown in cartoon helices; elements of the structure not related by symmetry are colored gray. Note that ligands are not included in this representation. (B) Sequence alignment between repeats extracted from the structural alignment in the regions contributing residues to the Na2 binding site. The Na1 site residues are Ala147, Met150 from repeat 1 (orange arrows) and Phe464, Thr467 and Ser468 from repeat 2 (blue arrows). Residues in a similar region of the other repeat (black arrows) are candidates for the Na1 binding site. (C) Close-up of the Na2 and putative Na1-binding site regions in the area outlined with a black box in (A). Sodium ions are shown as purple spheres, and residues discussed in the main text are shown in ball-and-stick format. (D) Structural relationships between BetP and all other structures in EncoMPASS with TM-scores <0.6. Structures of another BCCT family-member, CaiT are closely related (TM-score ∼0.85), while structures of other LeuT-fold family members are more distantly related. Example LeuT-fold family members are labelled. Each point is colored according to the difference in number of transmembrane segments from the chain in the center, according to the provided scale. (E) Clicking on the entry for ‘All Neighbors’ on the ‘Structure Relationships’ section of the chain page for 4AIN chain B brings up a table of structural homologues, which lists several relevant features and their structural and sequence similarity. The results shown were additionally filtered by the term ‘Chain A’ and sorted by TM-score to reveal the least similar structures. (F) Structure-based sequence alignments of representative structural homologues relative to BetP allow interrogation of putative sodium binding sites or sodium-independent mechanisms. Alignments were extracted from pairwise structure alignments provided in EncoMPASS, and combined using pyali (https://github.com/christang/pyali). The positions of Na1 and Na2 site residues in BetP are indicated by arrows colored by repeat.
To understand further the principles of sodium-coupled substrate transport, we can use EncoMPASS to investigate the conservation of the BetP sodium binding sites in related structures. In the polar plot provided for 4AIN chain B (Figure 3d), two clusters are immediately visible near the origin of the graph, and these clusters correspond to structures of two betaine/carnitine/choline transporter (BCCT) family members, BetP and CaiT. On the edges of the plot, we identify more distantly-related structures such as those of LeuT, ApcT, AdiC and Mhp1. Then, using the table of ‘All Neighbors’ of 4AIN, chain B, we can scroll through a list of all related structures (Figure 3E), select a representative for each protein and extract the relevant structural alignments from the downloadable Superpositions file. Using these alignments (Figure 3F), we can examine which proteins contain sodium-coordinating residues (e.g., LeuT and Mhp1), and which do not (e.g., CaiT), and correlate those structural features with their known coupling mechanisms (12).
These analyses of BetP provide a clear demonstration of the value of EncoMPASS for delving into the structure-function relationships of membrane proteins, including by leveraging information relating to their symmetry and their structural neighbors.
CONCLUSION
The online EncoMPASS database curates a wide variety of membrane protein structural data, which we expect will appeal to diverse communities. For example, users interested in finding template structures for homology modeling should find particularly useful the visual analysis of structure similarity networks; the analysis of symmetric regions provides complementary information to infer functionality; the complete set of accurate pairwise structure alignments and their corresponding structure-based sequence alignments constitutes a robust benchmark for membrane protein sequence alignment programs; and symmetric region classification could be used to trace evolutionary sequence-structure relationships. For this reason, the website has been designed to be easily searchable by different criteria, to provide intuitive interfaces to access the results of our analyses, and to allow access to all data for postprocessing.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the system administration teams of NINDS and of the LOBOS cluster at the National Heart, Lung and Blood Institute, NIH for computational support. We also thank members of the CSB lab for beta-testing of the website.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Division of Intramural Research of the National Institute of Neurological Disorders and Stroke (NINDS), National Institutes of Health. Funding for open access charge: National Institute of Neurological Disorders and Stroke Intramural Research Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hubbard T.J.P., Murzin A.G., Brenner S.E., Chothia C.. SCOP: a structural classification of proteins database. Nucleic Acids Res. 1997; 25:236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Orengo C., Michie A., Jones S., Jones D., Swindells M., Thornton J.. CATH – a hierarchic classification of protein domain structures. Structure. 1997; 5:1093–1109. [DOI] [PubMed] [Google Scholar]
- 4. Neumann S., Fuchs A., Mulkidjanian A., Frishman D.. Current status of membrane protein structure classification. Proteins Struct. Funct. Bioinf. 2010; 78:1760–1773. [DOI] [PubMed] [Google Scholar]
- 5. Sojo V., Dessimoz C., Pomiankowski A., Lane N.. Membrane proteins are dramatically less conserved than water-soluble proteins across the tree of life. Mol Biol Evol. 2016; 33:2874–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olivella M., Gonzalez A., Pardo L., Deupi X.. Relation between sequence and structure in membrane proteins. Bioinformatics. 2013; 29:1589–1592. [DOI] [PubMed] [Google Scholar]
- 7. Choi S., Jeon J., Yang J.-S., Kim S.. Common occurrence of internal repeat symmetry in membrane proteins. Proteins Struct. Funct. Bioinf. 2008; 71:68–80. [DOI] [PubMed] [Google Scholar]
- 8. Myers-Turnbull D., Bliven S.E., Rose P.W., Aziz Z.K., Youkharibache P., Bourne P.E., Prlić A.. Systematic detection of internal symmetry in proteins using CE-Symm. J. Mol. Biol. 2014; 426:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forrest L.R. Structural symmetry in membrane proteins. Annu. Rev. Biophys. 2015; 44:311–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robinson A.J., Overy C., Kunji E.R.S.. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:17766–17771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Radestock S., Forrest L.R.. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J. Mol. Biol. 2011; 407:698–715. [DOI] [PubMed] [Google Scholar]
- 12. Khafizov K., Perez C., Koshy C., Quick M., Fendler K., Ziegler C., Forrest L.R.. Investigation of the sodium-binding sites in the sodium-coupled betaine transporter BetP. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E3035–E3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tai C.H., Paul R., Dukka K.C., Shilling J.D., Lee B.. SymD webserver: a platform for detecting internally symmetric protein structures. Nucleic Acids Res. 2014; 42:296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rose P.W., Prlić A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J. et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015; 43:D345–D356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stamm M., Forrest L.R.. Structure alignment of membrane proteins: accuracy of available tools and a consensus strategy. Proteins Struct. Funct. Bioinf. 2015; 83:1720–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rego N., Koes D.. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2015; 31:1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lomize M.A., Pogozheva I.D., Joo H., Mosberg H.I., Lomize A.L.. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012; 40:D370–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kozma D., Simon I., Tusnády G.E.. PDBTM: Protein Data Bank of transmembrane proteins after 8 years. Nucleic Acids Res. 2012; 41:D524–D529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lomize A.L., Pogozheva I.D., Mosberg H.I.. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J. Chem. Inf. Model. 2011; 51:930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pandit S., Skolnick J.. Fr-TM-align: a new protein structural alignment method based on fragment alignments and the TM-score. BMC Bioinformatics. 2008; 9:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Skolnick J.. Scoring function for automated assessment of protein structure template quality. Proteins Struct. Funct. Genet. 2004; 57:702–710. [DOI] [PubMed] [Google Scholar]
- 22. Xu J., Zhang Y.. How significant is a protein structure similarity with TM-score = 0.5. Bioinformatics. 2010; 26:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim C., Basner J., Lee B.. Detecting internally symmetric protein structures. BMC Bioinformatics. 2010; 11:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farwick M., Siewe R.M., Krämer R.. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J. Bacteriol. 1995; 177:4690–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perez C., Koshy C., Yildiz Ö., Ziegler C.. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature. 2012; 490:126–130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.