Abstract
DNA methylation is an important epigenetic mechanism for regulating gene expression. Aberrant DNA methylation has been observed in various human diseases, including cancer. Single-nucleotide polymorphisms can contribute to tumor initiation, progression and prognosis by influencing DNA methylation, and DNA methylation quantitative trait loci (meQTL) have been identified in physiological and pathological contexts. However, no database has been developed to systematically analyze meQTLs across multiple cancer types. Here, we present Pancan-meQTL, a database to comprehensively provide meQTLs across 23 cancer types from The Cancer Genome Atlas by integrating genome-wide genotype and DNA methylation data. In total, we identified 8 028 964 cis-meQTLs and 965 050 trans-meQTLs. Among these, 23 432 meQTLs are associated with patient overall survival times. Furthermore, we identified 2 214 458 meQTLs that overlap with known loci identified through genome-wide association studies. Pancan-meQTL provides a user-friendly web interface (http://bioinfo.life.hust.edu.cn/Pancan-meQTL/) that is convenient for browsing, searching and downloading data of interest. This database is a valuable resource for investigating the roles of genetics and epigenetics in cancer.
INTRODUCTION
The interpretation of the function of genomic variants, particularly in non-coding regions, is a major challenge for the genetic dissection of complex diseases such as cancer (1). Genome-wide association studies (GWAS) have identified numerous genetic loci that influence the risk of human cancer (2,3), but most of these loci are located in non-coding regions and are without clear molecular mechanisms that contribute to the phenotypic outcome. Previous studies considered a diverse set of functional regions, including miRNA binding sites, protein modification sites and transcription factor binding sites (4,5). However, the link between variants and epigenetic signals involved in the regulation of key biological processes has been largely overlooked.
As a major epigenetic mechanism that directs gene expression, DNA methylation plays a key role in the regulation of crucial biological and pathological processes (6). Aberrant DNA methylation is frequently observed in various cancers (7) and represents an attractive biomarker and therapeutic target (8,9). Increasing evidence indicates that single-nucleotide polymorphisms (SNPs) contribute to tumor initiation, progression and prognosis by influencing DNA methylation levels (10,11). Therefore, DNA methylation may be an important molecular-level phenotype that links a genotype with the trait of a complex disease. It is fundamentally vital to build a public data repository to identify SNPs that significantly affect DNA methylation levels, i.e. methylation quantitative trait loci (meQTL). Recent methodological advances allow for genome-wide screening of meQTLs in different tissues, including blood (12), lung (13) and brain (14). However, no database has been developed to systematically analyze meQTLs across multiple cancer types.
The Cancer Genome Atlas (TCGA) provides genome-wide genotype data and DNA methylation data for ∼10 000 cancer samples, making it possible to systematically conduct meQTL analysis across cancer types. We identified millions of meQTLs across 23 cancer types and have made them available for browsing, searching and downloading through Pancan-meQTL (http://bioinfo.life.hust.edu.cn/Pancan-meQTL/), a database for systematic evaluation of the effects of genetic variants on DNA methylation across diverse cancer types.
DATA COLLECTION AND PROCESSING
Genotype data collection, imputation and processing
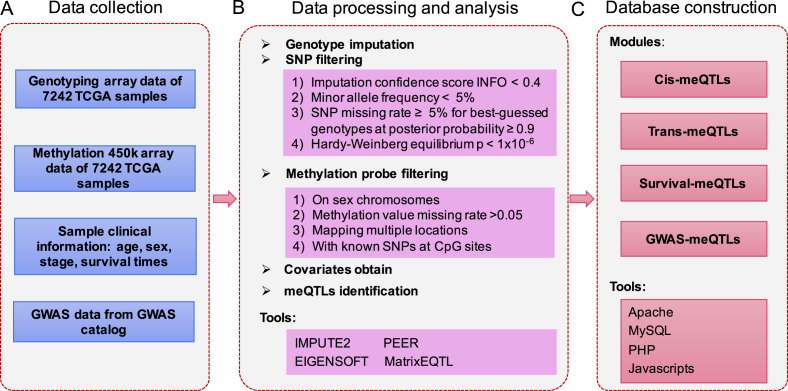
We downloaded genotype data (level 2) from TCGA data portal (https://portal.gdc.cancer.gov/) (Figure 1A). We kept 7735 samples with both genotype data and methylation data. We then combined colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) as colorectal cancer (CRC) (15) and removed cancer types with sample size <100 primary tumor samples. Thus, for further analysis, we had 7242 samples across 23 cancer types. We performed genotype imputation and filtering per cancer type as described in our previous study (16). After imputation and quality filtering, on average, 4 318 218 genotypes per cancer type were included in the meQTL analysis.
Figure 1.
Data collection, processing and database construction. Pancan-meQTL collected the genotype, methylation and clinical data from TCGA to evaluate the effects of SNPs on methylation levels. For each cancer type, the data were processed using a series of filtering and quality control steps. Four datasets, cis-meQTLs, trans-meQTLs, survival-meQTLs and GWAS-meQTLs, are included in the database.
Methylation data collection and processing
Methylation beta values (level 3) obtained from TCGA data portal (https://gdc-portal.nci.nih.gov/) were measured by the Illumina Infinium HumanMethylation450 BeadChip array, which contained 485 512 probes for each sample. Due to the specific nature of methylation patterns on sex chromosomes (17), we focused on autosomes. In each cancer type, probes were filtered by the following criteria: (i) methylation beta value missing rate > 0.05, (ii) mapping to multiple locations on the genome (18) and (iii) containing known SNP (1000 Genome Phase3 (19), MAF > 0.01) at CpG sites (20,21) (Figure 1B). On average, 369 244 high-quality methylation probes per cancer type were used for analyses. To minimize the effects of outliers on the regression scores, the values for each probe across samples per cancer type were transformed into a standard normal distribution based on rank (17,22,23).
Covariates
To correct for known and unknown confounders and increase the sensitivity of our analyses, we included several covariates. The top five principal components calculated by smartpca in the EIGENSOFT program (24) were included to control for ethnicity differences. To remove hidden batch effects and other confounders in the methylation data, we used PEER software (25) to select the first 15 PEER factors from the methylation data as covariates. We included the common confounders of age, sex, and tumor stage as additional covariates (21,23,26).
Identification of meQTLs
Identification of meQTL is straightforward that to test whether individuals carrying different genotypes show different methylation levels (17,20,27,28). For each cancer type, we performed linear regression with MatrixEQTL (29) to determine the effects of genetic variation on methylation levels. We calculated the pairwise associations between each SNP and CpG site. We used HumanMethylation450 BeadChip array annotation to extract the location (hg19) of methylation probes. We downloaded the SNP location (hg19) from dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP/). We defined meQTLs as SNPs with false discovery rates (FDRs) calculated by MatrixEQTL <0.05 and the absolute value of correlation coefficient (r) ≥0.3. If the SNP was within 1 Mb from the probe location (22), we denoted it as cis-meQTL; and if the SNP was beyond that point, we denoted it as trans-meQTL.
Identification of survival-associated meQTLs
To prioritize promising meQTLs, we identified meQTLs that may be associated with patient survival times. We downloaded the clinical data, including patient overall survival times, from TCGA data portal (https://gdc-portal.nci.nih.gov/). For each meQTL, we used the log-rank test to examine the association between genotypes and patient overall survival times. We plotted Kaplan–Meier (KM) curves to show the survival times for individuals carrying different genotypes. We defined meQTLs with FDR <0.05 as survival-meQTLs.
Identification of meQTLs in GWAS regions
Risk SNPs identified in GWAS studies were downloaded from GWAS catalog (http://www.ebi.ac.uk/gwas/) (2). GWAS linkage disequilibrium (LD) regions were extracted from SNAP (https://personal.broadinstitute.org/plin/snap/ldsearch.php) (30) with parameters (SNP data set: 1000 Genomes Pilot 1; LD r2 threshold: 0.5; population panel: CEU; distance limit: 500 kb). The meQTLs that overlapped with GWAS tagSNPs and LD SNPs were identified as GWAS-meQTLs.
Enrichment analysis
To assess the enrichment of meQTLs in DNA regulatory elements and GWAS loci, we generated a control data set of non-meQTL SNPs with minor allele frequency (MAF) and distance matched to the set of meQTLs for each cancer type. We downloaded transcription factor binding sites (TFBSs) of related ENCODE cell lines from the UCSC genome browser, and extracted cancer-specific GWAS loci from GWAS catalog. Enrichment analyses of meQTLs were performed by two-tailed Fisher's exact test with the following 2 × 2 table: columns; meQTL SNPs and non-meQTL SNPs, rows; SNPs within and not within the TFBSs/GWAS loci.
DATABASE CONTENT AND USAGE
Samples in Pancan-meQTL
Pancan-meQTL included 23 cancer types with sample size ≥100 in TCGA, covering 7242 tumor samples. The sample size of each cancer type ranged from 103 in skin cutaneous melanoma (SKCM) to 664 in breast invasive carcinoma (BRCA) (Table 1). For the genotype data, we obtained a median of 4 487 756 SNPs for each cancer type after imputation and quality control, ranging from 2 721 411 for BRCA to 5 121 896 for acute myeloid leukemia (LAML). For the methylation data, there were, on average, 384 903 probes for each cancer type after probe filtering and quality control. The range was from 380 594 for CRC to 385 618 for testicular germ cell tumors (TGCT).
Table 1.
Overview of samples and meQTLs in Pancan-meQTL
| Cis | Trans | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cancer type | Disease full name | No. of samples | No. of methylation probes | No. of genotypes | Pairs | meProbesa | cis_meQTLs | Pairs | meProbesa | meQTLs |
| BLCA | Bladder urothelial carcinoma | 405 | 384903 | 4182865 | 502774 | 8136 | 301392 | 216341 | 46165 | 51295 |
| BRCA | Breast invasive carcinoma | 664 | 384084 | 2721411 | 340881 | 7289 | 203391 | 47355 | 1804 | 31118 |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma | 290 | 383425 | 4289322 | 531458 | 11284 | 318082 | 212025 | 48741 | 46185 |
| CRC | Colon adenocarcinoma + Rectum adenocarcinoma | 354 | 380594 | 3879590 | 555556 | 10781 | 316517 | 185943 | 40872 | 55402 |
| ESCA | Esophageal carcinoma | 173 | 382712 | 4357977 | 408020 | 15726 | 260629 | 21790 | 5427 | 13297 |
| HNSC | Head and neck squamous cell carcinoma | 501 | 385146 | 4245789 | 706915 | 10590 | 392427 | 100579 | 16599 | 51195 |
| KIRC | Kidney renal clear cell carcinoma | 306 | 384916 | 4487756 | 827668 | 14184 | 455287 | 598701 | 95277 | 82497 |
| KIRP | Kidney renal papillary cell carcinoma | 271 | 384786 | 4827856 | 830528 | 14986 | 486170 | 232041 | 52484 | 58017 |
| LAML | Acute myeloid leukemia | 122 | 385529 | 5121896 | 408043 | 10758 | 283314 | 22473 | 606 | 17201 |
| LGG | Lower grade glioma | 487 | 385198 | 4611830 | 789126 | 11092 | 454090 | 151792 | 24257 | 61737 |
| LIHC | Liver hepatocellular carcinoma | 367 | 383962 | 4152031 | 449509 | 9378 | 272962 | 127824 | 28610 | 46149 |
| LUAD | Lung adenocarcinoma | 448 | 384670 | 4343043 | 525439 | 8848 | 297745 | 110266 | 19790 | 52156 |
| LUSC | Lung squamous cell carcinoma | 359 | 385034 | 3676672 | 598237 | 10039 | 334937 | 191909 | 44240 | 60597 |
| PAAD | Pancreatic adenocarcinoma | 177 | 382517 | 4989296 | 903193 | 24432 | 513599 | 120777 | 41820 | 34727 |
| PCPG | Pheochromocytoma and paraganglioma | 175 | 385235 | 4701955 | 589160 | 19388 | 385289 | 34905 | 2175 | 23719 |
| PRAD | Prostate adenocarcinoma | 479 | 385067 | 4801718 | 1129044 | 16366 | 574577 | 188046 | 46160 | 66084 |
| SARC | Sarcoma | 257 | 382095 | 4083674 | 458278 | 14059 | 287992 | 64709 | 22198 | 24734 |
| SKCM | Skin cutaneous melanoma | 103 | 384364 | 4838338 | 200763 | 6689 | 150232 | 8399 | 399 | 6332 |
| STAD | Stomach adenocarcinoma | 367 | 383290 | 4265676 | 687004 | 10723 | 382962 | 77711 | 9612 | 49489 |
| TGCT | Testicular germ cell tumors | 148 | 385618 | 4807863 | 470576 | 14871 | 309018 | 36183 | 3791 | 25912 |
| THCA | Thyroid carcinoma | 498 | 385449 | 4842972 | 830377 | 11908 | 477575 | 248274 | 36261 | 72317 |
| THYM | Thymoma | 120 | 385609 | 4940146 | 562478 | 17015 | 352356 | 28228 | 1981 | 22009 |
| UCEC | Uterine corpus endometrial carcinoma | 171 | 385407 | 4961809 | 314774 | 14181 | 218421 | 17953 | 2911 | 12881 |
ameProbes: methylation probes regulated by meQTLs.
meQTLs in Pancan-meQTL
Per cancer type, the associations between each SNP and methylation probe were analyzed for cis- and trans-meQTL mapping by linear regression with adjusted covariates. In the cis-meQTL analysis, we identified a total of 13 619 801 meQTL-CpG pairs at FDR < 0.05 and |r| ≥0.3 in 23 cancer types, which corresponded to a median P-value <4.34 × 10−7. We found a total of 8 028 964 cis-meQTLs across the cancer types, with a median of 318 082 meQTLs per cancer type, and a range from 150 232 in SKCM to 574 577 in prostate adenocarcinoma (PRAD) (Table 1). These meQTLs control DNA methylation at a median of 11 284 CpG sites per cancer type. On average, 35.1% cis-meQTLs are associated with multiple CpG sites. In trans-meQTL analysis, we identified 3 044 224 meQTL-CpG pairs at FDR < 0.05 and |r| ≥0.3 in 23 cancer types, which corresponded to a median of P-value < 1 × 10−9. There are 965 050 trans-meQTLs, with a median of 46 185 meQTLs per cancer type, and a range from 6332 in SKCM to 82 497 in kidney renal clear cell carcinoma (KIRC) (Table 1).
We further identified 23 432 meQTLs associated with patient overall survival times across different cancer types at FDR < 0.05. The number of survival-meQTLs ranged from 218 in BRCA to 11 212 in PRAD. We compared meQTL results to GWAS data and found 2 214 458 meQTLs that overlap with GWAS linkage disequilibrium regions of one or multiple traits. Enrichment analyses showed that meQTLs are significant enriched in most of TFBSs, such as CTCF, SIN3AK20 and NRSF (Supplementary Figure S1) and GWAS loci (Supplementary Table S1).
WEB DESIGN AND INTERFACE
The database was built based on Apache, MySQL, PHP and Javascripts (Figure 1C). A user-friendly web interface is provided to facilitate searching, browsing and downloading.
Querying meQTLs
We provide several entries for querying. Pancan-meQTL contains four datasets: cis-meQTLs, trans-meQTLs, survival-meQTLs and GWAS-meQTLs (Figure 2A and B). We developed four corresponding pages to display each dataset (Figure 2A). Users can enter each page from the browser bar to query SNPs, methylation data, and methylation-related genes of interest. On the ‘home’ page, we set a search section for users to query the four meQTL datasets. Also from the home page, users can search results by cancer type from the corresponding cancer type diagram. A quick search option is available on each page (top right) for searching by SNP ID, methylation probe or methylation-related gene symbol. Moreover, we provide a batch search page on the ‘Pancan-meQTLs’ page (Figure 2C). On this page, users can input multiple SNPs, methylation probes or genes of interest. A heatmap will show all the r values across all analyzed cancer types. For example, with inputs of ‘rs11047888’ and ‘rs4975682’, our results show that rs11044788 has significant correlations with cg11559192 and cg25763538 in 17 and 16 cancer types, respectively; whereas rs4975682 correlates with cg14565270 and cg03265642 in two and six cancer types, respectively (Figure 2D).
Figure 2.
Overview of Pancan-meQTL database. (A) Browser bar, with a quick search box on the right. (B) Four modules in Pancan-meQTL: cis-meQTLs, trans-meQTLs, survival-associated meQTLs and GWAS-related meQTLs. (C) Batch search box allows users to input multiple SNPs, methylation probes and genes of interest. (D) Example of heatmap in pancan-meQTLs page showing the correlation coefficient across cancer types. (E) Records of cis-meQTLs on cis-meQTLs page. (F) Example of meQTL boxplot. (G) Records of survival-meQTLs on survival-meQTLs page. (H) Example of a Kaplan–Meier plot on survival-meQTL page.
Querying on the cis/trans-meQTL page, a table with SNP ID, SNP genomic position, SNP alleles, methylation probe, methylation position, methylation-related gene symbol, gene position, beta value (effect size of SNP on gene expression), r value and P-value of meQTL will be returned (Figure 2E). For each record, a vector diagram of boxplot is provided to display the association between SNP genotypes and methylation levels (Figure 2F).
Querying on the survival-meQTL page, details with SNP ID, SNP genomic position, SNP alleles, log-rank test P-value and median survival times of different genotypes will be displayed (Figure 2G). A vector diagram of the KM plot is embedded in each record to display the association between SNP genotypes and overall survival times (Figure 2H).
Querying on the GWAS-meQTLs page will return the SNP information, related methylation, gene information and related GWAS traits. Search boxes are designed for retrieving specific cancer types, SNP, methylation probe and gene.
Downloading data and figures
All the cis/trans-meQTLs for each cancer type can be downloaded from the ‘Download’ page. The queried results can be downloaded from the query page by clicking the ‘Download’ button. The r values of the batch search can be downloaded from the ‘Pancan-meQTL’ page. The vector diagrams of the boxplot and KM plot can be downloaded from the cis/trans-meQTL and survival-meQTL pages, respectively.
Help section
The ‘Help’ page provides information for data collection, processing, result summary and contact. Pancan-meQTL welcomes any feedback by email to the address provided in the ‘Contact us’ section.
CONCLUSION
We developed Pancan-meQTL as a resource that provides millions of meQTLs in multiple cancer types. Pancan-meQTL is the first public database that focuses on cancer-specific meQTLs. Our comprehensive analyses across 23 cancer types provide a great opportunity to investigate the patterns of meQTLs among cancer types. Among meQTLs, we also identified abundant meQTLs associated with patient survival time or located in known GWAS loci. These meQTLs are potentially promising candidates for genetic research. The Pancan-meQTL database will be continually updated as large-scale genotypic and methylation data become available. As a comprehensive database that characterizes meQTLs across multiple cancer types, Pancan-meQTL will be valuable for improving the interpretation of cancer risk-associated SNPs identified in genetic studies. It represents an important resource for cancer and epigenetic research.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Xianchun Tu for helping design and debug the website. We thank LeeAnn Chastain for editorial assistance. We thank the support from the Cancer Prevention & Research Institute of Texas (CPRIT RR150085).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Prevention & Research Institute of Texas [RR150085 to L.H.]. Funding for open access charge: Cancer Prevention & Research Institute of Texas [RR150085].
Conflict of interest statement. None declared.
REFERENCES
- 1. Frazer K.A., Murray S.S., Schork N.J., Topol E.J.. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009; 10:241–251. [DOI] [PubMed] [Google Scholar]
- 2. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al. . The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L. et al. . The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014; 42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gong J., Liu C., Liu W., Wu Y., Ma Z., Chen H., Guo A.Y.. An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools. Database (Oxford). 2015; 2015:bav029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gong J., Tian J., Lou J., Wang X., Ke J., Li J., Yang Y., Gong Y., Zhu Y., Zou D. et al. . A polymorphic MYC response element in KBTBD11 influences colorectal cancer risk, especially in interaction with an MYC-regulated SNP rs6983267. Ann. Oncol. 2018; 29:632–639. [DOI] [PubMed] [Google Scholar]
- 6. Schubeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–326. [DOI] [PubMed] [Google Scholar]
- 7. Nebbioso A., Tambaro F.P., Dell’Aversana C., Altucci L.. Cancer epigenetics: moving forward. PLoS Genet. 2018; 14:e1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Hoesel A.Q., Sato Y., Elashoff D.A., Turner R.R., Giuliano A.E., Shamonki J.M., Kuppen P.J., van de Velde C.J., Hoon D.S.. Assessment of DNA methylation status in early stages of breast cancer development. Br J. Cancer. 2013; 108:2033–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li W., Li Q., Kang S., Same M., Zhou Y., Sun C., Liu C.C., Matsuoka L., Sher L., Wong W.H. et al. . CancerDetector: ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res. 2018; doi:10.1093/nar/gky423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heyn H. Quantitative trait loci identify functional noncoding variation in cancer. PLos Genet. 2016; 12:e1005826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sung H., Yang H.H., Zhang H., Yang Q., Hu N., Tang Z.Z., Su H., Wang L., Wang C., Ding T. et al. . Common genetic variants in epigenetic machinery genes and risk of upper gastrointestinal cancers. Int. J. Epidemiol. 2015; 44:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhernakova D.V., Deelen P., Vermaat M., van Iterson M., van Galen M., Arindrarto W., van ’t Hof P., Mei H., van Dijk F., Westra H.J. et al. . Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017; 49:139–145. [DOI] [PubMed] [Google Scholar]
- 13. Shi J., Marconett C.N., Duan J., Hyland P.L., Li P., Wang Z., Wheeler W., Zhou B., Campan M., Lee D.S. et al. . Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat. Commun. 2014; 5:3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J. et al. . Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLos Genet. 2010; 6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gong J., Mei S., Liu C., Xiang Y., Ye Y., Zhang Z., Feng J., Liu R., Diao L., Guo A.Y. et al. . PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 2018; 46:D971–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McClay J.L., Shabalin A.A., Dozmorov M.G., Adkins D.E., Kumar G., Nerella S., Clark S.L., Bergen S.E., Swedish Schizophrenia C., Hultman C.M. et al. . High density methylation QTL analysis in human blood via next-generation sequencing of the methylated genomic DNA fraction. Genome Biol. 2015; 16:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R.. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013; 8:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Genomes Project, C. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A. et al. . A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andrews S.V., Ellis S.E., Bakulski K.M., Sheppard B., Croen L.A., Hertz-Picciotto I., Newschaffer C.J., Feinberg A.P., Arking D.E., Ladd-Acosta C. et al. . Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat. Commun. 2017; 8:1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schulz H., Ruppert A.K., Herms S., Wolf C., Mirza-Schreiber N., Stegle O., Czamara D., Forstner A.J., Sivalingam S., Schoch S. et al. . Genome-wide mapping of genetic determinants influencing DNA methylation and gene expression in human hippocampus. Nat. Commun. 2017; 8:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. GTEx Consortium The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaunt T.R., Shihab H.A., Hemani G., Min J.L., Woodward G., Lyttleton O., Zheng J., Duggirala A., McArdle W.L., Ho K. et al. . Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016; 17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D.. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006; 38:904–909. [DOI] [PubMed] [Google Scholar]
- 25. Stegle O., Parts L., Piipari M., Winn J., Durbin R.. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012; 7:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ongen H., Andersen C.L., Bramsen J.B., Oster B., Rasmussen M.H., Ferreira P.G., Sandoval J., Vidal E., Whiffin N., Planchon A. et al. . Putative cis-regulatory drivers in colorectal cancer. Nature. 2014; 512:87–90. [DOI] [PubMed] [Google Scholar]
- 27. Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J. et al. . Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017; 49:131–138. [DOI] [PubMed] [Google Scholar]
- 28. Jaffe A.E., Gao Y., Deep-Soboslay A., Tao R., Hyde T.M., Weinberger D.R., Kleinman J.E.. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016; 19:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012; 28:1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O’Donnell C.J., de Bakker P.I.. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008; 24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.