Abstract
BitterDB (http://bitterdb.agri.huji.ac.il) was introduced in 2012 as a central resource for information on bitter-tasting molecules and their receptors. The information in BitterDB is frequently used for choosing suitable ligands for experimental studies, for developing bitterness predictors, for analysis of receptors promiscuity and more. Here, we describe a major upgrade of the database, including significant increase in content as well as new features. BitterDB now holds over 1000 bitter molecules, up from the initial 550. When available, quantitative sensory data on bitterness intensity as well as toxicity information were added. For 270 molecules, at least one associated bitter taste receptor (T2R) is reported. The overall number of ligand–T2R associations is now close to 800. BitterDB was extended to several species: in addition to human, it now holds information on mouse, cat and chicken T2Rs, and the compounds that activate them. BitterDB now provides a unique platform for structure-based studies with high-quality homology models, known ligands, and for the human receptors also data from mutagenesis experiments, information on frequently occurring single nucleotide polymorphisms and links to expression levels in different tissues.
INTRODUCTION
Bitter taste is one of the basic taste modalities, along with sweet, sour, salty and umami (1,2). The taste modalities are needed to interpret the nutritional value of food, and play a major role in food choice and intake. Bitter taste is recognized by a subfamily of G-protein coupled receptors (GPCRs), called the T2Rs or TAS2Rs. The number of subtypes varies in different species (from 3 in chicken to 50 in frog (3)). Even mouse and human have a different number of T2R subtypes, where orthologs are difficult or impossible to assign (4). The connection to the number (and breadth of tuning) of the bitter taste receptors in the species and the habitat of the species is among the questions explored in the chemosensory field of studies (5,6). Additional questions include the potential existence of different types of bitter taste, the level of overlap between the compounds recognized as bitter by different species, the quest for endogenous ligands for extra-orally expressed bitter taste receptors (7–10) and the chemical features associated with extreme bitterness. The data on bitter compounds and the associations with the receptors that were gathered in the BitterDB (11) enabled us (12) and others (13–15) to develop bitterness predictors. The BitterPredict bitterness predictor (12) has been used to evaluate bitterness of toxic compounds, leading to the conclusion that the link between bitterness and toxicity may not be as strong as previously suggested (16).
The evaluation of bitterness of molecules is important in drug discovery, particularly for pediatric drugs where the compliance of bitter-tasting drug is a major obstacle (17). Current practice mainly relies on human panels, BATA tests using rats and electronic tongues (18). Thus, comparison between human and rodent responses and analysis of physicochemical and molecular properties of bitterants is of great importance. Further development of in silico predictors has practical implications and holds potential to minimize the use of animals and to reduce costs (19).
The study of interactions between bitterants and their cognate T2Rs can provide additional tools for rational discovery of bitter taste agonists and antagonists. T2Rs have low similarity to Class A GPCRs and lack some of the typical Class A features (20). Nevertheless, modeling T2R structures on Class A templates have been successful when integrated with experimental techniques (21–23), and validated homology models are available in the current BitterDB edition.
Interestingly, multiple genetic polymorphisms in T2Rs, which affect individual bitter taste perception, exist in the population with high frequencies (24,25). The single nucleotide polymorphisms (SNPs) in human TAS2R38 gene have been widely investigated: differences in three amino acids determine the ability to taste the bitterness of the substances phenylthiocarbamide, 6-n-propylthiouracil (26) and other related compounds. Many additional functionally important SNPs are known (27). Combining the data on SNPs, site-directed mutagenesis and related T2Rs from other species provide a broad view on structure–activity of T2Rs.
BitterDB is currently the only freely electorally searchable repository, which holds bitter taste compounds and receptors. It is widely used by academia and industry as a handy electronic resource to check for compound bitterness, and obtains the relevant citations and activity data. BitterDB offers a large representative dataset of bitter compounds to employ in chemoinformatics analyses. The database is routinely updated with data from the literature and is open to uploads from users. In addition to the continuous growth, several new features have been added in the current BitterDB edition, as detailed below.
DATABASE ADDITIONS AND IMPROVEMENTS
Bitter compounds
BitterDB in 2019 contains 1041 compounds (increase of 2-fold from original BitterDB), including 50 short peptides, which were cited in the literature as bitter for human and/or were shown to activate at least one bitter taste receptor (in one of the four organisms—human, chicken, mouse or cat). The compounds were collected from over 100 publications (3-fold increase from previous version) that have been manually reviewed. The molecular properties, identifiers (SMILES, InChIKey, IUPAC name, CAS number and AA sequences for peptides), cross-links, bitter category (bittersweet, extremely bitter etc.) and several file formats for download as described in (11) are available for each compound. Experimental rat LD50 values extracted from the Acute Oral Toxicity Database (28) (see ‘Materials and Methods’ section) were added when available, as estimates for compounds’ toxicity.
In addition to the existing qualitative bitterness category, quantitative sensory data about compounds bitterness were added as a new feature. The quantitative sensory data were collected for more than 200 molecules from over 33 publications in which sensory tests to detect compound bitterness were performed. For such compounds, BitterDB now provides recognition threshold concentration, EC50 or recognition threshold concentration per kg of body weight and reference to the publication in which the sensory test was performed.
An important part of BitterDB is the associations between bitter compounds and their corresponding bitter taste receptors. BitterDB now holds over 800 interactions based on in vitro experimental activity measures such as EC50 or effective threshold, usually detected using HEK cells transfected to heterologously expressed T2Rs, where activation by ligands is monitored by calcium imaging (23,29) or other cell-based methods (30).
Finally, when available, BitterDB provides a link to the compound entry in PubChem (31), ZINC (32), DrugBank (33) and IUPHAR/BPS Guide to PHARMACOLOGY database (34).
Not only human
While originally the BitterDB included data related to human only, it is expanded now to additional species and includes data for chicken (Gallus gallus), mouse (Mus musculus) and cat (Felis catus) 3, 35 and 12 bitter taste receptors, respectively, and their currently known ligands (22 81 17 ligands, respectively). The BitterDB interface was updated to support the multi-species addition, and it offers the users two modes: organism-specific mode (human, mouse chicken or cat) and all-species mode. In the organism-specific mode, database functionally is reduced to the available data relevant for the selected organism only. For example, if the user switches to ‘mouse’ mode in the browse tables, only mouse bitter taste receptors or compounds that activate mouse bitter taste receptors will be shown.
Bitter taste receptors
For each receptor, the BitterDB provides known ligands, sequence, molecular weight (MW) and genome location as previously described (11). The current version also contains new features for structure-based studies:
3D homology models for the human, chicken and part of the more studied mouse receptors are now available for view and download. These models are a result of an extensive modeling integrated with experimental validations (6,22,23)). For the human bitter taste receptors, we added data about common SNPs. For each receptor, the related SNPs are summarized in a table with links to the relevant dbSNP page (35). Furthermore, we expanded the data regarding mutagenesis experiments from ∼20 publications. For each receptor that such data exist, we generated a table that summarizes the residues that were mutated and references to the related publications. The residues involved in SNPs or mutagenesis experiments are also marked on the 2D protein diagram.
The bitter taste receptors in BitterDB are cross-linked with UniProt (36), GPCRDB (37) and UCSC (38) (when possible). The human bitter receptors are linked also with the Human Protein Atlas (www.proteinatlas.org) (39), which provides important data about the tissues that express bitter taste receptors and to GeneCards (40).
Table 1 summarizes the increase in content and features in the current edition of BitterDB, in comparison to the 2012 version.
Table 1.
Comparison of data in original (2012) and new (2019) BitterDB versions
| BitterDB | 2012 | 2019 |
|---|---|---|
| # of bitter molecules | 551 | 1041 |
| # of molecules associated with at least one T2R | ∼100 | ∼260 |
| # of scientific publication used | ∼30 | ∼140 |
| # of species | 1 | 4 |
| # of receptors | 25 | 75 |
| # of ligand–receptor associations | 250 | >800 |
| # of 3D models | 0 | 43 |
| # of SNPs | 0 | 47 |
| # of receptors with data about mutagenesis | 6 | 11 |
| # of cross-linked databases for compounds | 1 | 4 |
| # of cross-linked databases for receptors | 2 | 5 |
| # of natural/synthetic/unassigned | 63/39/448 | 306/58/677 |
| # peptides | 0 | 51 |
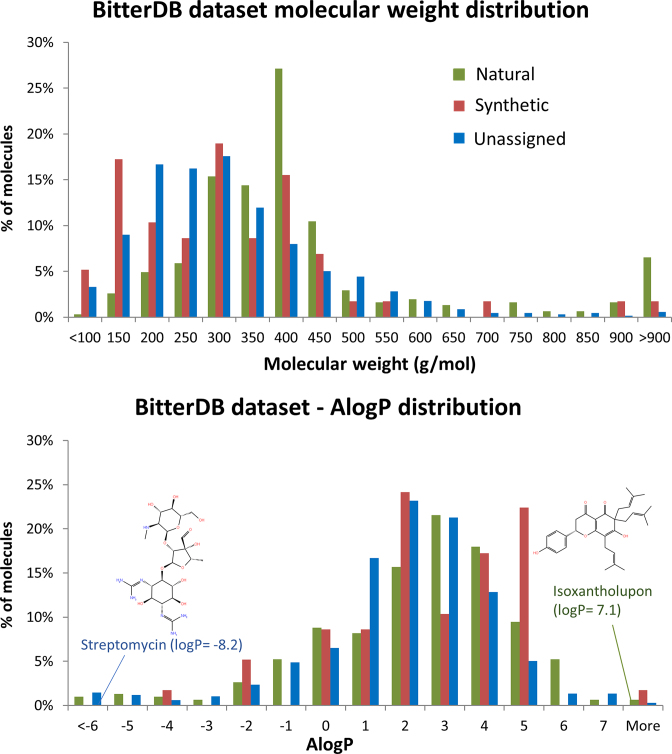
Since the data on natural and synthetic compounds have grown significantly, it is interesting to assess the distribution of physicochemical properties in these two groups (see Figure 1). The histograms indicate similar distribution of molecular weights and hydrophobicity values, as represented by AlogP. It should be noted that while most of the bitter compounds are hydrophobic, some polar bitter compounds (AlogP lower than −3) exist as well.
Figure 1.
Distribution of AlogP and molecular weight values for different subgroups in the current BitterDB dataset.
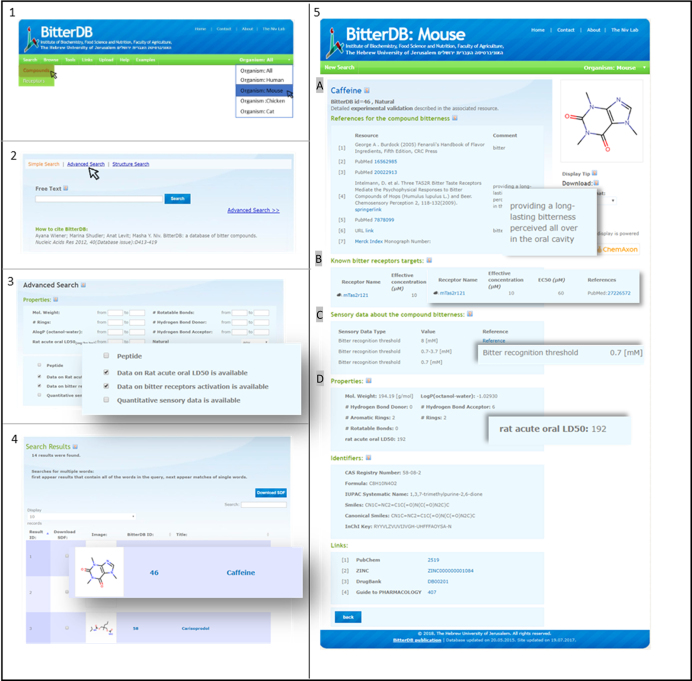
In Figure 2, we show a step-by-step example of a usage scenario. Specifically, in the main window we selected the specific organism (mouse) mode and chose search→compounds (panel 1). On the compound search form, we selected advanced search (panel 2). In the advanced search page, the options ‘data on Rat acute oral LD50 is available’ and ‘data on bitter receptors is available’ were chosen (panel 3). This search returned 14 molecules that fit the criteria. SDF file of these 14 molecules can be downloaded. From the results, caffeine was selected (panel 4). On caffeine entry page (panel 5), bitterness description, quantitative in vitro, sensory and toxicity data are shown in A till D, respectively.
Figure 2.
BitterDB possible usage: Panel 1+2: Compounds advanced search was chosen from menu, in mouse mode. Panel 3: In advanced search page, the options for compounds with specific data on rat acute oral LD50 and data on associated receptors were chosen. Panel 4: Search result panel, 14 compounds that fit the criteria were found, caffeine was selected for detailed view. Panel 5: Caffeine page in BitterDB: (A) Detailed description of the resources about caffeine bitterness and description of specific bitter taste when available. (B) Detailed data, including receptor activation measures from in vitro studies of associated receptor in mice (when moving to ‘All species’ mode, more targets will be shown). (C) Quantitative sensory data: detailed information about the sensory bitter recognition thresholds from three different publications is represented. (D) The toxicity measure (rat acute LD50) can be found in the compound properties.
MATERIALS AND METHODS
Compound data acquisition
The new compounds added to BitterDB, the bitter intensity sensory values and the ligand bitter receptors interaction details were extracted manually from dozens of publications.
When the exact molecule structure was available in a publication (including stereochemistry speciation), it was searched in PubChem (31), and SMILES representation of the molecule was extracted. When only the name of the compound appeared in a publication, the structural data SMILES representation was extracted from the PubChem entry with the same name. Molecules that were not found in PubChem were drawn manually using ChemSketch (ACD/Structure Elucidator, 2016.1.1, Advanced Chemistry Development, Inc., Toronto, ON, Canada, www.acdlabs.com, 2015) or using https://chemicalize.com ChemAxon (https:/www.chemaxon.com), and SMILES representation was generated using these tools. The SMILES representation of the bitter peptides was generated using the CycloPs server (41).
The new bitter compound data was collected from: (4,42–46). The data about ligand–bitter receptor interactions was collected for human bitter receptors activation from (4,42,44), for mouse bitter receptors activation from (4), for chicken bitter receptors activation from (3,22), and for cat bitter receptors activation from (47). The sensory bitter intensity data was collected from (43,45,46,48,49).
Compound identifiers, CAS number, Canonical SMILES, Isomeric SMILES, IUPAC name and InChIKey, were extracted from PubChem. For compounds that are not present in PubChem, the identifiers were computed using RDKit (Open-source cheminformatics; http://www.rdkit.org). Physicochemical properties (MW, AlogP, H-bond donor, H-bond acceptor, number of rings and number of aromatic rings) were calculated using RDKit. SDF files available for download were also generated using RDKit.
The Acute Oral Toxicity (rat acute oral LD50) values, where extracted using the OECD QSAR Toolbox v3.4.0.17 (50), and Acute Oral Toxicity Database (28) as described in (16). The toxicity LD50 value is represented as ‘mg/kg bw’, meaning amount of compound in milligrams for each kilogram body weight of exposed subject.
Conversion between database IDs was done using the Fiehnlab conversion tool (http://cts.fiehnlab.ucdavis.edu/batch).
Receptors data acquisition
The receptors’ numbering in the receptor pages titles appears as in the reference publication about bitter receptors activation in that species, namely, human (29), mouse (4), cat (47) and chicken (3). Receptors sequences and properties were extracted from UniProt (36) and UCSC (38). For cat (Felis catus), due to the lack of data or inconsistency in the different protein databases, sequences were extracted from (47) and searched using NCBI protein BLAST (51) to be linked with protein entries in UniProt and NCBI RefSeq (52). Gene names and genome locations were taken from NCBI’s Genome Data Viewer (version 4.4).
Homology Modeling: hT2R14 receptor model generated by (53) was used as a template for modeling all human and mouse bitter taste receptors loaded in the database, whereas the chicken bitter taste receptors were prepared using the ggT2R1structure modeled and validated in (6,22) as template.
SNPs: The SNP data for the bitter receptors were extracted from dbSNP (35). Only SNPs with Minor Allele Frequency values >0.05, considered commonly occurring, were added to BitterDB.
Tools
The Alignment page was updated (http://bitterdb.agri.huji.ac.il/dbbitter.php#ReceptorAlignment) to contain a Multiple Sequence Alignment (MSA) of the 25 human bitter taste receptors. This MSA was generated using structural information from 11 sequence distinct Class A GPCRs, for which a crystal structure has been solved. The 25 human bitter taste receptors and the 11 Class A GPCRs were aligned using the PROMALS3D webserver (54), which allows construction of MSAs for distantly related proteins using 3D structural information.
The BitterPredict page shares data and results of our recent studies performed using BitterDB. The code for BitterPredict, an machine learning classifier, which predicts whether a compound is bitter or not based on its chemical structure, can be download. BitterPredict prediction results on different datasets (ZINC, drugBank, ChEBI and FooDB) as well as the external validation sets used in (12) can be explored or downloaded.
The BitterToxic page shares links and data related to our recent study that explored the connection between bitterness and toxicity (16).
Visual improvements and updates
2D images for BitterDB 2.0 were generated using: http://hulab.rxnfinder.org/smi2img/. Homology model visualization is provided with NGL Viewer (55) and the alignment is presented using BioJS (56).
SUMMARY AND OUTLOOK
The number of bitter molecules can now be estimated as tens of thousands (16), harboring dramatic structural diversity. All these molecules are recognized by a limited number of bitter taste receptors. The primary goal in established BitterDB was to create a platform that provides a realistic overview of the nature of the numerous bitter molecules, as well as molecular features of specific bitter taste receptors.
The extended literature search that was performed for the updated edition of BitterDB is a significant step toward illuminating the sensory chemical space. The current knowledge may help design rational experiments, in which compounds in under-studied regions of the chemical space may be tested.
To facilitate this goal, future development of BitterDB will involve organization within BitterDB by connecting between similar molecules, investigating specific chemical families and gathering deeper knowledge on bitter compounds stereochemistry.
We encourage the scientific community to contribute to BitterDB the structures of the compounds found to activate T2Rs, which can be done via the ‘upload’ link or by direct correspondence with the BitterDB team.
Over 19 000 of users, with ∼300 new users added per month are routinely using BitterDB (see Google Analytics Report http://bitterdb.agri.huji.ac.il/additionalFiles/BitterDB_Report_7_years.pdf). The current edition will help these and new users, from academia and from industry, to carry out cross-species comparative studies and structure-based studies, as well as develop tools for bitterness prediction and modification. Due to expression of bitter receptors in extra-oral tissues and increasing interest in these receptors as novel drug targets, a wealth of information is likely to be obtained by groups working in both food-related and drug discovery related fields.
Fundamental questions as well as potential therapeutic applications of the modulation of bitter taste receptors continue to motivate further development of the BitterDB database. Future extensions will include data from new publications (such as more 3D receptor structure models and more mutagenesis data), inclusion of additional species, information on bitter taste maskers and inhibitors, classifications of compounds based on chemotypes, spectroscopic data on compounds and more.
ACKNOWLEDGEMENTS
The authors thank Rina Moyal, Rita Zlotnikov and Gregory Pasternak for their contribution to the development of BitterDB website, John Irwin for his help with connection between BitterDB and the ZINC database and Louise Slade for helpful discussions.
FUNDING
Israel Science Foundation [ISF 494/16, ISF-NSFC 2463/16 to M.Y.N.]; Lady Davis Fellowship (to A.D.P.); PBC Fellowship (to M.S.B.). Funding for open access charge: Israel Science Foundation [ISF-NSFC 2463/16 to M.Y.N.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chandrashekar J., Hoon M.A., Ryba N.J.P., Zuker C.S.. The receptors and cells for mammalian taste. Nature. 2006; 444:288–294. [DOI] [PubMed] [Google Scholar]
- 2. Chaudhari N., Roper S.D.. The cell biology of taste. J. Cell Biol. 2010; 190:285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Behrens M., Korsching S.I., Meyerhof W.. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol. Biol. Evol. 2014; 31:3216–3227. [DOI] [PubMed] [Google Scholar]
- 4. Lossow K., Hubner S., Roudnitzky N., Slack J.P., Pollastro F., Behrens M., Meyerhof W.. Comprehensive analysis of mouse bitter taste receptors reveals different molecular receptive ranges for orthologous receptors in mice and humans. J. Biol. Chem. 2016; 291:15358–15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Behrens M., Meyerhof W.. Vertebrate bitter taste receptors: keys for survival in changing environments. J. Agric. Food Chem. 2018; 66:2204–2213. [DOI] [PubMed] [Google Scholar]
- 6. Xue A.Y., Di Pizio A., Levit A., Yarnitzky T., Penn O., Pupko T., Niv M.Y.. Independent evolution of strychnine recognition by bitter taste receptor subtypes. Front. Mol. Biosci. 2018; 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jaggupilli A., Howard R., Upadhyaya J.D., Bhullar R.P., Chelikani P.. Bitter taste receptors: novel insights into the biochemistry and pharmacology. Int. J. Biochem. Cell Biol. 2016; 77:184–196. [DOI] [PubMed] [Google Scholar]
- 8. Shaik F.A., Singh N., Arakawa M., Duan K., Bhullar R.P., Chelikani P.. Bitter taste receptors: extraoral roles in pathophysiology. Int. J. Biochem. Cell Biol. 2016; 77:197–204. [DOI] [PubMed] [Google Scholar]
- 9. Behrens M., Meyerhof W.. Gustatory and extragustatory functions of mammalian taste receptors. Physiol. Behav. 2011; 105:4–13. [DOI] [PubMed] [Google Scholar]
- 10. Avau B., Depoortere I.. The bitter truth about bitter taste receptors: beyond sensing bitter in the oral cavity. Acta Physiol. 2016; 216:407–420. [DOI] [PubMed] [Google Scholar]
- 11. Wiener A., Shudler M., Levit A., Niv M.Y.. BitterDB: a database of bitter compounds. Nucleic Acids Res. 2012; 40:D413–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dagan-Wiener A., Nissim I., Ben Abu N., Borgonovo G., Bassoli A., Niv M.Y.. Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 2017; 7:12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang W., Shen Q., Su X., Ji M., Liu X., Chen Y., Lu S., Zhuang H., Zhang J.. BitterX: a tool for understanding bitter taste in humans. Sci. Rep. 2016; 6:23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng S., Jiang M., Zhao C., Zhu R., Hu Z., Xu Y., Lin F.. e-Bitter: Bitterant prediction by the consensus voting from the machine-learning methods. Front. Chem. 2018; 6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banerjee P., Preissner R.. BitterSweetForest: a random forest based binary classifier to predict bitterness and sweetness of chemical compounds. Front. Chem. 2018; 6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nissim I., Dagan-Wiener A., Niv M.Y.. The taste of toxicity: a quantitative analysis of bitter and toxic molecules. IUBMB Life. 2017; 69:938–946. [DOI] [PubMed] [Google Scholar]
- 17. Mennella J.A., Spector A.C., Reed D.R., Coldwell S.E.. The bad taste of medicines: overview of basic research on bitter taste. Clin. Ther. 2013; 35:1225–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohamed-Ahmed A.H., Soto J., Ernest T., Tuleu C.. Non-human tools for the evaluation of bitter taste in the design and development of medicines: a systematic review. Drug Discov. Today. 2016; 21:1170–1180. [DOI] [PubMed] [Google Scholar]
- 19. Bahia M.S., Nissim I., Niv MY.. Bitterness prediction in-silico: a step towards better drugs. Int. J. Pharm. 2018; 536:526–529. [DOI] [PubMed] [Google Scholar]
- 20. Di Pizio A., Levit A., Slutzki M., Behrens M., Karaman R., Niv M.Y.. Comparing Class A GPCRs to bitter taste receptors: Structural motifs, ligand interactions and agonist-to-antagonist ratios. Methods Cell Biol. 2016; 132:401–427. [DOI] [PubMed] [Google Scholar]
- 21. Yarnitzky T., Levit A., Niv M.Y.. Homology modeling of G-protein-coupled receptors with X-ray structures on the rise. Curr. Opin. Drug Discov. Dev. 2010; 13:317–325. [PubMed] [Google Scholar]
- 22. Di Pizio A., Kruetzfeldt L.M., Cheled-Shoval S., Meyerhof W., Behrens M., Niv M.Y.. Ligand binding modes from low resolution GPCR models and mutagenesis: chicken bitter taste receptor as a test-case. Sci. Rep. 2017; 7:8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nowak S., Di Pizio A., Levit A., Niv M.Y., Meyerhof W., Behrens M.. Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochim. Biophys. Acta. 2018; 1862:2162–2173. [DOI] [PubMed] [Google Scholar]
- 24. Roudnitzky N., Behrens M., Engel A., Kohl S., Thalmann S., Hubner S., Lossow K., Wooding S.P., Meyerhof W.. Receptor polymorphism and genomic structure interact to shape bitter taste perception. PLoS Genet. 2015; 11:e1005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pronin A.N., Xu H., Tang H., Zhang L., Li Q., Li X.. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr. Biol. 2007; 17:1403–1408. [DOI] [PubMed] [Google Scholar]
- 26. Bufe B., Breslin P.A., Kuhn C., Reed D.R., Tharp C.D., Slack J.P., Kim U.K., Drayna D., Meyerhof W.. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 2005; 15:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hayes J.E., Wallace M.R., Knopik V.S., Herbstman D.M., Bartoshuk L.M., Duffy V.B.. Allelic variation in TAS2R bitter receptor genes associates with variation in sensations from and ingestive behaviors toward common bitter beverages in adults. Chem. Senses. 2011; 36:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomasulo P. ChemIDplus-super source for chemical and drug information. Med. Ref. Serv. Q. 2002; 21:53–59. [DOI] [PubMed] [Google Scholar]
- 29. Meyerhof W., Batram C., Kuhn C., Brockhoff A., Chudoba E., Bufe B., Appendino G., Behrens M.. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2010; 35:157–170. [DOI] [PubMed] [Google Scholar]
- 30. Riedel K., Sombroek D., Fiedler B., Siems K., Krohn M.. Human cell-based taste perception - a bittersweet job for industry. Nat. Prod. Rep. 2017; 34:484–495. [DOI] [PubMed] [Google Scholar]
- 31. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sterling T., Irwin J.J.. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 2015; 55:2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harding S.D., Sharman J.L., Faccenda E., Southan C., Pawson A.J., Ireland S., Gray A.J.G., Bruce L., Alexander S.P.H., Anderton S. et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to immunopharmacology. Nucleic Acids Res. 2018; 46:D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. The UniProt, C UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pandy-Szekeres G., Munk C., Tsonkov T.M., Mordalski S., Harpsoe K., Hauser A.S., Bojarski A.J., Gloriam D.E.. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018; 46:D440–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 40. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. et al. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 41. Duffy F.J., Verniere M., Devocelle M., Bernard E., Shields D.C., Chubb A.J.. CycloPs: generating virtual libraries of cyclized and constrained peptides including nonnatural amino acids. J. Chem. Inf. Model. 2011; 51:829–836. [DOI] [PubMed] [Google Scholar]
- 42. Roland W.S., van Buren L., Gruppen H., Driesse M., Gouka R.J., Smit G., Vincken J.P.. Bitter taste receptor activation by flavonoids and isoflavonoids: modeled structural requirements for activation of hTAS2R14 and hTAS2R39. J. Agric. Food Chem. 2013; 61:10454–10466. [DOI] [PubMed] [Google Scholar]
- 43. Stark T., Hofmann T.. Structures, sensory activity, and dose/response functions of 2,5-diketopiperazines in roasted cocoa nibs (Theobroma cacao). J. Agric. Food Chem. 2005; 53:7222–7231. [DOI] [PubMed] [Google Scholar]
- 44. Brockhoff A., Behrens M., Massarotti A., Appendino G., Meyerhof W.. Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium. J. Agric. Food Chem. 2007; 55:6236–6243. [DOI] [PubMed] [Google Scholar]
- 45. Dresel M., Dunkel A., Hofmann T.. Sensomics analysis of key bitter compounds in the hard resin of hops (Humulus lupulus L.) and their contribution to the bitter profile of Pilsner-type beer. J. Agric. Food Chem. 2015; 63:3402–3418. [DOI] [PubMed] [Google Scholar]
- 46. Dresel M., Vogt C., Dunkel A., Hofmann T.. The bitter chemodiversity of hops (Humulus lupulus L.). J. Agric. Food Chem. 2016; 64:7789–7799. [DOI] [PubMed] [Google Scholar]
- 47. Lei W., Ravoninjohary A., Li X., Margolskee R.F., Reed D.R., Beauchamp G.K., Jiang P.. Functional analyses of bitter taste receptors in domestic cats (Felis catus). PLoS One. 2015; 10:e0139670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Intelmann D., Batram C., Kuhn C., Haseleu G., Meyerhof W., Hofmann T.. Three TAS2R bitter taste receptors mediate the psychophysical responses to bitter compounds of hops (Humulus lupulus L.) and beer. Chemosens Percept. 2009; 2:118–132. [Google Scholar]
- 49. Degenhardt A.G., Hofmann T.. Bitter-tasting and kokumi-enhancing molecules in thermally processed avocado (Persea americana Mill.). J. Agric. Food Chem. 2010; 58:12906–12915. [DOI] [PubMed] [Google Scholar]
- 50. Dimitrov S.D., Diderich R., Sobanski T., Pavlov T.S., Chankov G.V., Chapkanov A.S., Karakolev Y.H., Temelkov S.G., Vasilev R.A., Gerova K.D. et al. QSAR Toolbox - workflow and major functionalities. SAR QSAR Environ. Res. 2016; 27:203–219. [DOI] [PubMed] [Google Scholar]
- 51. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L.. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008; 36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thawabteh A., Lelario F., Scrano L., Bufo S.A., Nowak S., Behrens M., Di Pizio A., Niv M.Y., Karaman R.. Bitterless guaifenesin prodrugs – design, synthesis, characterization, in vitro kinetics and bitterness studies. Chem. Biol. Drug Des. 2018; doi:10.1111/cbdd.13409. [DOI] [PubMed] [Google Scholar]
- 54. Pei J., Kim B.H., Grishin N.V.. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008; 36:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rose A.S., Bradley A.R., Valasatava Y., Duarte J.M., Prlic A., Rose P.W.. NGL Viewer: Web-based molecular graphics for large complexes. Bioinformatics. 2018; doi:10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yachdav G., Wilzbach S., Rauscher B., Sheridan R., Sillitoe I., Procter J., Lewis S.E., Rost B., Goldberg T.. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics. 2016; 32:3501–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]