Abstract
Advances in proteomics and sequencing have highlighted many non-annotated open reading frames (ORFs) in eukaryotic genomes. Genome annotations, cornerstones of today's research, mostly rely on protein prior knowledge and on ab initio prediction algorithms. Such algorithms notably enforce an arbitrary criterion of one coding sequence (CDS) per transcript, leading to a substantial underestimation of the coding potential of eukaryotes. Here, we present OpenProt, the first database fully endorsing a polycistronic model of eukaryotic genomes to date. OpenProt contains all possible ORFs longer than 30 codons across 10 species, and cumulates supporting evidence such as protein conservation, translation and expression. OpenProt annotates all known proteins (RefProts), novel predicted isoforms (Isoforms) and novel predicted proteins from alternative ORFs (AltProts). It incorporates cutting-edge algorithms to evaluate protein orthology and re-interrogate publicly available ribosome profiling and mass spectrometry datasets, supporting the annotation of thousands of predicted ORFs. The constantly growing database currently cumulates evidence from 87 ribosome profiling and 114 mass spectrometry studies from several species, tissues and cell lines. All data is freely available and downloadable from a web platform (www.openprot.org) supporting a genome browser and advanced queries for each species. Thus, OpenProt enables a more comprehensive landscape of eukaryotic genomes’ coding potential.
INTRODUCTION
An ever-increasing number of studies relate the discovery of functional yet non-annotated open reading frames (ORFs) across eukaryotic genomes (1–8). These are usually small ORFs encoded in currently annotated non-coding RNAs (ncRNAs) (9–11). However, a substantial number are present in mRNAs, either overlapping the CDS or within the 5′ or 3′ ‘untranslated’ regions (UTRs) (6,12–17). They have been found involved in numerous cellular functions, from calcium or insulin regulation to mitochondrial biogenesis (6,7,10,11,16). These examples highlight both the underestimation of coding potential in eukaryotic genomes relayed by current annotations, and the polycistronic nature of eukaryotic genes (6). Since genome annotations lay the foundation for proteomics and sequencing explorations, such underestimation has consequences on most of today's research.
Recent efforts for a more comprehensive view of eukaryotic genomes’ coding potential have focused on annotation of small ORFs, defined as any ORF between 10 and 100 codons, alongside associated evidence from conservation, ribosome profiling and/or mass spectrometry (18–20). Yet, these databases suffer limitations, notably a maximum length threshold that forbids detection of ORFs longer than 100 codons, and they do not account for the polycistronic nature of eukaryotic genomes. In parallel, proteogenomics strategies are emerging to offer an unbiased approach to the study of eukaryotic proteomes, yet they remain the expertise of a few and still depend on sample preparation adapted to the identification of small proteins (21–24). Despite these significant studies, we still lack a systematic approach to fathom the deepest parts of eukaryotic proteomes.
Here, we present OpenProt (www.openprot.org), the first database upholding a polycistronic model of eukaryotic genes to date. OpenProt distinguishes three ORF categories: already annotated ones (RefProts), novel RefORF isoforms (Isoforms, II_ accessions) and novel alternative ORFs (AltProts, IP_ accessions). We define as AltProt the product of any unannotated ORF, anywhere on transcripts (ncRNAs and mRNAs), that do not display protein sequence similarity with a RefProt from the same gene (otherwise categorized as novel isoform: product from an unannotated ORF with a significant sequence similarity to a RefProt from the same gene). OpenProt currently offers deep annotation for 10 species, cumulating supporting evidence of protein orthology, translation and expression. Moreover, through custom downloads and a user-friendly web platform, OpenProt enables wide applications, making this ‘hidden’ proteome easily accessible to the wider scientific community. OpenProt thus aims to foster discoveries of functional yet currently non annotated proteins.
MATERIALS AND METHODS
Open reading frames (ORFs) prediction
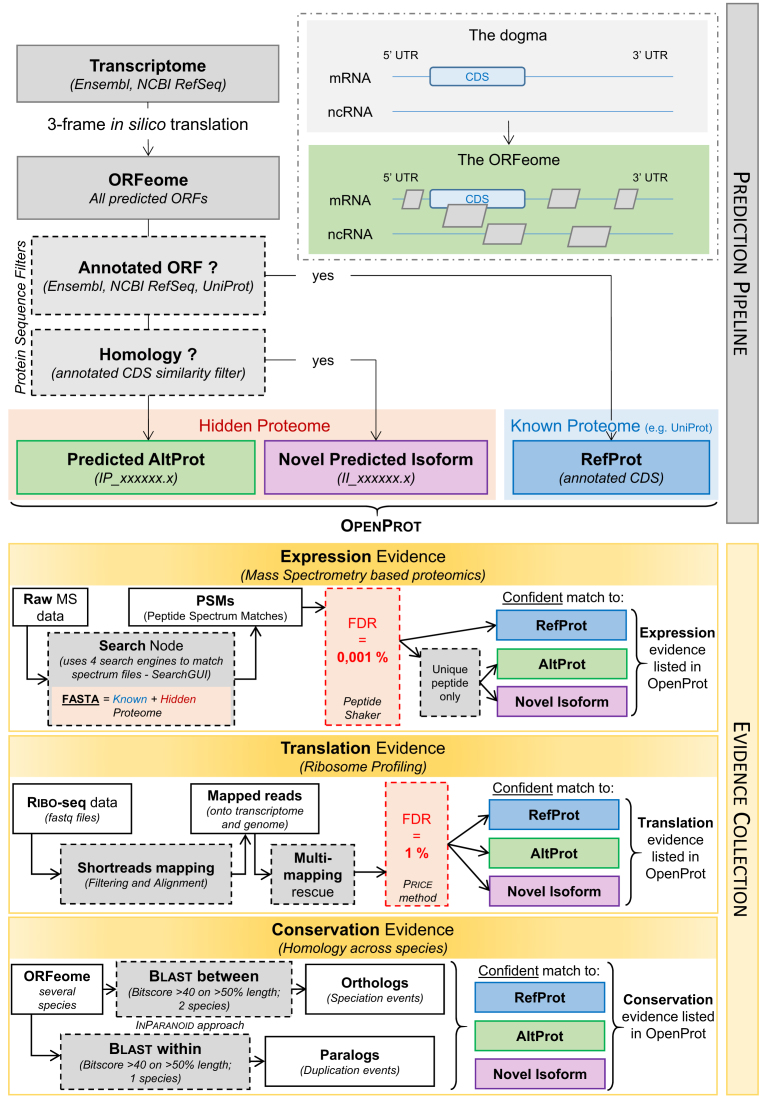
The first step of OpenProt pipeline is the ORF prediction (Figure 1). First, we retrieve an exhaustive transcriptome by combining two well-used annotations (NCBI RefSeq (25) and Ensembl (26)). Annotations overlap is not whole because of variations in algorithms and information sources. In a context of exploration and discovery, a more complex annotation is preferable (27). Hence, we retrieve NCBI RefSeq and Ensembl annotations and compile them into a more exhaustive one. For example in human, NCBI RefSeq (GRCh38.p7) contains 109 077 mRNAs and 29 484 ncRNAs, while Ensembl (GRCh38.83) contains 93 855 mRNAs and 105 150 ncRNAs; only 7578 RNAs are common to both annotations. The source annotation is associated with each ORF prediction so that users can look at predictions from either annotation alone if preferred. Genome assemblies and annotation releases currently supported by OpenProt are listed for each species in Table 1. We then perform a 3-frames in silico translation using EMBOSS Transeq (28) to predict all possible ORFs with an ATG start codon and a minimum length of 30 codons. This constitutes the OpenProt ORFeome.
Figure 1.
OpenProt pipeline graphical representation. OpenProt pipeline contains two main features: prediction and evidence collection. OpenProt enforces a polycistronic model of eukaryotic genes contrary to the actual dogma of one CDS per transcript. The protein sequence similarity filter (Homology) holds two arguments as described in the material and methods. The hidden proteome consists of currently non-annotated ORFs highlighted by OpenProt. These ORFs are either novel isoforms of known CDS (II_accessions) or novel alternative proteins (IP_ accessions). All evidence collection parameters are described in the material and methods section.
Table 1.
OpenProt (1.3) prediction pipeline output
| Annotations | ORFeome (both annotations) | ||||||
|---|---|---|---|---|---|---|---|
| Species | Genome assembly | NCBI RefSeq | Ensembl | Total # | Ref # | II_ # | IP_ # |
| Homo sapiens | GRCh38.p5 | GRCh38.p7 | GRCh38.83 | 646 403 | 129 888 | 55 053 | 461 462 |
| Pan troglodytes | CHIMP2.1.4 | CHIMP2.1.4 | CHIMP2.1.4.87 | 227 950 | 37 059 | 16 402 | 174 489 |
| Mus musculus | GRCm38.p4 | GRCm38.p4 | GRCm38.84 | 486 198 | 82 477 | 30 220 | 373 501 |
| Rattus norvegicus | Rnor_6.0 | Rnor_6.0 | Rnor_6.0.84 | 289 077 | 51 423 | 6718 | 230 936 |
| Bos taurus | UMD_3.1 | UMD_3.1 | UMD_3.1.86 | 220 483 | 49 026 | 6942 | 164 515 |
| Ovis aries | Oar_v3.1 | Oar_v3.1 | Oar_v3.1.89 | 340 974 | 40 000 | 19 022 | 281 952 |
| Danio rerio | GRCz10 | GRCz10 | GRCz10.84 | 257 534 | 56 247 | 14 523 | 186 764 |
| Drosophila melanogaster | Release 6 plus ISO1 MT | BDGP6 | BDGP6.84 | 97 934 | 22 204 | 2148 | 73 582 |
| Caenorhabditis elegans | WBcel235 | WBcel235 | WBcel235.84 | 94 087 | 29 563 | 2340 | 62 184 |
| Saccharomyces cerevisiae S288c | R64 | R64 | R64.83 | 16 865 | 6613 | 28 | 10 224 |
Ref = currently annotated protein (RefProt), II_ = novel isoforms of known protein, IP_ = novel protein from alternative ORF (AltProt).
ORF product classification: reference proteins, novel isoforms, and alternative proteins
This exhaustive ORFeome is then filtered using NCBI RefSeq, Ensembl and UniProt (29) protein entries to identify annotated proteins, called RefProts (Figure 1). We add UniProt entries at this level as UniProt contains proteins with supporting experimental evidence that are not present in either NCBI RefSeq or Ensembl annotations. For example in human, 11,860 proteins annotated in UniProt (UniProtKB-SwissProt, 2017-09-27) are not present in either NCBI RefSeq or Ensembl annotations. Once known ORFs are filtered out (RefProt category), we are left with currently unannotated ORFs. A similarity filter is then implemented to identify and annotate Novel RefProt Isoforms (Figure 1). The similarity filter targets ORFs from a same gene and contains two arguments: (a) over 80% of protein sequence identity over 50% of the length (Basic Local Alignment Search Tool (BLAST) (30)), and (b) identical genomic coordinates of start or end codon with a protein sequence identity (EMBOSS Matcher PAM10 matrix score ≤ 100) over 20% of the length (28). If any of the similarity filter argument is met, the unannotated ORF is categorized as a novel predicted isoform (II_ accessions). The leftover predicted ORFs constitute the alternative proteins (AltProt category, IP_ accessions) and can be summarized by a simple equation: AltORFs = ORFeome - RefORFs - Novel Isoforms. All predictions for each species are present on the OpenProt website and can be downloaded, queried or visualized using the Genome Browser. A comprehensive guide for all three is provided under the Help section of the OpenProt website (www.openprot.org/p/help).
Mass spectrometry data analysis pipeline
In order to gather protein expression evidence, OpenProt retrieves publicly available mass spectrometry (MS) based proteomics studies from ProteomeXchange (31), PRIDE archive (32) and collaborators. Such studies are re-analyzed using the OpenProt protein FASTA containing all RefProts, AltProts and Isoforms. The OpenProt MS pipeline was developed using PeptideShaker software (version 1.13.4) (33) configured to systematically run 4 search engines on raw MS files (X!Tandem, MS-GF+, Comet and OMSSA) via SearchGUI (version 3.1.0) (34). SearchGUI general parameters were set as previously described and then individually inferred based on studies specifics (7). Classical MS analyses use a false discovery rate (FDR) of 1%. However, adding all AltProts and Isoforms leads to a substantial increase of the search space (about six times bigger for human). To only account for highly confident identifications, we set the FDR at 0.001% (Figure 1). Initial validations included: (a) 80% minimum overlap of RefProts identifications with the original MS study, and (b) manual validation of randomly selected spectra (supporting materials S1). Admittedly, false positives may still be and we strongly encourage seeking ORFs with evidence across multiple datasets as false-positive identifications would differ across datasets. Moreover, a novel predicted protein (AltProt or Isoform) will be identified only if it is recognized by a unique peptide. In the case where a peptide matches a novel predicted protein and a RefProt, it will always be assigned to the RefProt only. The identification results are then implemented to the OpenProt database and can be downloaded or queried from the Genome Browser or the Search page. More information can be found on the web platform Help page (www.openprot.org/p/help) and in supporting materials S1.
Ribosome profiling data analysis pipeline
In order to gather ORF translation evidence, OpenProt retrieves publicly available ribosome profiling (Ribo-seq) data. Ribosomal footprints raw data are re-analyzed using the PRICE workflow (version 1.0.2) (35). PRICE is an entropy-based model for identification of translated ORFs from Ribo-seq data. PRICE is run with default parameters (except for the FDR) using the rescue mode (Figure 1), and fed with both NCBI RefSeq and Ensembl annotations (run separately). Briefly, reads mapping to ribosomal RNAs are filtered out and remaining footprints are mapped onto the genome and transcriptome with up to three mismatches (35). Multi-mapped reads are fractionated across all possible sites unless uniquely mapped reads to nearby loci allow confident identification of the footprint coordinates (35). PRICE reconstitutes the set of codons most likely to give the observed reads, creating a list of ORF candidates. These are filtered according to a stringent 1% FDR (usually set at 10%) to focus on highly confident translation events (35). The identification results are then implemented to the OpenProt database and can be downloaded or queried from the Genome Browser or the Search page. More information can be found on the web platform Help page (www.openprot.org/p/help) and in supporting materials S2.
Conservation analysis pipeline
In order to gather protein conservation evidence, OpenProt computes orthology relationships from the 10 currently supported species. Protein sequence homology is evaluated using an InParanoid-like approach and separates orthologs (homologous sequences from different species) from paralogs (homologous sequences from the same species but different genes) (36). To identify orthologs, protein sequences from two different species are compared using an all-vs-all BLAST (36). For example, all protein sequences from Homo sapiens are BLAST searched against all protein sequences from Pan troglodytes. All orthology relationships identified are available on OpenProt (one-to-one; one-to-many; many-to-one and many-to-many). In parallel, the same pipeline is run within one species to identify paralogs (Figure 1). OpenProt uses a significance filter set at a bitscore of 40 for an overlap over 50% of the query sequence, as previously published (7, 37). The results are then implemented to the OpenProt database and can be downloaded or queried from the Search page. More information can be found on the web platform Help page (www.openprot.org/p/help) and in supporting materials S3.
Protein functional domain prediction
All predicted proteins are also scanned to identify known functional domains. All protein sequences are run through the InterProScan algorithm (version 5.14–53.0) using the default parameters (38). Domain predictions as well as gene ontology (GO) and pathway annotations are reported if significant (e-value < 10−3). The results are inserted into the OpenProt database and can be downloaded or queried from the Search page.
Database and website development
All data are generated using in-house Perl (version 5.18.2) and Python (version 2.7.6) scripts and stored in a PostgreSQL database (version 9.6). All re-analyzed mass spectrometry and ribosome profiling studies are accessible from the Help page (supporting materials S1 and S2). The OpenProt web platform was built using the Flask framework (version 1.0.2) and developed using HTML, SQL and JavaScript.
DATABASE CONTENT AND USER INTERFACE
ORFeome: numbers and classification
ORF predictions can be queried from either one or both of the annotations or both from the search or browser page (Figure 2A). Numbers of identified RefProts, AltProts and Isoforms are displayed in Table 1 for each species. For example in human to date, OpenProt (release 1.3) predicts 55,053 novel isoforms. These are currently non-annotated in NCBI RefSeq or Ensembl, they originate from transcripts present in current annotations and display significant similarity with the gene annotated CDS (see similarity filter arguments).
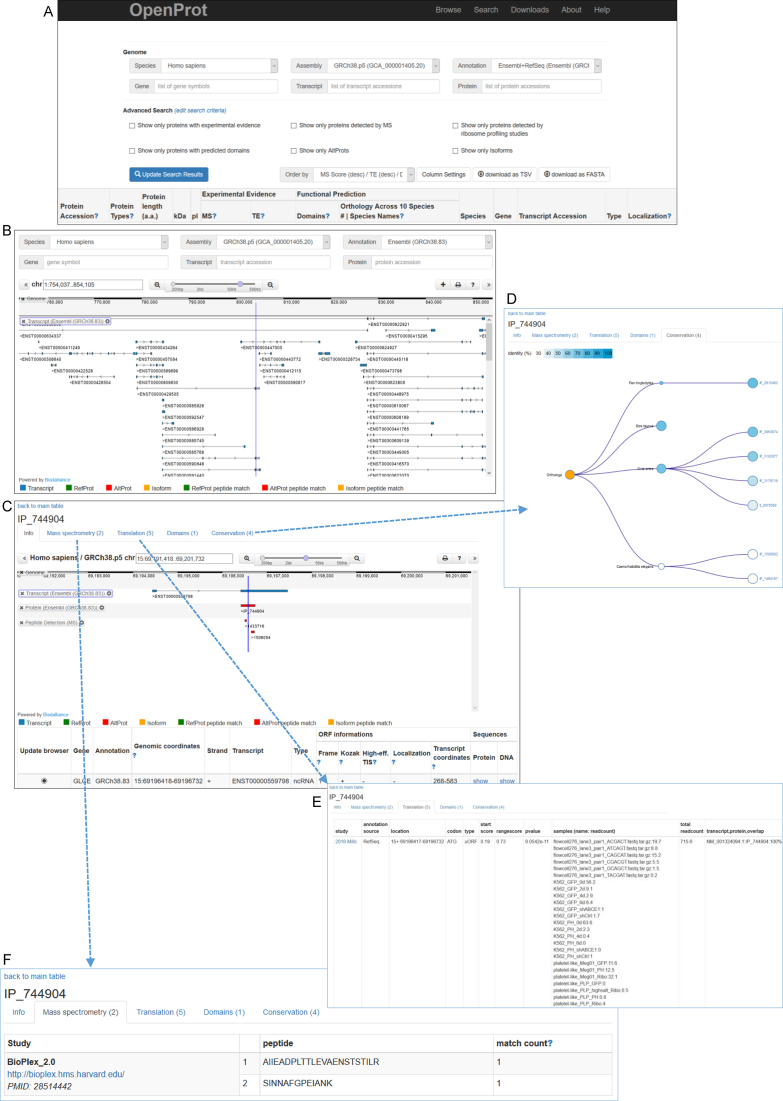
Figure 2.
OpenProt features. (A) OpenProt Search page. All OpenProt pages are accessible from the top menu. The Search page displays advanced query settings (species, annotation, gene, transcript and protein) and allows to filter according to supporting evidence and other characteristics. The result table displays the main protein characteristics, supporting evidence, and link to a Details page (shown in C). A getting started tutorial from the Search page is available from the help menu (www.openprot.org/p/help). (B) OpenProt Genome Browser page. The Browser page displays advanced query settings (species, annotation, gene, transcript, protein and genomic coordinates) and a genome browser with customable tracks. By default, the genome browser shows tracks of transcripts, proteins and peptide detection (from re-analyzed mass spectrometry studies). A getting started tutorial from the Browser page is available from the help menu (www.openprot.org/p/help). (C) Each ORF has an individual ‘Details’ page. The page holds 5 tabs, the first one shown here displays information such as genomic and transcript coordinates, and other characteristics. Protein and DNA sequences are available from the details link. (D) The ‘Conservation’ tab displays the ORF orthologs and paralogs. The size of the nodes represents the number of related ORFs and the colour shows the identity percentage. The conservation score (top of the tab) corresponds to the number of species in which an ortholog was identified out of the 10 currently supported by OpenProt. (E.)The ‘Translation’ tab displays PRICE analysis results with the associated P-value and the readcount per sample. The TE score (top of the tab) corresponds to the number of studies in which this ORF was detected. (F) The ‘Mass spectrometry’ tab displays the identified peptides with the study name and a link to the original data. The match count corresponds to the number of PSM (peptide spectrum match) per peptide, per study. The MS score (top of the tab) corresponds to the sum of unique peptides per study. More information and tutorials are available from the Help page (www.openprot.org/p/help).
All ORF predictions can be visualized using the genome browser where transcripts, ORFs, and MS-based peptides are displayed. For easy visualization, RefProts are coloured in blue, AltProts are in green, and Isoforms are in orange (Figure 2B).
Moreover, all predictions can be downloaded as TSV, BED or FASTA (DNA or Protein) files. These include accession numbers, gene name, nucleotide and protein sequences, and other characteristics. Every downloadable file format is detailed in the attached readme documents.
Protein characteristics annotation
The result table from a query also contains additional information on the predicted ORF (Figure 2A). The predicted protein characteristics, such as the length (in amino acids), the isoelectric point and molecular weight, are displayed. Each protein annotation is linked to all source transcripts for which accession numbers and type (mRNA or ncRNA) are reported. Localization within the transcript (CDS, 5′ or 3′ UTR) is also displayed for ORF predictions from mRNAs. Each ORF can be inspected individually in the details page (Figure 2C). The details page contains ORF information, such as genomic and transcript coordinates, the presence of a simplified Kozak motif (RNNATGG where R stands for A or G) (39) or a high-efficiency translation initiation motif (RYMRMVAUGGC where R stands for A or G, Y for U or C, M for A or C, and V for A, C or G) (40) and access to protein and DNA sequences. Other tabs display specifics of lines of evidence: protein conservation, MS, translation events and protein domain prediction.
Supporting evidence annotation
OpenProt first predicts potential ORFs and then collects diverse types of evidence, such as functional predictions (protein conservation and predicted domains) and experimental evidence (translation event and protein expression detection). Numbers of predicted ORFs supported by conservation, translation or expression evidence are displayed in Table 2 per ORF category for each species.
Table 2.
OpenProt (1.3) evidence collection output
| Conservation evidence | Translation evidence (Ribo-seq) | Protein evidence (MS) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Sp # | Ref # | II_ # | IP_ # | St # | Ref # | II_ # | IP_ # | St # | Ref # | II_ # | IP_ # |
| Homo sapiens | 9 | 189 319 | 38 325 | 239 394 | 33 | 17 435 | 2048 | 5 696 | 62 | 113 006 | 1455 | 28 641 |
| Pan troglodytes | 9 | 63 408 | 28 930 | 148 989 | 0 | n/a | n/a | n/a | 0 | n/a | n/a | n/a |
| Mus musculus | 9 | 131 130 | 21 245 | 121 890 | 22 | 14 607 | 1 088 | 3081 | 28 | 61 440 | 165 | 2 877 |
| Rattus norvegicus | 9 | 82 951 | 5354 | 81 600 | 2 | 6661 | 202 | 870 | 8 | 21 282 | 19 | 410 |
| Bos taurus | 9 | 70 697 | 6086 | 88 550 | 0 | n/a | n/a | n/a | 1 | 12 778 | 5 | 37 |
| Ovis aries | 9 | 56 331 | 28 247 | 107 900 | 0 | n/a | n/a | n/a | 1 | 1 466 | 18 | 69 |
| Danio rerio | 9 | 81 958 | 19 560 | 8 965 | 2 | 9 | 1 | 0 | 7 | 26 114 | 263 | 386 |
| Drosophila melanogaster | 9 | 39 246 | 763 | 452 | 3 | 2453 | 39 | 113 | 3 | 9783 | 20 | 113 |
| Caenorhabditis elegans | 9 | 28 429 | 861 | 450 | 5 | 8142 | 161 | 84 | 0 | n/a | n/a | n/a |
| Saccharomyces cerevisiae S288c | 9 | 5842 | 5 | 38 | 20 | 5357 | 4 | 283 | 4 | 4028 | 0 | 20 |
Sp = Number of species evaluated for orthology relationships (not counting the queried species); St = number of studies re-analyzed by OpenProt; Ref = currently annotated CDS (RefORF); II_ = novel isoforms of known CDS; IP_ = novel CDS from alternative ORF (AltORF); n/a = when no dataset has been re-analysed for this species yet (OpenProt release 1.3). Conservation evidence = all proteins with at least one ortholog in at least one species. Translation evidence = all ORFs detect in at least one detection by PRICE analysis of Ribo-seq data. Protein evidence = all proteins with at least one unique peptide in at least one study.
Protein orthology and paralogy relationships can be visualized for each protein annotated in OpenProt (Figure 2D). Under the Conservation tab of each protein Details page, orthologs and paralogs are listed per species. Similarly, predicted functional domains, gene ontology and pathway terms can be browsed under the Domains tab (Details page).
Experimental evidence is accessible under the Translation and Mass spectrometry tabs (Details page). The Translation tab collects PRICE (35) reports of identifications with the associated P-value and read count per sample (Figure 2E). The Mass spectrometry tab contains all identified unique peptides alongside the associated peptide spectrum match (PSM) count within each dataset (Figure 2F). For transparency purposes, all original datasets are accessible by clicking on the study name. At this time, the database re-analyzed 87 ribosome profiling and 114 mass spectrometry studies across several species and diverse cell lines and tissues (Table 2). A complete list of all studies implemented in OpenProt is available from the Help section (supplementary materials S1 and S2).
Applications and downloads
Expanded databases, such as OpenProt, are invaluable tools for functional proteomics discoveries (2,4,6,7). OpenProt is tailored for every need of all researchers, giving them the required tools for a more comprehensive view of eukaryotic genomes’ coding potential. That is why in addition to the genome Browser and the advanced query page, the Downloads page allows users to download custom databases. For example, one may choose to focus on only the most confident annotations (previously unannotated proteins detected with at least two unique peptides in mass spectrometry experiments), when another may want to focus on discovery of novel functional proteins (all predictions). Moreover, personalized database generation and download based on custom RNA-seq results is also supported, as detailed under the Help section (supplementary materials S1). The results table from specific queries can also be shared or downloaded as a TSV file or as a protein FASTA file. Overall, OpenProt allows (1) advanced search and download of results table, (2) genome browsing with visualization of MS evidence, and (3) personalized downloads (data and file format) for any endeavour.
DISCUSSION AND COMPARISON TO EXISTING RESOURCES
OpenProt annotates thousands of novel predicted proteins supported by experimental evidence and functional predictions. As more Ribo-seq and MS datasets are constantly added to the database, we expect this number to rise. The out-of-focus resolution at which we currently look at eukaryotic coding potential and proteomes is gradually being acknowledged (1,3,5,18,20), yet we still lack systematic approaches to the problem and this could impede on our understanding of basic biology questions (6).
To the best of our knowledge, OpenProt is the first database that fully endorses a polycistronic model of mammalian genome annotation. OpenProt differs from other smORFs databases in that it does not uphold a maximum length threshold (below 100 codons for smORFs). OpenProt also allows for multiple ORFs per transcript, and supports two transcriptome annotations. Furthermore, the OpenProt pipeline allows for the identification and detection of novel isoforms. Thus, OpenProt reaches a deeper ORF annotation throughout the genome.
In addition, OpenProt distinguishes itself from UniProt as it provides a graphical interface that allows the user to browse all predicted ORFs, in addition to providing functional annotations (conservation, translation, expression and presence of functional domains). OpenProt does not currently allow for deep functional annotation, such as UniProt does. However, when a novel protein is discovered and sufficiently characterized to meet UniProt annotation requirements, it will then become a RefProt in OpenProt database (with a UniProt accession). Thus, OpenProt is a modern tool that fills a major gap in the field of functional annotation of proteins by fostering less serendipitous discoveries of novel proteins.
FUTURE DIRECTIONS
The OpenProt pipeline is automated so that new releases of NCBI RefSeq and/or Ensembl will automatically be taken into account. Following iGenomes update, data will be updated at the beginning of the following year for computational resources access reasons. OpenProt is a release-based platform, developed in accordance to the FAIR guiding principles for scientific data management and stewardship (41). This ensures an up-to-date, continuous availability of all OpenProt data through time.
As the field progresses, OpenProt is expected to elaborate its pipeline, notably taking in account ORFs shorter than 30 codons or non-ATG start codons. For statistical and computational reasons, OpenProt current release (1.3) still holds these cut-offs. Simultaneously, OpenProt will continue to increase the number of datasets for supporting evidence, as well as supported species. OpenProt can be contacted through the Contact page for request of species annotations and/or dataset analyses (http://www.openprot.org/p/about). New tools and metrics will also be added following developments in the field. For instance, a pipeline is being developed to allow AltORFs and novel Isoforms consideration in genome or exome sequencing studies.
The quantity and quality of data provided by OpenProt along with its ease of use and transparent data availability hold potential to make it a popular tool.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our collaborators for providing additional mass spectrometry (MS) datasets. We thank Mark Vaudel for the support on our MS identification pipeline (PeptideShaker), and Florian Erhard for his help implementing the PRICE pipeline on Ribo-seq data. We also thank Felix Primeau, Christian Landry, Darel Hunting, and Alan A. Cohen for helpful discussions and contributions. F.M.B., M.S. and X.R. are members of the Fonds de Recherche du Québec Santé (FRQS)-supported Centre de Recherche du Centre Hospitalier Universitaire de Sherbrooke. We thank the team at Calcul Québec and Compute Canada for their support with the use of the supercomputer mp2 from Université de Sherbrooke.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canada Research Chair in Functional Proteomics and Discovery of Novel Proteins (to X.R.); CIHR grants [MOP-137056 and MOP-136962] (to X.R.). Operation of the mp2 supercomputer is funded by the Canada Foundation of Innovation (CFI), le ministère de l’Économie, de la science et de l’innovation du Québec (MESI); les Fonds de Recherche du Québec – Nature et technologies (FRQ-NT). Funding for open access charge: Canada Research Chairs.
Conflict of interest statement. None declared.
REFERENCES
- 1. Albuquerque J.P., Tobias-Santos V., Rodrigues A.C., Mury F.B., da Fonseca R.N.. small ORFs: a new class of essential genes for development. Genet. Mol. Biol. 2015; 38:278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Delcourt V., Staskevicius A., Salzet M., Fournier I., Roucou X.. Small proteins encoded by unannotated ORFs are rising stars of the proteome, confirming shortcomings in genome annotations and current vision of an mRNA. Proteomics. 2018; 18:e1700058. [DOI] [PubMed] [Google Scholar]
- 3. Hellens R.P., Brown C.M., Chisnall M.A.W., Waterhouse P.M., Macknight R.C.. The emerging world of small ORFs. Trends Plant Sci. 2016; 21:317–328. [DOI] [PubMed] [Google Scholar]
- 4. Saghatelian A., Couso J.P.. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 2015; 11:909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pueyo J.I., Magny E.G., Couso J.P.. New peptides under the s(ORF)ace of the genome. Trends Biochem. Sci. 2016; 41:665–678. [DOI] [PubMed] [Google Scholar]
- 6. Brunet M.A., Levesque S.A., Hunting D.J., Cohen A.A., Roucou X.. Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Res. 2018; 28:609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Samandi S., Roy A.V., Delcourt V., Lucier J.-F., Gagnon J., Beaudoin M.C., Vanderperre B., Breton M.-A., Motard J., Jacques J.-F. et al. . Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLife. 2017; 6:e27860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andrews S.J., Rothnagel J.A.. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014; 15:193–204. [DOI] [PubMed] [Google Scholar]
- 9. Matsumoto A., Clohessy J.G., Pandolfi P.P.. SPAR, a lncRNA encoded mTORC1 inhibitor. Cell Cycle. 2017; 16:815–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pauli A., Norris M.L., Valen E., Chew G.-L., Gagnon J.A., Zimmerman S., Mitchell A., Ma J., Dubrulle J., Reyon D. et al. . Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. 2014; 343:1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson D.M., Anderson K.M., Chang C.-L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R. et al. . A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015; 160:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014; 15:205–213. [DOI] [PubMed] [Google Scholar]
- 13. Ingolia N.T. Ribosome footprint profiling of translation throughout the genome. Cell. 2016; 165:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vanderperre B., Staskevicius A.B., Tremblay G., McCoy M., O’Neill M.A., Cashman N.R., Roucou X.. An overlapping reading frame in the PRNP gene encodes a novel polypeptide distinct from the prion protein. FASEB J. 2011; 25:2373–2386. [DOI] [PubMed] [Google Scholar]
- 15. Bergeron D., Lapointe C., Bissonnette C., Tremblay G., Motard J., Roucou X.. An out-of-frame overlapping reading frame in the ataxin-1 coding sequence encodes a novel ataxin-1 interacting protein. J. Biol. Chem. 2013; 288:21824–21835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Autio K.J., Kastaniotis A.J., Pospiech H., Miinalainen I.J., Schonauer M.S., Dieckmann C.L., Hiltunen J.K.. An ancient genetic link between vertebrate mitochondrial fatty acid synthesis and RNA processing. FASEB J. 2008; 22:569–578. [DOI] [PubMed] [Google Scholar]
- 17. Andreev D.E., O’Connor P.B.F., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., Baranov P.V.. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015; 4:e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olexiouk V., Van Criekinge W., Menschaert G.. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2018; 46:D497–D502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hao Y., Zhang L., Niu Y., Cai T., Luo J., He S., Zhang B., Zhang D., Qin Y., Yang F. et al. . SmProt: a database of small proteins encoded by annotated coding and non-coding RNA loci. Brief. Bioinform. 2018; 19:636–643. [DOI] [PubMed] [Google Scholar]
- 20. Hsu P.Y., Benfey P.N.. Small but Mighty: Functional peptides encoded by small ORFs in plants. Proteomics. 2018; 18:e1700038. [DOI] [PubMed] [Google Scholar]
- 21. Ma J., Diedrich J.K., Jungreis I., Donaldson C., Vaughan J., Kellis M., Yates J.R., Saghatelian A.. Improved identification and analysis of small open reading frame encoded polypeptides. Anal. Chem. 2016; 88:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nesvizhskii A.I. Proteogenomics: concepts, applications and computational strategies. Nat. Methods. 2014; 11:1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olexiouk V., Menschaert G.. Identification of small novel coding sequences, a proteogenomics endeavor. Adv. Exp. Med. Biol. 2016; 926:49–64. [DOI] [PubMed] [Google Scholar]
- 24. Wright J.C., Mudge J., Weisser H., Barzine M.P., Gonzalez J.M., Brazma A., Choudhary J.S., Harrow J.. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat. Commun. 2016; 7:11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. . Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. et al. . Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu P.-Y., Phan J.H., Wang M.D.. Assessing the impact of human genome annotation choice on RNA-seq expression estimates. BMC Bioinformatics. 2013; 14:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R.. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015; 43:W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R. et al. . UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 31. Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S. et al. . The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017; 45:D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. . 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaudel M., Burkhart J.M., Zahedi R.P., Oveland E., Berven F.S., Sickmann A., Martens L., Barsnes H.. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015; 33:22–24. [DOI] [PubMed] [Google Scholar]
- 34. Vaudel M., Barsnes H., Berven F.S., Sickmann A., Martens L.. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 2011; 11:996–999. [DOI] [PubMed] [Google Scholar]
- 35. Erhard F., Halenius A., Zimmermann C., L’Hernault A., Kowalewski D.J., Weekes M.P., Stevanovic S., Zimmer R., Dölken L.. Improved Ribo-seq enables identification of cryptic translation events. Nat. Methods. 2018; 15:363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sonnhammer E.L.L., Östlund G.. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015; 43:D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Östlund G., Schmitt T., Forslund K., Köstler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L.L.. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010; 38:D196–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M. et al. . InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002; 299:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L.. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014; 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E. et al. . The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.