Abstract
DoriC, a database of replication origins, was initially created to present the bacterial oriCs predicted by Ori-Finder or determined by experiments in 2007. DoriC 5.0, an updated database of oriC regions in both bacterial and archaeal genomes, was published in the 2013 Nucleic Acids Research database issue. Now, the latest release DoriC 10, a large-scale update of replication origins in prokaryotic genomes including chromosomes and plasmids, has been presented with a completely redesigned user interface, which is freely available at http://tubic.org/doric/ and http://tubic.tju.edu.cn/doric/. In the current release, the database of DoriC has made significant improvements compared with version 5.0 as follows: (i) inclusion of oriCs on more bacterial chromosomes increased from 1633 to 7580; (ii) inclusion of oriCs on more archaeal chromosomes increased from 86 to 226; (iii) inclusion of 1209 plasmid replication origins retrieved from NCBI annotations or predicted by in silico analysis; (iv) inclusion of more replication origin elements on bacterial chromosomes including DnaA-trio motifs. Now, DoriC becomes the most complete and scalable database of replication origins in prokaryotic genomes, and facilitates the studies in large-scale oriC data mining, strand-biased analyses and replication origin predictions.
INTRODUCTION
In all living organisms, DNA replication is regulated precisely at the assembly stage of the replication machinery (1). Replication origins are the particular genomic loci, where double-stranded DNA unwinds to form single-stranded DNA templates to initiate synthesis of new strands. In most bacteria, the replication origin (oriC) contains several DnaA box motifs recognized by the principal initiator protein DnaA, and a region with high AT content, namely, the DNA unwinding element (DUE), where single-stranded DNA is also recognized by DnaA (2–5). Similarly, AT-rich DNA unwinding element is also found to be essential in archaeal replication origin, which is flanked by the origin recognition boxes (ORBs) that serve as binding sites for origin recognition proteins (6,7). In a large number of plasmids, the origin of vegetative replication (oriV) often consists of direct repeats or iteron DNA sequences, which interact with Rep proteins to form the initial complex during the process of replication initiation (8). There is also an AT-rich region near the location of iterons in oriV, which serves as the DNA unwinding element (9).
It is interesting that the replication origin is usually next to the replication-related genes, such as dnaA, orc1/cdc6 and rep genes. The similar structures of prokaryotic replication origins on chromosomes and plasmids provide the opportunity to design algorithms for origin prediction based on the same framework. Initially, Ori-Finder was developed to identify the oriC regions on bacterial chromosomes (10).
As a separate domain in the three-domain system, most archaea exist in various extreme environments on earth, and the particular habits make it difficult to identify their replication origins by experimental methods (11). Therefore, the web-based tool Ori-Finder 2 was developed to predict the oriC regions in archaeal genomes in silico, and the predicted results can be helpful in the identification of archaeal origins in the laboratory (12).
Plasmids are extrachromosomal auto-replicating genetic elements, which are widespread in bacteria, archaea, yeast and some higher eukaryotic cells (8). Plasmids often carry genes that bring some special features to the host cells, such as antibiotic resistance and toxin–antitoxin system (13). Therefore, autonomous DNA replication of plasmids is critical for cell survival. The origin of vegetative replication is one of the most important elements in plasmid. Up to now, the location and characteristic of oriVs were well understood in a broad range of plasmids, such as RK2, F, P1, R6K and pPS10 plasmids (14–18). However, bioinformatics tools are urgently needed to identify oriVs automatically on plenty of sequenced plasmids.
The predictions of Ori-Finder system were organized into an online database to facilitate the related research on replication origins (19–21). In 2007, DoriC, a database of oriC regions, was first publicly available to present the bacterial oriCs, and in 2013, DoriC 5.0 included replication origins in both bacterial and archaeal genomes (22,23). In the past six years, the rapid progress in next-generation sequencing technologies and the accumulation of sequenced genomes from various microbial genome projects have promoted the expansion of DoriC, and this expanded database is presented here as DoriC 10.0, which includes the replication origins of plasmids for the first time. DoriC database and Ori-Finder system ensure a better understanding of the structure and function of replication origins, and provide new insights into the regulatory mechanisms of the initiation step in DNA replication. So far, many of the predictions stored in DoriC are now verified in the laboratory, and more applications based on DoriC database and Ori-Finder system in the past years have been reviewed in our recent article (24).
DATABASE UPDATES
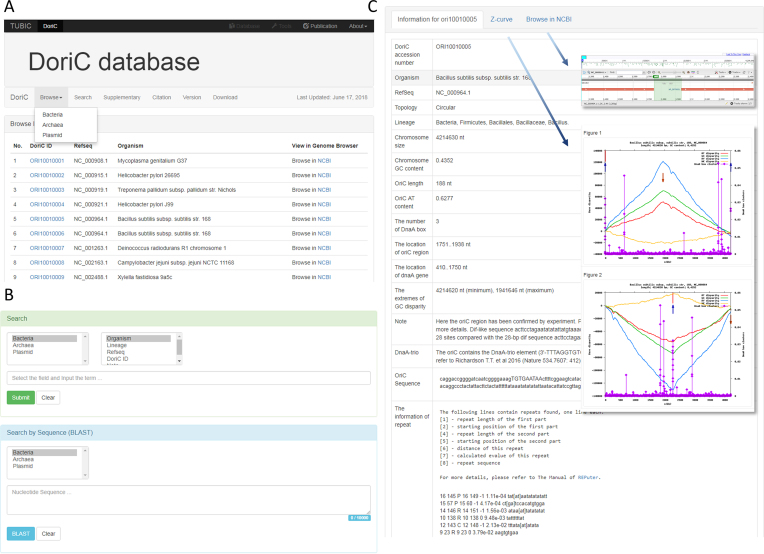
In the current release, the content of DoriC is significantly improved compared with version 5.0 as follows: (i) the oriCs on bacterial chromosomes have increased fourfold from 1633 to 7580; (ii) the oriCs on more archaeal chromosomes have increased from 86 to 226; (iii) 1209 plasmid replication origins are presented for the first time, including 348 annotated origins retrieved from NCBI records and 861 predicted origins by a modified Ori-Finder system; (iv) more sequence elements in bacterial replication origins are incorporated, including DnaA-trio element, a new repeating trinucleotide motif important for origin function. DnaA-trios play a role in origin unwinding and DNA helicase loading, which are highly conserved throughout the bacterial kingdom by bioinformatics analysis with DoriC (20). The DnaA-trio-like sequence was searched in DoriC database, and then the information was supplemented to the corresponding oriC record. Furthermore, we redesigned the user interface of the database to make it more convenient and intuitive (Figure 1).
Figure 1.
Snapshot of the redesigned user interface of DoriC. (A) Database browse interface for accessing the whole DoriC records of bacteria, archaea or plasmids. (B) Database search interface including record query and BLAST search. (C) A representative view of the record in DoriC displaying the information of replication origin, such as the location, sequence, DnaA-trio element and repeats. It can switch over to the Z-curve figures or the NCBI genome browser by Tabs.
DATABASE DESCRIPTION
Replication origins on bacterial and archaeal chromosomes
Except those collected from scientific publications, the oriCs on bacterial chromosomes were predicted by Ori-Finder, which has also been used in the annotation of more than 120 newly sequenced bacteria in their genome reports. In general, the oriC predictions by Ori-Finder can be integrated into DoriC database directly. In some cases, manual curation based on the information from DoriC is required for the questionable predictions by Ori-Finder. For example, two potential oriCs were predicted in some Gammaproteobacteria genomes, but only the one next to the gidA gene was added to DoriC finally according to the enrichment of GATC motifs in oriC regions (21).
This large-scale update of DoriC provides new insights into the replication origins on bacterial chromosomes. For example, the oriC in cyanobacteria is usually adjacent to dnaN gene, and contains the species-specific DnaA boxes (TTTTCCACA) (25). However, the oriC with a cluster of DnaA boxes (TTTTCCACA) is located far from dnaA and dnaN genes in Synechococcus lividus PCC 6715. Besides that, adenine or thymine in some positions of the DnaA box motif ‘TTTTCCACA’ tends to offset to guanine or cytosine in some GC-rich genomes of cyanobacteria, such as Cyanobium gracile PCC 6307 (GC content: 0.6871).
The oriCs on archaeal chromosomes were predicted by Ori-Finder 2.0, which is mainly used to predict the replication origins adjacent to some replication-related genes, such as cdc6 gene (12). The ORB motifs used by Ori-Finder 2.0 were summarized based on the available experimentally determined oriCs stored in DoriC, and the understanding of the ORB motifs is still quite limited at the present stage. Therefore, some potential orc1/cdc6-adjacent replication origins without known ORB motifs were also included in DoriC if they are located around the extremes of disparity curves with significant repeats.
Replication origins on plasmids
In this release, the oriVs on plasmids retrieved from NCBI annotations or predicted by in silico analysis have been collected into DoriC. For the oriVs retrieved from NCBI annotations, the intergenic regions, which have an overlap with the annotated replication origins according to NCBI records, are presented as potential oriVs in order to include as many repeat features as possible. The original location of oriV in the NCBI records was added as a note. The direct repeats or iterons DNA sequences in oriVs frequently conform to a consensus motif, but they are rarely identical in different plasmid sequences (26). In addition, the Rep proteins interact with inverted repeated (IR) sequences, causing transcriptional auto-repression (9,27). These repeat sequences were discovered in a great deal of oriVs according to NCBI annotations, and the information of repeats identified by REPuter pipeline was displayed in DoriC (28). Based on the characteristics of known replication origins on plasmids, oriVs were also predicted by a modified Ori-Finder system, and the intergenic regions, which are adjacent to rep genes with highly significant iterons DNA sequences and inverted repeats, were predicted as oriVs. The AT-rich regions in oriVs are usually followed by one or two DnaA-boxes (29), so the number of DnaA boxes identified in oriVs is also presented.
CONCLUSION
With the significant advancements in sequencing technology, the number of sequenced microbial genomes has continued to increase dramatically, which presents both challenges and opportunities for the studies of replication origins. In this latest release, DoriC has stored over 9,928 records of prokaryotic chromosomal origins and included 1209 records of plasmid replication origins for the first time. As an essential database in microbial genomics, DoriC has been used in a large number of studies associated with the replication origins. In the past dozen years, the development of DoriC has promoted a better understanding of replication origins, and some records of replication origins in DoriC were confirmed by experiments. Despite significant progress in DoriC, there are still many issues needed to be addressed urgently. In the future, the database will be further improved with the continuous update by taking into account more scientific discoveries in the field of DNA replication and the predicted replication origins in draft genome sequences.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Chun-Ting Zhang for the invaluable assistance and inspiring discussions.
FUNDING
National Natural Science Foundation of China [31571358, 21621004, 31171238, 11626250, 91746119]. Funding for open access charge: National Natural Science Foundation of China [31571358].
Conflict of interest statement. None declared.
REFERENCES
- 1. Marczynski G.T., Rolain T., Taylor J.A.. Redefining bacterial origins of replication as centralized information processors. Front. Microbiol. 2015; 6:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leonard A.C., Mechali M.. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013; 5:a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leonard A.C., Grimwade J.E.. The orisome: structure and function. Front. Microbiol. 2015; 6:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sakiyama Y., Kasho K., Noguchi Y., Kawakami H., Katayama T.. Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 2017; 45:12354–12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mackiewicz P., Zakrzewska-Czerwinska J., Zawilak A., Dudek M.R., Cebrat S.. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004; 32:3781–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barry E.R., Bell S.D.. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 2006; 70:876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelman L.M., Kelman Z.. Archaeal DNA replication. Annu. Rev. Genet.. 2014; 48:71–97. [DOI] [PubMed] [Google Scholar]
- 8. Konieczny I., Bury K., Wawrzycka A., Wegrzyn K.. Iteron plasmids. Microbiol.Spectrum. 2014; 2:doi:10.1128/microbiolspec.PLAS-0026-2014. [DOI] [PubMed] [Google Scholar]
- 9. del Solar G., Giraldo R., Ruiz-Echevarria M.J., Espinosa M., Diaz-Orejas R.. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998; 62:434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao F., Zhang C.T.. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics. 2008; 9:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woese C.R., Kandler O., Wheelis M.L.. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo H., Zhang C.T., Gao F.. Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes. Front. Microbiol. 2014; 5:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerdes K., Christensen S.K., Lobner-Olesen A.. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005; 3:371–382. [DOI] [PubMed] [Google Scholar]
- 14. Doran K.S., Helinski D.R., Konieczny I.. Host-dependent requirement for specific DnaA boxes for plasmid RK2 replication. Mol. Microbiol. 1999; 33:490–498. [DOI] [PubMed] [Google Scholar]
- 15. Kawasaki Y., Wada C., Yura T.. Roles of Escherichia coli heat shock proteins DnaK, DnaJ and GrpE in mini-F plasmid replication. Mol. Gen. Genet.: MGG. 1990; 220:277–282. [DOI] [PubMed] [Google Scholar]
- 16. Park K., Chattoraj D.K.. DnaA boxes in the P1 plasmid origin: the effect of their position on the directionality of replication and plasmid copy number. J. Mol. Biol. 2001; 310:69–81. [DOI] [PubMed] [Google Scholar]
- 17. McEachern M.J., Filutowicz M., Helinski D.R.. Mutations in direct repeat sequences and in a conserved sequence adjacent to the repeats result in a defective replication origin in plasmid R6K. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:1480–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nieto C., Giraldo R., Fernandez-Tresguerres E., Diaz R.. Genetic and functional analysis of the basic replicon of pPS10, a plasmid specific for Pseudomonas isolated from Pseudomonas syringae patovar savastanoi. J. Mol. Biol. 1992; 223:415–426. [DOI] [PubMed] [Google Scholar]
- 19. Korem T., Zeevi D., Suez J., Weinberger A., Avnit-Sagi T., Pompan-Lotan M., Matot E., Jona G., Harmelin A., Cohen N. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015; 349:1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richardson T.T., Harran O., Murray H.. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature. 2016; 534:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bendall M.L., Luong K., Wetmore K.M., Blow M., Korlach J., Deutschbauer A., Malmstrom R.R.. Exploring the roles of DNA methylation in the metal-reducing bacterium Shewanella oneidensis MR-1. J. Bacteriol. 2013; 195:4966–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao F., Zhang C.T.. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics. 2007; 23:1866–1867. [DOI] [PubMed] [Google Scholar]
- 23. Gao F., Luo H., Zhang C.T.. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 2013; 41:D90–D93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo H., Quan C.L., Peng C., Gao F.. Recent development of Ori-Finder system and DoriC database for microbial replication origins. Brief Bioinform. 2018; doi:10.1093/bib/bbx174. [DOI] [PubMed] [Google Scholar]
- 25. Gao F., Zhang C.T.. Origins of replication in Cyanothece 51142. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:E125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lilly J., Camps M.. Mechanisms of theta plasmid replication. Microbiol.Spectrum. 2015; 3:doi:10.1128/microbiolspec.PLAS-0029-2014. [PMC free article] [PubMed] [Google Scholar]
- 27. Pearson C.E., Zorbas H., Price G.B., Zannis-Hadjopoulos M.. Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. J. Cell Biochem. 1996; 63:1–22. [DOI] [PubMed] [Google Scholar]
- 28. Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R.. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001; 29:4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rajewska M., Wegrzyn K., Konieczny I.. AT-rich region and repeated sequences - the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 2012; 36:408–434. [DOI] [PubMed] [Google Scholar]