Abstract
BloodSpot is a gene-centric database of mRNA expression of haematopoietic cells. The web-based interface to the database includes three concomitant levels of visualization for a gene query; foremost is the expression across hematopoietic cell types, second is analysis of survival of Acute Myeloid Leukaemia patients based on gene expression, and lastly, the expression visualized in an interactive developmental tree. With the introduction of single cell data we have now also included an unbiased dimensionality reduction method to show gene expression over the continuum of haematopoiesis. The webserver includes a few select analysis functionalities, like Student's t-test, identification of correlating genes and lookup of whole genetic signatures, with the aim of making generation and testing of hypotheses quick and intuitive. The visualizations have been updated to accommodate new datatypes and the database has been largely expanded with RNA-sequencing datasets, both purified in bulk and at single cell resolution, increasing the number of single samples more than 10 fold, while keeping simplicity in presentation. The database should be of interest for any researcher within leukaemia, haematopoiesis, cellular development, or stem cells. The database is freely available at www.bloodspot.eu
INTRODUCTION
BloodSpot (1) is a database of haematopoietic cells in health and disease. The database and interface have been built with the aim of providing quick access for hypothesis testing and generation, via gene-centric lookup of mRNA expression throughout the course of haematopoiesis as well as in expanded leukemic blasts. The interface is, importantly, a one-click, no scroll access to relevant information (on the majority of screens). Uniquely for collected databases, BloodSpot provides detailed information on the definition and inclusion-criteria for each cell type, allowing researchers to draw conclusions without scavenging through supplementary material from original papers.
In the initial versions (2,3) of BloodSpot, microarray was the standard high-throughput technique to assess gene expression in haematopoietic cell types, and large and comprehensive studies delineated the full constitution of the haematopoietic system (4), as well large cohorts of patients with aberrant and leukemic blasts (5,6) with intricate fluorescence-activated cell sorting (FACS) schemes.. Microarrays have now almost entirely been replaced by short read RNA-sequencing. Recently, it has also become possible to investigate haematopoiesis at single cell resolution (7), either in combination with FACS (8) or as an unbiased outline of the full constitution of the bone marrow (9,10). This has allowed a glimpse into the full continuum of haematopoiesis, independently of surface exposed marker proteins used for FACS. Quality assessment and filtering are important steps when processing single cell RNA-sequencing data and several methods have been developed for this purpose, e.g. (11,12).
A number of other hematopoietic expression databases, each filling a niche, have existed alongside BloodSpot, as reviewed by (13). Most notably are stem cell specific databases like Stemformatics (14) and SyStemCell (15) both also including cells from the hematopoietic stem cell compartment; their interfaces are built for creating analysis workflows rather than accessing processed data. Hematopoietic specific databases are found in ErythronDB (16) (specifically erythropoiesis) and Haemosphere (17), both providing multi-click access to analysis and data, with focus on in-house data. The latter is specifically useful for the use of multidimensional scaling plots to outline problematic quality and cell types of the included data. The ambitious Leukemia Gene Atlas (18) and Gene Expression Commons (19) are no longer updated (last data addition from 2013) and dedicated mouse database BloodExpress (20) has been retired.
With this update of BloodSpot we embrace the newest available techniques and data, both from bulk sequencing of highly-purified FACS sorted cells and single cell RNA-seq, to quickly visualize expression of genes or signatures across hematopoietic cells, in the most informative way, to assist researchers and clinicians within the fields of leukaemia, stem cells, and development, to test and generate hypotheses.
MATERIALS AND METHODS
In-house single cell data was processed as described in (21) and external single cell data was obtained either as deposited in github (Setty, M., Kiseliovas, V., Levine, J., Gayoso, A., Mazutis, L. and Pe’er, D. (2018) Palantir characterizes cell fate continuities in human hematopoiesis. bioRxiv, https://doi.org/10.1101/385328), Unique Molecular Identifiers (UMIs) acquired and processed though a standard workflow utilizing 10× genomics cellranger (10), or as normalized and filtered read counts (8, 22). Blueprint data was downloaded at the processing level ‘gene_quantification.rsem_grape2_crg.GRCh38’ (23). Purified FACS sorted early human progenitor data from Notta et al. (24) was trimmed for NEXTERA adaptors using trim_galore (version 0.4.0, with additional parameters: -q 15 –stringency 3 –length 36) and aligned and quantified using star- 2.5.2b.
Single cell RNA sequencing data visualizations and dimensionality reduction was performed using a recent manifold learning technique, Uniform Manifold Approximation and Projection (UMAP) (McInnes, L., Healy, J. (2018) UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction, arXiv, https://arxiv.org/abs/1802.03426). In essence UMAP optimizes towards retaining local structure of the data, while preserving the global structure. It was applied both for visualization (reducing the dimensionality to two) and as a pre-processing step to the clustering algorithm (reducing the dimensionality to 10). Furthermore, k-means were used for clustering the single cell datasets. The elbow method was used to determine the final number of clusters, k. Briefly, plotting the inertia (within-cluster sum of squares) for varying values of k allows for a sensible k to be set, i.e. large enough that adding a new cluster would not improve the inertia (Supplementary Figure S1). By choosing a clustering algorithm and dimensionality so that clusters in the 2D plot apparently become split into separate clusters, it is possible not only to appreciate the continuum of haematopoietic development, and assess expression at different stages, but also to include relevant information from dimensions which do not appear on the two-dimensional plot. In the single cell data the abundant zero-count values were excluded from the main expression SinaPlot (26), as it greatly slowed the loading of the page, without adding information, but have been retained for calculations and visualizations on the UMAPs.
Signatures from DMAP (4) where calculated from the processed and normalized expression matrix. Samples included were common myeloid progenitor, megakaryocyte and pre-B-cell. Differential testing was performed with Limma (27) creating contrasts for each cell type against all other (weighted) and requiring genes to have P > 0.05 and log2-foldchange above 1 to be included in the signature. The intensity of the expression levels of cells was used to colour samples in the UMAP. The intensity is computed as the mean of an expression score function across all genes of the signatures. The function is given by the logarithm of the expression multiplied by the expression score function (x log x).
RESULTS AND DISCUSSION
Single cell RNA-sequencing of haematopoietic stem and progenitor cells
Development of new and sensitive library preparation protocols have made single cell resolution expression profiling possible. In particular in the hematopoietic stem cell compartment these advances provide an unprecedented opportunity to investigate early blood development in an unbiased manner. We have included several recent unique datasets for the study of hematopoietic progenitors at the single cell level in mouse (21,22) and human (8, 10, Setty et al.), and devised a new interface window for investigating their gene expression. Every single cell is visualized as one dot in a dimensionality reduced UMAP plot, such that the full continuum of differentiating cells can be assessed and addressed in an antibody-independent manner. This in effect means that the UMAP plots are a result of expression from all the genes and the cells, in such a way that cells that are similar are close together, and cells that are dissimilar are further apart. As in a principal component analysis (PCA) genes that are more informative are weighted higher in the assessment of similarity, (higher variance, and in this case also higher correlation, over cells). Importantly for single cell sequencing of haematopoietic cells, UMAP offers meaningful organization of cell clusters and also preserves cellular continuums, unlike the popular t-SNE plot (Becht, E., Dutertre, C.-A., Kwok, I.W.H., Ng, L.G., Ginhoux, F. and Newell, E.W. (2018) Evaluation of UMAP as an alternative to t-SNE for single-cell data. bioRxiv, https://doi.org/10.1101/298430); this advantage comes, at times, at the cost of increased white space and overlapping dots in the plots.
Clustering of single cells in a UMAP space
With use of k-means clustering in a 10D UMAP space we clustered unlabelled single cells, and colour coded the clusters for interpretability, and ease of interpretation in the SinaPlot (26). The expression of a query gene will appear as the intensity of colours on the UMAP, and is independent of the clustering. The clustering serves to evaluate the expression quantitatively over the continuum, and also helps to discover cellular connections that are not apparent in a 2D plot.
Validation of UMAP visualization
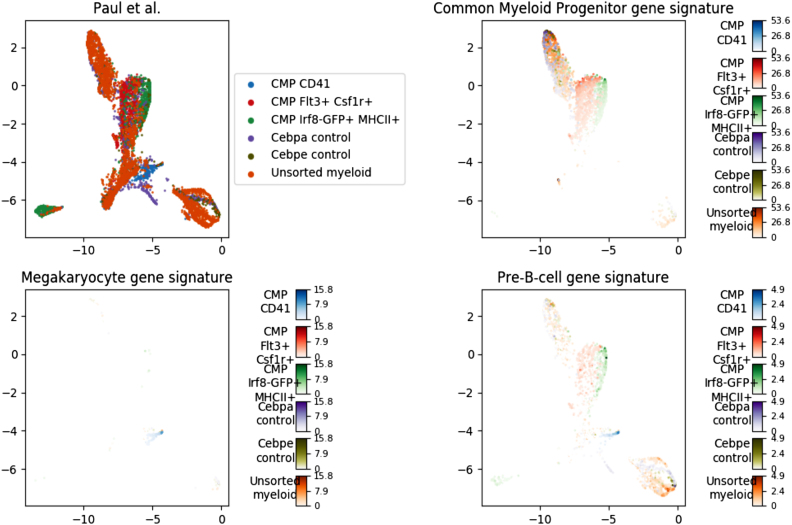
Expression of hematopoietic signatures created from DMAP(4) was used to assess the validity of the visualization and clustering. In Figure 1 single cell data from Paul et al. (22) is seen showing mean expression of DMAP gene signatures. Figures for remaining cell types and single cell datasets can be found in Supplementary Figures S2–S5. Whereas distinct separation of each cell type is not to be expected, it is clear that UMAP clusters and map regions that are dominated by, and in some cases only contain, a single classically defined cell type or its progenitor state.
Figure 1.
UMAP embeddings of the expression levels of the cells from Paul et al. study visualized on two dimensions. (A) all cells are visualized, colour corresponds to the type, as can be seen on legend. (B–D) The intensity of the expression levels of cells is computed as the mean of an expression score function across all genes of the signatures Common Myeloid Progenitor (B), Megakaryocyte (C) and Pre-B-cell (D). As it is shown in the colour bar, more intense colour corresponds to higher expression levels. Colour intensities are logarithm of the expression multiplied by expression (x log x) and was chosen for visualization of expression, to help differentiate between regions with different expression levels.
Inclusion criteria
We have included large studies of FACS sorted cells which broadly cover hematopoietic compartments, as well as single cell datasets, which in an unbiased way represent haematopoietic cells, independent of surface markers. We included newly published data, which analysed >1000 cells and where we could re-find priming of cells which have known precursors in the HCS compartment (as shown in Figure 1 and Supplement Figures S2–S5).
RNA-sequencing of FACS purified cells
BloodSpot is now expanded with high quality RNA-seq of FACS purified bulk sequencing data (23,24,28). Noteworthy is data from the BLUEPRINT epigenetics consortium: further to the epigenetics assays the consortium provided a conspectus of expression profiles from sorted populations of the human hematopoietic system. This task was first performed in microarrays by the DMAP (4) project, who conducted this task with a sorting resolution and with a completeness of cell types that yet remains to be exceeded.
The BloodSpot database update
The BloodSpot webserver is updated with curated high quality RNA-sequencing data from both single cell and FACS sorted purified cells. It now includes >25 000 samples, that are presented in an easy-to-navigate manner, and requires only a gene name as input for results. The database interface continues to be a one-click service, even if modifications to data inclusion and statistical tests can be performed, if required for publication purposes. On a gene query a plot of expression will be shown along with survival data, or UMAP for single cell data, and a hierarchical display based on the hematopoietic development or sample correlation. A dropdown can display correlating genes or pathways and can be useful for hypothesis generation. The database has a steady growing userbase and fills a niche within existing databases. With this update we ensure that the BloodSpot remains a resource at the forefront of the hematopoietic field. New data will continuously be curated and added to the database. Furthermore, biannual meetings with a user group and developers will systematically review new data releases since the last update, to ensure data is up to date. The database should be relevant for all researchers and clinicians within haematopoiesis, cellular development and stem cells.
DATA AVAILABILITY
Umap is available in the GitHub repository https://github.com/lmcinnes/umap
The Following data was acquired from Gene Expression Omnibus (GEO): GSE75478 (human single cells HSC), GSE60101 (Mouse purified bulk), GSE108155 and GSE72857 (Mouse single cell HSC). GSE76234 (Human purified bulk)
Blueprint data was acquired from http://dcc.blueprint-epigenome.eu and cd34+ (13) can be found at http://support.10xgenomics.com/single-cell/datasets.
DMAP data was downloaded from http://www.broadinstitute.org/dmap/home
Human HSC 10x genomics data was acquired from https://github.com/dpeerlab/Palantir/
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Lundbeck Foundation [R193-2015-1611, R182-2014-3881 to F.O.B.]; NovoNordisk Foundation [NNF15CC0027852 to N.R.]. Funding for open access charge: Rigshospitalet, Denmark.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bagger F.O., Sasivarevic D., Sohi S.H., Laursen L.G., Pundhir S., Sønderby C.K., Winther O., Rapin N., Porse B.T.. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016; 44:D917–D924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagger F.O., Rapin N., Theilgaard-Mönch K., Kaczkowski B., Thoren L.A., Jendholm J., Winther O., Porse B.T.. HemaExplorer: a database of mRNA expression profiles in normal and malignant haematopoiesis. Nucleic Acids Res. 2013; 41:D1034–D1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagger F.O., Rapin N., Theilgaard-Mönch K., Kaczkowski B., Jendholm J., Winther O., Porse B.. HemaExplorer: a Web server for easy and fast visualization of gene expression in normal and malignant hematopoiesis. Blood. 2012; 119:6394–6395. [DOI] [PubMed] [Google Scholar]
- 4. Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T. et al. . Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011; 144:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohlmann A., Kipps T.J., Rassenti L.Z., Downing J.R., Shurtleff S.A., Mills K.I., Gilkes A.F., Hofmann W.-K., Basso G., Dell’orto M.C. et al. . An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br. J. Haematol. 2008; 142:802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haferlach T., Kohlmann A., Wieczorek L., Basso G., Kronnie G. Te, Béné M.-C., De Vos J., Hernández J.M., Hofmann W.-K., Mills K.I. et al. . Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010; 28:2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laurenti E., Göttgens B.. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018; 553:418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Velten L., Haas S.F., Raffel S., Blaszkiewicz S., Islam S., Hennig B.P., Hirche C., Lutz C., Buss E.C., Nowak D. et al. . Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 2017; 19:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. et al. . Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016; 167:1398–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. et al. . Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ilicic T., Kim J.K., Kolodziejczyk A.A., Bagger F.O., McCarthy D.J., Marioni J.C., Teichmann S.A.. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016; 17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lun A.T.L., McCarthy D.J., Marioni J.C.. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor [version 2; referees: 3 approved, 2 approved with reservations]. F1000Research. 2016; 5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Q., Ding N., Zhang L., Zhao X., Yang Y., Qu H., Fang X.. Biological databases for hematology research. Genomics Proteomics Bioinforma. 2016; 14:333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wells C.A., Mosbergen R., Korn O., Choi J., Seidenman N., Matigian N.A., Vitale A.M., Shepherd J.. Stemformatics: visualisation and sharing of stem cell gene expression. Stem Cell Res. 2013; 10:387–395. [DOI] [PubMed] [Google Scholar]
- 15. Yu J., Xing X., Zeng L., Sun J., Li W., Sun H., He Y., Li J., Zhang G., Wang C. et al. . Systemcell: a database populated with multiple levels of experimental data from stem cell differentiation research. PLoS One. 2012; 7:e35230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kingsley P.D., Greenfest-Allen E., Frame J.M., Bushnell T.P., Malik J., McGrath K.E., Stoeckert C.J., Palis J.. Ontogeny of erythroid gene expression. Blood. 2013; 6:e5–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Graaf C.A., Choi J., Baldwin T.M., Bolden J.E., Fairfax K.A., Robinson A.J., Biben C., Morgan C., Ramsay K., Ng A.P. et al. . Haemopedia: An expression atlas of murine hematopoietic cells. Stem Cell Rep. 2016; 7:571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hebestreit K., Gröttrup S., Emden D., Veerkamp J., Ruckert C., Klein H.-U., Müller-Tidow C., Dugas M.. Leukemia gene atlas - a public platform for integrative exploration of genome-wide molecular data. PLoS One. 2012; 7:e39148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seita J., Sahoo D., Rossi D.J., Bhattacharya D., Serwold T., Inlay M.A., Ehrlich L.I.R., Fathman J.W., Dill D.L., Weissman I.L.. Gene expression commons: An open platform for absolute gene expression profiling. PLoS One. 2012; 7:e40321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miranda-Saavedra D., De S., Trotter M.W., Teichmann S.A., Göttgens B.. BloodExpress: a database of gene expression in mouse haematopoiesis. Nucleic Acids Res. 2009; 37:D873–D879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lauridsen F.K.B., Jensen T.L., Rapin N., Aslan D., Wilhelmson A.S., Pundhir S., Rehn M., Paul F., Giladi A., Hasemann M.S. et al. . Differences in cell cycle status underlie transcriptional heterogeneity in the HSC compartment. Cell Rep. 2018; 24:766–780. [DOI] [PubMed] [Google Scholar]
- 22. Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A. et al. . Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015; 163:1663–1677. [DOI] [PubMed] [Google Scholar]
- 23. Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. et al. . Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016; 167:1398–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Notta F., Zandi S., Takayama N., Dobson S., Gan O.I., Wilson G., Kaufmann K.B., McLeod J., Laurenti E., Dunant C.F. et al. . Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016; 351:aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sidiropoulos N., Sohi S.H., Pedersen T.L., Porse B.T., Winther O., Rapin N., Bagger F.O.. SinaPlot: an enhanced chart for simple and truthful representation of single observations over multiple classes. J. Comput. Graph. Stat. 2018; 27:673–676. [Google Scholar]
- 27. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S. et al. . Chromatin state dynamics during blood formation. Science. 2014; 345:943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Umap is available in the GitHub repository https://github.com/lmcinnes/umap
The Following data was acquired from Gene Expression Omnibus (GEO): GSE75478 (human single cells HSC), GSE60101 (Mouse purified bulk), GSE108155 and GSE72857 (Mouse single cell HSC). GSE76234 (Human purified bulk)
Blueprint data was acquired from http://dcc.blueprint-epigenome.eu and cd34+ (13) can be found at http://support.10xgenomics.com/single-cell/datasets.
DMAP data was downloaded from http://www.broadinstitute.org/dmap/home
Human HSC 10x genomics data was acquired from https://github.com/dpeerlab/Palantir/