Abstract
The CRISPR/Cas9 system, as a revolutionary genome editing tool for all areas of molecular biology, provides new opportunities for research on lncRNA’s function. However, designing a CRISPR/Cas9 single guide RNA (sgRNA) for lncRNA is not easy with an unwarrantable effectiveness. Thus, it is worthy of collecting validated sgRNAs, to assist in efficiently choosing sgRNA with an expected activity. CRISPRlnc (http://www.crisprlnc.org or http://crisprlnc.xtbg.ac.cn) is a manually curated database of validated CRISPR/Cas9 sgRNAs for lncRNAs from all species. After manually reviewing more than 200 published literature, the current version of CRISPRlnc contains 305 lncRNAs and 2102 validated sgRNAs across eight species, including mammalian, insect and plant. We handled the ID, position in the genome, sequence and functional description of these lncRNAs, as well as the sequence, protoacceptor-motif (PAM), CRISPR type and validity of their paired sgRNAs. In CRISPRlnc, we provided the tools for browsing, searching and downloading data, as well as online BLAST service and genome browse server. As the first database against the validated sgRNAs of lncRNAs, CRISPRlnc will provide a new and powerful platform to promote CRISPR/Cas9 applications for future functional studies of lncRNAs.
INTRODUCTION
Long non-coding RNAs (lncRNAs), which were once regarded as junk sequences, are defined as non-protein-coding transcripts longer than 200 nt (1–3). Instead of being translated into proteins, lncRNAs exert their functions in cellular processes (4–6), organismal development (7,8) and diseases (9,10) directly in the shape of RNA. With an enormous range of applications of next generation sequencing, a large number of lncRNAs have been found in human (11,12), mouse (13,14), zebrafish (7,15,16), etc. At the same time, a large number of lncRNA databases have been created, and committed to large-scale collection and annotation of lncRNAs for various species. For example, NONCODE contained more than 350 000 lncRNA genes across 17 species (17). lncRNAdb manually collected and annotated about 300 functional lncRNAs that have been studied to date (18). However, due to the extremely complicated and inconstant mechanisms when compared with protein-coding genes, most of lncRNAs have not been well studied, despite that a host of researches have extensively demonstrated the significance and diversity of lncRNAs in regulatory functions.
CRISPR/Cas9 system, as a revolutionary gene editing tool for all the areas of molecular biology, can induce site-specific DNA cleavage by an RNA-guided DNA-endonuclease. CRISPR/Cas9 system can be used in the unmodified form of ‘molecular scissors’ (wtCas9) to edit parts of the genome by removing, adding or altering sections of the DNA sequence or to create a knockout genotype (CRISPRko) (19,20). After inactivating the nuclease domain to create a dead Cas9 (dCas9) (21), accessional function elements could conduce to branching out the applications of CRISPR/Cas9 system, such as transcriptional activation (CRISPRa) (22), transcriptional interference (CRISPRi) (23,24), gene editing (CRISPRedit) (25) and so on.
Unlike protein-coding genes, many lncRNAs are confined to nucleus, and some exert their molecular functions in a transcript-independent mode, meaning that the transcribing event of lncRNA in itself could affect target genes. Therefore, there are probably a lot of restrictions to use RNAi method for performing loss-of-function studies of lncRNAs (26). Contrastively, CRISPR/Cas9 has a huge advantage in lncRNA researches, resulting from its in-cis regulative function in cell nucleus. Hence, CRISPR/Cas9 provides new opportunities for deeply researching lncRNA’s functions, and is receiving increasing attention in the field of lncRNA studies.
The first step of CRISPR/Cas9 gene editing is to design a single guide RNA (sgRNA) to target your gene of interest. However, because sgRNAs vary widely in their activity and action models, designing a sgRNA is not easy due to an unwarrantable effectiveness. Thus, it is worthy of collecting validated sgRNA sequences, to assist in efficiently choosing sgRNA with an expected activity. For example, Varshney et al. had constructed CRISPRz to collect validated sgRNAs for zebrafish coding-genes (27). However, CRISPR/Cas9 applications for lncRNAs are much different from coding-genes, as indicated by many known works (26,28,29). For instance, it is not necessary for lncRNA to maintain an intact open reading frame for functioning. Besides, lncRNA as well as their surrounding coding/noncoding neighbors had complicated genomic architecture, like sense/antisense, intergenic/intragenic etc. Therefore, a validated sgRNA database specifically for lncRNAs is profoundly valuable for the relevant academic community.
In this study, we constructed CRISPRlnc (http://www.crisprlnc.org or http://crisprlnc.xtbg.ac.cn)—a manually curated database of validated sgRNAs for lncRNAs. After manually reviewing more than 200 published literature, the current version of CRISPRlnc contains 305 lncRNAs and 2102 validated sgRNAs across eight species, including mammalian, insect and plant. We handled the ID, position in the genome, sequence and functional description of these lncRNAs, as well as the sequence, protoacceptor-motif (PAM), CRISPR type and validity of their corresponding sgRNAs. In CRISPRlnc, we also provided the tools for browsing, searching and downloading all of the data covered, as well as online BLAST service and genome browse server. As the first database against the validated sgRNAs of lncRNAs, CRISPRlnc will give an efficient assistance when employing CRISPR/Cas9 system as tools for lncRNA genome editing. Moreover, the high-quality integrated information of validated sgRNAs for lncRNAs could be used as a gold standard dataset to guide the design of sgRNAs themselves.
DATA COLLECTION AND DATABASE CONTENT
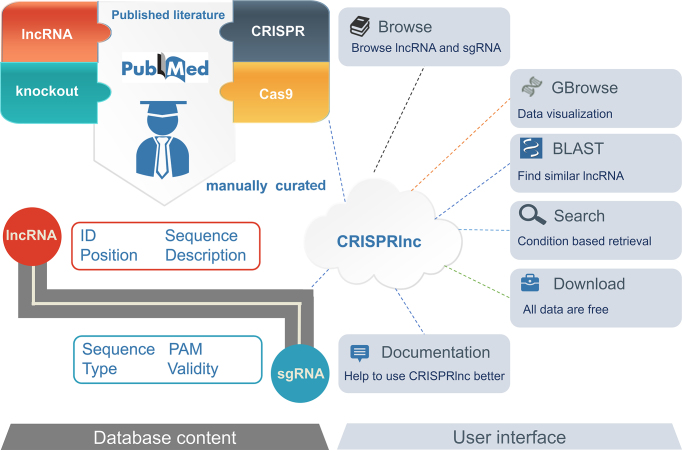
As shown in Figure 1, to exhaustively collect the validated data associated with CRISPR/Cas9 applications into lncRNA functional interrogation, first, we searched PubMed (30) to retrieve all relevant literature upon a list of keywords, such as ‘lncRNA’, ‘long noncoding RNA’ and ‘lincRNA’. Next, the preliminary screening results were further traversed in title or abstract via our own in-house python script to extract those containing ‘CRISPR’, ‘Cas9’, ‘Knockout’ or ‘Knockdown’. As a result, more than 200 articles are reserved. To ensure the high quality of our data, we manually checked and read over these papers. For each collected lncRNA, its ID, genomic position, nucleotide sequence and functional description are filed, along with the resources of its corresponding sgRNAs (including targeting sequence, PAM, CRISPR type and validity). Given the differential designation for the same gene between literatures, we normalized the gene ID and transcript ID of these lncRNAs in line with NCBI and Ensembl. The position of sgRNAs and lncRNAs in genome was obtained by aligning the consensus sequence to the reference genome using BLAT (31). Besides, in consideration of the positions of sgRNA that often link its functional effect—for example, the sgRNAs for CRISPRa or CRISPRi usually located near the transcription start sites of lncRNA (32,33)—and the expressional specificity of lncRNA in different tissues as well as the varying activity of CRISPR across different cell lines (32), we also integrated the information of ‘the location of sgRNA relative to lncRNA’ and ‘the effective cell line of sgRNA’ in our database.
Figure 1.
Overview of the CRISPRlnc database.
Based on the original CRISPR/Cas9 system, researchers had developed a variety of technological innovation to satisfy differentiated intervention on gene function. Therefore, in this work, we collected data upon different CRISPR/Cas9-based systems, including CRISPR knockout (CRISPRko), CRISPR activation (CRISPRa), CRISPR interference (CRISPRi) and CRISPR edit (CRISPRedit). To assist in efficiently choosing sgRNA with an expected activity, we further divided our data into three types according to the sgRNA validity as stated in the corresponding literature: (i) recommended high-activity sgRNAs, referring to those that had been either clearly demonstrated as powerful executors in one study or, multi-employed by several studies; (ii) experimentally validated sgRNAs, which has some low-throughput experiments to verify effectiveness; (iii) sgRNAs with partial experimental validation are those designed by experts with high-throughput experimental validation—for example, using a high-throughput genomic deletion strategy, 51 lncRNAs were found to be capable of regulating human cancer cell growth when knocked out via a CRISPR system which constructs thousands of targeted sgRNAs in a lentiviral paired-guide RNA library (34).
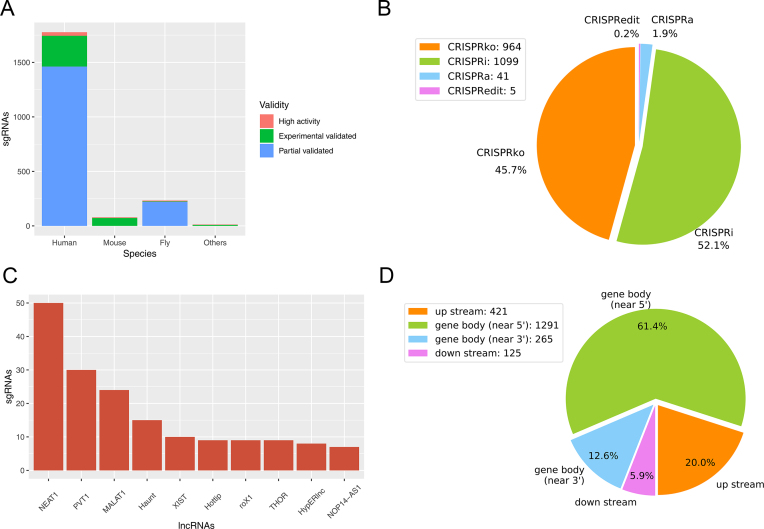
The current version of CRISPRlnc contains 305 lncRNAs and 2102 validated sgRNAs across 8 species, including mammalian, insect and plant. The statistics of CRISPRlnc is summarized in Figure 2. Figure 2A shows the sgRNA distribution in various species. The collected sgRNA data are mainly from human (Homo sapiens, 1777 in total), mouse (Mus musculus, 79 in total) and fly (Drosophila melanogaster, 233 in total). Figure 2B shows the type of these data, most of which are CRISPR interference (CRISPRi, about 52.1%); while the minimum type is CRISPRedit (about 0.2%). According to the Top10 lncRNAs possessing the designed sgRNAs (Figure 2C, regardless of type III sgRNAs supported by high-throughput experiments), CRISPR/Cas9 applications for lncRNAs are currently focused on some potentially important lncRNAs in human, such as NEAT1 (Nuclear Enriched Abundant Transcript 1), PVT1 (Plasmacytoma variant translocation 1) and MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1). We also calculated the localization tendency of sgRNAs on lncRNAs, including up/downstream region of gene and gene body near 5′/3′ end (Figure 2D). Our results revealed that the collected sgRNAs were more likely to locate at gene body near 5′ end (61.4% in 5′ end versus 12.6% in 3′ end), as well as more likely at gene upstream as opposed to downstream (20.0% versus 5.9%).
Figure 2.
Statistics of the CRISPRlnc database. (A) The distribution of sgRNAs in various species. (B) The functional types of sgRNA. (C) The Top10 lncRNAs that possess known designed sgRNAs, without regard to those high-throughput experimental data. (D) The position distribution of sgRNAs on lncRNAs.
DATABASE ORGANIZATION AND WEB INTERFACE
Database structure and implementation
The CRISPRlnc database is running on an Ubuntu Linux server (version 18.04), while Nginx (version 1.14.0) as the web server and SQLite (version 3.22.0) as the database server. The website is developed using Python based on Django web-framework (version 1.11). The web-frontend is developed using the Bootstrap framework (version 4.1.1). The GBrowser function is developed using Biodalliance (version 0.13.8) (35). The BLAST function is developed using django-blastplus (version 0.4.0), which is a simple Django app to conduct web-based homology search with blast+ (https://pypi.python.org/pypi/django-blastplus/).
USER INTERFACE
For biologists to better access the information of CRISPRlnc, we established a user-friendly website. As shown in Figure 1, in CRISPRlnc, we provided the tools for browsing, searching and downloading all data, as well as online BLAST service and genome browse server. Each section contains sufficient help materials to ensure an easy use without any prerequisite knowledge or experience.
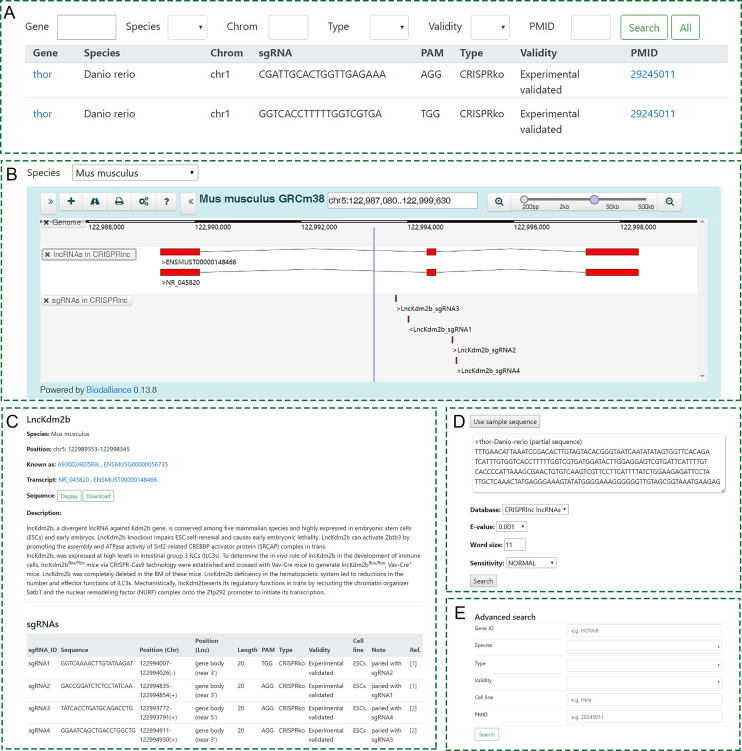
In ‘Browse’, user can retrieve all lncRNAs and sgRNAs in CRISPRlnc (Figure 3A). User can find the basic annotations on the ‘Browse’ page, such as gene ID, sgRNA sequence, PAM motif, CRISPR type and validity. The detailed information page for a specific gene can be accessed by clicking on the gene ID. The linked page is mainly containing lncRNA’s information (Gene ID, transcript ID, genome location, sequence and function description), and a sgRNA table which contains all validated sgRNA information related to this lncRNA (Figure 3C). In the sgRNA table, each sgRNA is marked with a source of literature, so you can backtrack to the original documents.
Figure 3.
User interface of the CRISPRlnc database. (A) The ‘Browse’ page. (B) ‘GBrowse’, the genome browser. (C) The ‘Details’ page. (D) ‘Online BLAST’ search. (E) The ‘Advanced Search’ page.
In CRISPRlnc, we also provided some user-friendly web services. In ‘GBrowse’, you can view lncRNAs, sgRNAs and other genomic annotation according to their locations in genome (Figure 3B). The sgRNAs from some genome-scale CRISPR/Cas9 screens (32–34) were placed in a GBrowse track. For a lncRNA with a given genomic location, we recommend you use ‘GBrowse’ to find out whether it has validated sgRNAs. In addition, you can find a lncRNA or its homologs through BLAST sequence similarity search in CRISPRlnc (Figure 3D). Maybe the lncRNA in one species you are interested in does not have validated sgRNAs, but if you are lucky, you may find its homolog has. Moreover, you can find the lncRNAs or sgRNAs you are interested in using ‘Advanced Search’, by providing/selecting a list of keywords, such as gene ID, species, CRISPR type, validity, cell line and PMID (Figure 3E).
DISCUSSION AND FUTURE DEVELOPMENT
Until now, tools with various characteristics for ab initio designing sgRNA are crowded, whereas systematic collection of validated sgRNAs is rare. Thus, we developed CRISPRlnc, a database that provides a comprehensive list of validated CRISPR/Cas9 sgRNAs for lncRNAs from all species. CRISPRlnc will provide not only an efficient assistance for genome editing of lncRNAs, but also a gold standard dataset that is crucial for subsequent development of sgRNA design tools of lncRNAs. We believed that CRISPRlnc will thrive in the fields related to lncRNA and CRISPR/Cas9 studies.
In the future, to respond to the rapid growth of researches in lncRNA and CRISPR/Cas9, we will continue to manually curate newly validated sgRNAs for lncRNAs and update the database every 3 months. In addition, we will continue to integrate more online tools and data sources to make CRISPRlnc more resourceful and more usable. As the first database specifically targeting to the validated sgRNAs of lncRNAs, we sincerely hope to get the support and advice from the academic community to help us improve it. And it is greatly appreciated for submitting data to us.
FUNDING
National Natural Science Foundation of China [31471220, 91440113]; Xishuangbanna Tropical Botanical Garden Start-up Fund; ‘Top Talents Program in Science and Technology' from Yunnan Province; Fund of State Key Laboratory of Developmental Biology of Freshwater Fish [2017KF003]; Scientific Research Fund of Hunan Provincial Education Department [15CY006]; Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province [20134486]. Funding for open access charge: National Natural Science Foundation of China [91440113].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wang K.C., Chang H.Y.. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011; 43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ulitsky I., Bartel D.P.. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013; 154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fatica A., Bozzoni I.. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014; 15:7–21. [DOI] [PubMed] [Google Scholar]
- 5. Devaux Y., Zangrando J., Schroen B., Creemers E.E., Pedrazzini T., Chang C.P., Dorn G.W. 2nd, Thum T., Heymans S. Cardiolinc network . Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015; 12:415–425. [DOI] [PubMed] [Google Scholar]
- 6. Greco C.M., Condorelli G.. Epigenetic modifications and noncoding RNAs in cardiac hypertrophy and failure. Nat. Rev. Cardiol. 2015; 12:488–497. [DOI] [PubMed] [Google Scholar]
- 7. Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P.. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011; 147:1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin N., Chang K.Y., Li Z., Gates K., Rana Z.A., Dang J., Zhang D., Han T., Yang C.S., Cunningham T.J. et al. . An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 2014; 53:1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015; 21:1253–1261. [DOI] [PubMed] [Google Scholar]
- 10. Ling H., Vincent K., Pichler M., Fodde R., Berindan-Neagoe I., Slack F.J., Calin G.A.. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015; 34:5003–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. . The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Blass G., Kellis M., Werber M. et al. . The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013; 24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M. et al. . Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013; 2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A. et al. . Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012; 22:577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaushik K., Leonard V.E., Kv S., Lalwani M.K., Jalali S., Patowary A., Joshi A., Scaria V., Sivasubbu S.. Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS One. 2013; 8:e83616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., Zhao L., Li X., Teng X., Sun X. et al. . NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018; 46:D308–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E.. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015; 43:D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. . Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M.. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013; 31:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. et al. . CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goyal A., Myacheva K., Gross M., Klingenberg M., Duran Arque B., Diederichs S.. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017; 45:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Varshney G.K., Zhang S., Pei W., Adomako-Ankomah A., Fohtung J., Schaffer K., Carrington B., Maskeri A., Slevin C., Wolfsberg T. et al. . CRISPRz: a database of zebrafish validated sgRNAs. Nucleic Acids Res. 2016; 44:D822–D826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ho T.T., Zhou N., Huang J., Koirala P., Xu M., Fung R., Wu F., Mo Y.Y.. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2015; 43:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghosh S., Tibbit C., Liu J.L.. Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Res. 2016; 44:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coordinators N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018; 46:D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D., Attenello F.J., Villalta J.E., Cho M.Y., Chen Y. et al. . CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017; 355:eaah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joung J., Engreitz J.M., Konermann S., Abudayyeh O.O., Verdine V.K., Aguet F., Gootenberg J.S., Sanjana N.E., Wright J.B., Fulco C.P. et al. . Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017; 548:343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu S., Li W., Liu J., Chen C.H., Liao Q., Xu P., Xu H., Xiao T., Cao Z., Peng J. et al. . Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 2016; 34:1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Down T.A., Piipari M., Hubbard T.J.. Dalliance: interactive genome viewing on the web. Bioinformatics. 2011; 27:889–890. [DOI] [PMC free article] [PubMed] [Google Scholar]