Abstract
The Cucurbitaceae family (cucurbit) includes several economically important crops, such as melon, cucumber, watermelon, pumpkin, squash and gourds. During the past several years, genomic and genetic data have been rapidly accumulated for cucurbits. To store, mine, analyze, integrate and disseminate these large-scale datasets and to provide a central portal for the cucurbit research and breeding community, we have developed the Cucurbit Genomics Database (CuGenDB; http://cucurbitgenomics.org) using the Tripal toolkit. The database currently contains all available genome and expressed sequence tag (EST) sequences, genetic maps, and transcriptome profiles for cucurbit species, as well as sequence annotations, biochemical pathways and comparative genomic analysis results such as synteny blocks and homologous gene pairs between different cucurbit species. A set of analysis and visualization tools and user-friendly query interfaces have been implemented in the database to facilitate the usage of these large-scale data by the community. In particular, two new tools have been developed in the database, a ‘SyntenyViewer’ to view genome synteny between different cucurbit species and an ‘RNA-Seq’ module to analyze and visualize gene expression profiles. Both tools have been packed as Tripal extension modules that can be adopted in other genomics databases developed using the Tripal system.
INTRODUCTION
The Cucurbitaceae family (cucurbit) includes several economically important vegetable and fruit crops, such as cucumber (Cucumis sativus L.), melon (C. melo L.), watermelon (Citrullus lanatus (Thunb.) Matsum. & Nakai), pumpkin/squash (Cucurbita spp.), bottle gourd (Lagenaria siceraria) and bitter gourd (Momordica charantia). In 2016, the agricultural production of cucurbits utilized 11 million hectares of land, and yielded 256 million tons of vegetables and fruits (http://faostat.fao.org). In addition to being used as vegetables and fruits, cucurbit crops can also be used for medicine, containers, musical instruments and decoration (1). Moreover, several species such as pumpkin and bottle gourd have been used as rootstocks for other cucurbit crops in order to enhance tolerance to soil-borne diseases and abiotic stresses and to improve fruit yield and quality (2). Furthermore, the cucurbit species have long served as model systems for studies of fundamental biological processes such as fruit ripening (3), sex determination (4), and vascular development (5). In addition, both xylem and phloem sap of cucurbits can be readily collected for studies on long-distance signaling events (6).
Due to the rapid advances in sequencing technologies, high-quality reference genome sequences of a number of cucurbit crops have been generated and released (7–14). Together with large volumes of transcriptome and genetic data generated from a variety of studies, a database is needed to store, mine, analyze, and disseminate these large-scale datasets and to provide a central portal for the cucurbit research and breeding community. Several genomics and functional genomics databases have been developed for cucurbit crops including Cucumber Genome Database (http://cucumber.genomics.org.cn), MELONOMICS (https://www.melonomics.net/), MeloGene (http://melogene.upv.es), CucurbiGen (https://cucurbigene.upv.es/) and Melonet-DB (http://gene.melonet-db.jp; (15)). These databases, despite being very useful, are limited to a specific cucurbit species, e.g. cucumber or melon, or a specific data type, e.g., genome sequences or gene expression, and lack functions for comparative genomics and functional genomics, and comprehensive integration of different data types. To facilitate the usage and application of genomic resources for cucurbit research and breeding, we have developed a family-wide cucurbit genomics database (CuGenDB; http://cucurbitgenomics.org). CuGenDB was first released in 2007 with only expressed sequence tag (EST) and unigene data and has since integrated rich genomics and genetics resources of cucurbits including genetic maps, genomes, gene models, and functional annotations. Recently, CuGenDB has been rebuilt using Tripal (16), a toolkit for construction of online genomic and genetic databases by integrating the GMOD Chado database schema (17) and Drupal (https://www.drupal.org/), a popular Content Management Systems (CMS). Tripal has been used to implement a number of widely used databases such as Genome Database for Rosaceae (18), CottonGen (19) and Coffee Genome Hub (20). Furthermore, we have generated comprehensive functional annotations for all cucurbit gene models, identified synteny blocks and homologous genes among different cucurbits, incorporated expression profiles based on public RNA-Seq data, and developed new modules in CuGenDB to analyze and visualize comparative genomics and expression datasets of different cucurbit species.
DATABASE CONTENTS AND FEATURES
Genome sequences and gene annotations
Currently CuGenDB contains a total of 10 publicly available high-quality reference genome sequences of cucurbits including two cultivated cucumbers (C. sativus L. var. sativus cv. 9930 and cv. Gy14), one wild cucumber (C. sativus var. hardwickii PI 183967), one cultivated melon (C. melo L. cv. DHL92), two cultivated watermelons (C. lanatus subsp. vulgaris cv. 97103 and cv. Charleston Gray), three cultivated Cucurbita species (C. maxima cv. Rimu, C. moschata cv. Rifu, and C. pepo cv. MU-CU-16), and one cultivated bottle gourd (Lagenaria siceraria cv. USVL1VR-Ls) (7–14). Seven of the 10 genomes were first released in CuGenDB, while the genome sequences of cultivated cucumber Gy14, melon DHL92, and zucchini MU-CU-16 are downloaded from phytozome (https://phytozome.jgi.doe.gov), MELONOMICS (https://www.melonomics.net/), and CucurbiGen (https://cucurbigene.upv.es), respectively. CuGenDB provides a feature page for each cucurbit genome, which contains various sections of information and a submenu to access both the genome data and bioinformatics analysis tools. The information sections provide basic information regarding the sequenced material such as genus, species, cultivar and common name, and the genome such as the number of predicted genes, the description of genome assembly and the list of related publications, where available. The submenu provides links to a pathway database, a genome browser, an FTP site for downloading genome and gene sequences and annotations, and bioinformatics tools such as BLAST, batch query and basic search functions.
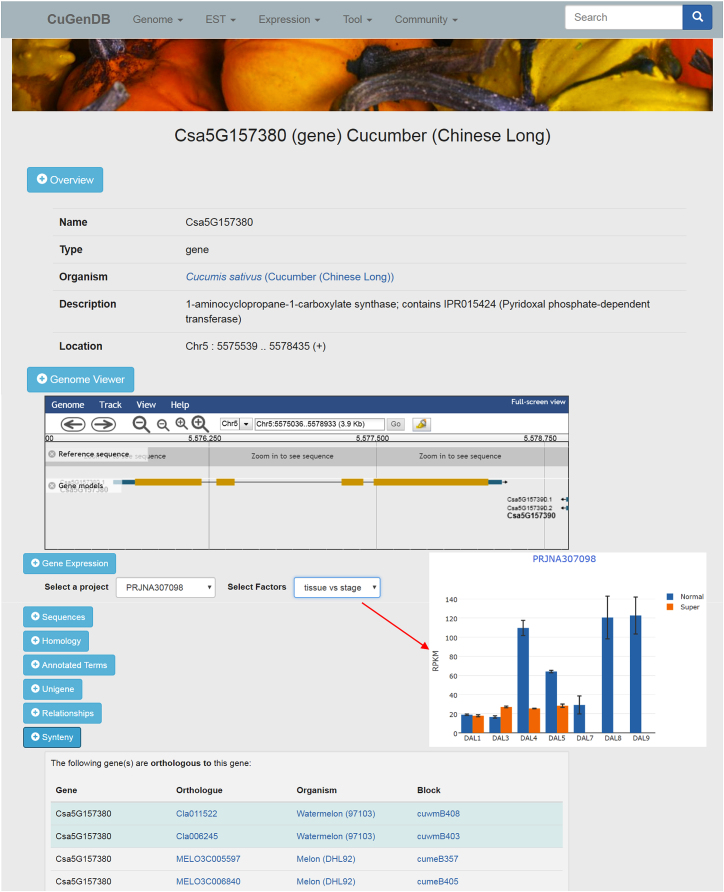
A total of 265 334 protein-coding genes have been predicted from these 10 genomes and included in the database. A standard and unified procedure has been developed to comprehensively annotate predicted protein-coding genes. First, protein sequences of the predicted genes are compared against the GenBank non-redundant protein (nr), UniProt (TrEMBL and SwissProt), and Arabidopsis protein databases using the BLAST program with an E-value cutoff of 1e-4. The protein sequences are further compared against the InterPro database using InterProScan (21) to identify functional protein domains. The BLAST results against the nr database and the identified InterPro domains are fed to the Blast2GO program (22) for assigning gene ontology (GO) terms to protein-coding genes, and the BLAST results against the UniProt and Arabidopsis protein databases are loaded into the AHRD program (https://github.com/groupschoof/AHRD) to assign concise, informative and precise functional descriptions of genes. The top BLAST hits (homologs), GO terms and InterPro domains assigned to each of the protein-coding genes have been imported into CuGenDB using Tripal Analysis Extension Modules (16). The functional descriptions generated by AHRD are loaded into the database using an in-house Perl script. Each gene has a detailed feature page in the database that contains all the related sequence and annotation information; the gene feature page is divided into different sections based on the content types (Figure 1).
Figure 1.
Gene feature page in CuGenDB. The page contains different sections with different content types. The inset histogram shows expression profiles of a specific gene under a selected project in the ‘Gene Expression’ section.
Synteny blocks and homologous genes
We have identified synteny blocks and homologous gene pairs within the synteny blocks for all pairwise comparisons of the cucurbit genomes (currently 45 comparisons between 10 different genomes), as well as within each genome. Protein sequences are first aligned against each other (pairwise comparisons) or against themselves (within each genome) using BLASTP with an E-value cutoff of 1e–05 and a maximum of five alignments. Based on the BLASTP results, synteny blocks are determined using MCScanX (23) with default parameters. In total, 36,051 synteny blocks and 1 106 351 homologous gene pairs have been identified and stored in CuGenDB. As shown in Figure 1, a ‘Synteny’ section has been included in the gene feature page to display all available synteny blocks and homologous gene pairs related to a specific gene. Each synteny block is further linked to the page that lists all genes, including homologous gene pairs located in the syntenic region, and contains an image to display homologous gene pairs, which is generated by the ‘SyntenyViewer’ module described below.
ESTs and unigenes
Approximately 1.74 million EST sequences have been collected in CuGenDB for four cucurbit species, among which 129,240, 513,801, 588,800, and 508,456 are from melon, cucumber, watermelon, and Cucurbita pepo, respectively. These EST sequences are first screened against the NCBI UniVec database (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html), Escherichia coli genome and rRNA sequences using SeqClean (http://sourceforge.net/projects/seqclean/) to remove possible contaminations. The cleaned EST sequences from each species are independently assembled into unigenes using the iAssembler program v1.3.3 (24), resulting in a total of 24 444, 93 903, 75 068 and 136 038 unigenes for melon, cucumber, watermelon, and C. pepo, respectively. Cleaned ESTs and assembled unigenes are mapped to the corresponding genomes of the same species using GMAP (25). The relationship between unigenes and gene models are established if they are mapped to same locations. The unigenes are comprehensively annotated using the same procedure for annotating predicted protein-coding genes as described above. As with the predicted protein-coding genes, CuGenDB also provides a feature page for each EST or unigene to list the related sequence and annotation information. In addition, similar to the genome feature page, for the EST and unigene collection of each cucurbit species, the database provides a feature page containing related information and a submenu to access the data and analysis tools.
Gene expression profiles
We have collected all available RNA-Seq data from NCBI Sequence Read Archive (SRA) for the cucurbit species that have available reference genome sequences. A unified pipeline for RNA-Seq data processing and analysis has been applied to these RNA-Seq datasets. Briefly, raw RNA-Seq reads are processed to remove adaptor and low-quality sequences using Trimmomatic v0.32 (26), and trimmed reads shorter than 80% of their original length are discarded. The remaining high-quality reads are aligned to the SILVA rRNA database (27) using Bowtie v1.1.2 (28) allowing up to 3 mismatches, and the mapped reads are removed. The final cleaned reads or read pairs are aligned to the corresponding genome using HISAT (29) allowing up to two mismatches. Raw counts are then derived for each predicted gene model and normalized to FPKM (fragments per kilobase of exon per million mapped fragments). Gene expression profiles for a specific gene can be accessed under the ‘Gene Expression’ section of the gene feature page described above (Figure 1), where after selecting an RNA-Seq project, a histogram showing expression profiles across different experimental conditions/tissues is displayed (Figure 1).
To import these expression data (raw counts and FPKM) and the associated meta-information into CuGenDB, we have developed two Tripal extension modules, ‘SRA’ and ‘RNA-Seq’. The ‘SRA’ module is a mimic of the SRA database but does not require the storage of raw sequencing files. The main purpose of this module is to provide a management system for the collected project, sample, and experiment information. The average expression values and the standard deviation from biological replicates are calculated for each gene and loaded into CuGenDB using the ‘RNA-Seq’ extension module. The home page of ‘RNA-Seq’ lists all collected projects with mouse-over descriptions.
Genetic maps
A total of 21 published genetic maps have been collected for cucurbit species, including 15 for melon, four for cucumber, and two for watermelon. The map view and search functions in the CuGenDB database have been developed using the CMap module from the GMOD project, which is a genetic extensible web-based comparative map viewer for displaying and comparing genetic and physical maps (30). Currently, we are in the process of implementing the TripalMAP extension module (https://tripal.info/extensions/modules/tripalmap) in CuGenDB to replace CMap for viewing and managing genetic maps.
Biochemical pathways
We use PathwayTools (31) to predict biochemical pathways for cucurbit species. For each cucurbit genome or unigene set, gene functional descriptions assigned by AHRD, GO terms assigned by Blast2GO, and enzyme commission (EC) numbers derived from the top hits in the UniProt database are integrated into a file in the PathoLogic format, which is used by PathwayTools for pathway prediction. A total of 320–430 biochemical pathways have been predicted from each genome or unigene set. A cucurbit biochemical pathway database (CucurbitCyc) has been implemented in CuGenDB using the PathwayTools web server (31). Users can search and browse the predicted pathways, as well as perform comparative and omics data analyses through the CucurbitCyc database.
DATABASE FUNCTIONS
Search
To facilitate the query for gene functional annotation data stored in CuGenDB, we have used the Apache Solr search engine (http://lucene.apache.org/solr/) to build the search index for different types of annotation data including AHRD descriptions, homologous genes, GO terms and InterPro domains. A basic search form is provided for each genome or unigene collection, which allows users to input a specific gene or unigene ID for retrieving the corresponding gene or unigene information, or to input a keyword for retrieving a list of genes whose annotations contain such keyword. In addition, a global search function, which queries against all the records stored in the database, is provided under the main menu of CuGenDB.
Batch query, which allows for retrieving sequences, annotations and other features for a list of user-provided genes, is an important function in genomics databases, e.g. the Batch Entrez tool in NCBI (https://www.ncbi.nlm.nih.gov/sites/batchentrez). The batch query function in CuGenDB is modified from the ‘Sequence Retrieval’ page of Tripal (16). In addition to sequences and functional descriptions assigned by AHRD, starting with a list of user input genes, the query also allows for retrieval of transcription factors and transcriptional regulators, which are predicted from all cucurbit genes stored in the database using the iTAK program (32).
Genome browser
In CuGenDB, we have implemented a genome browser using JBrowse (33) to display genome sequences, gene models, unigene and EST alignments, and expression profile data. Currently, all collected cucurbit genomes and gene models are imported into JBrowse. The tracks of reference sequence and gene models are also embedded in the gene feature page to provide a graphical and informative view of the gene structure (Figure 1). In addition, the genome browser has several supporting tracks including expression abundances derived from the RNA-Seq datasets and alignments of ESTs and unigenes. Other interesting data, such as single-base resolution genome variants that are being generated by the community, can be easily added to the genome browser for view in future updates.
BLAST
We have implemented an instance of NCBI’s BLAST tool in CuGenDB using the BLAST UI extension module in Tripal (16). We have modified the interface of the BLAST UI module to integrate the BLAST program and database options into one query interface. All genome, mRNA, CDS and protein sequences as well as EST and unigene sequences stored in CuGenDB are available for comparison through this BLAST interface. To prevent users from choosing incompatible BLAST programs and databases in the interface, the list of databases is automatically updated based on the selected BLAST programs. The BLAST UI module provides downloadable output files in three different formats, HTML, TSV and XML.
Enrichment analysis and gene functional classification
Genomic and functional genomic studies normally generate large lists of interesting genes, and translating such lists into biologically meaningful information is critical to understand the underlying regulatory mechanisms of the related biological processes. The enrichment analysis is a powerful method to identify classes of genes that are overrepresented in a list of genes, which represent highly affected biological processes or biochemical pathways under certain experimental conditions or developmental stages. In CuGenDB, we have developed two extension modules to identify significantly overrepresented GO terms and pathways, respectively. The ‘GO tool’ extension module has been implemented by wrapping the GO::TermFinder Perl module, which determines enriched GO terms using the hypergeometric distribution test (34). The ‘Pathway tool’ extension module has been developed based on the biochemical pathways predicted by PathwayTools (31), and also uses the hypergeometric distribution test to calculate the significance of enrichment. These two modules have been packed into Tripal, and can be implemented in other genomics databases built with Tripal.
Classifying a list of interesting genes into different functional categories is also important to help understand specific biological processes. In CuGenDB, we developed a tool which can functionally classify a list of genes based on a set of plant-specific GO subset (http://www.geneontology.org/page/go-subset-guide). This tool has been integrated into the ‘GO tool’ extension module.
SyntenyViewer
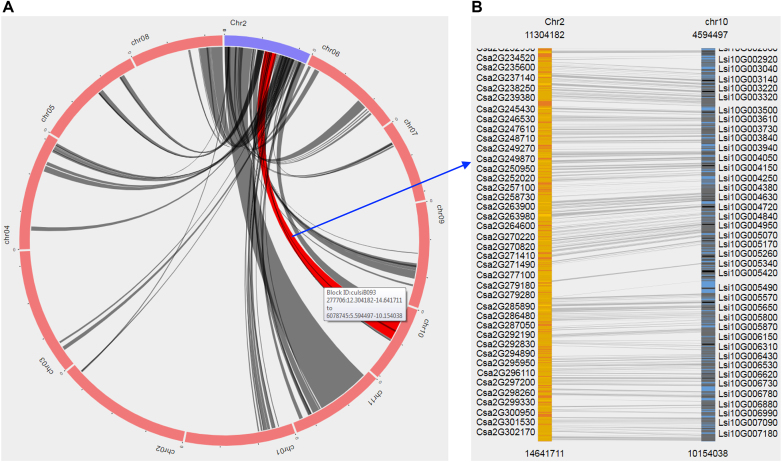
To view genome synteny and homologous gene pairs between different cucurbit species, we have developed ‘SyntenyViewer’ as an extension module of Tripal. The synteny blocks can be retrived in CuGenDB by selecting a query genome and one or more compared genomes, or by providing a specific block ID. SyntenyViewer draws circos plots to display synteny blocks for every pair of query and compared genomes (Figure 2A), and provides a full list of the synteny blocks. For a specific synteny block, SyntenyViewer generates an image to show the homologous gene pairs, and the image can be zoomed in or out for different views (Figure 2B). The full list of genes including the homologous gene pairs within the synteny block is also provided, with each gene linked to the gene feature page. In summary, the SyntenyViewer module can not only display synteny blocks for multiple genomes in an intuitive manner, but also connect homologous gene pairs among different cucurbit species. With this module, for a specific genome region of interest in one cucurbit species, features of homologous regions such as interesting genes can be easily identified and intuitively viewed in another cucurbit genome. It is worth noting that the SyntenyViewer has already been adopted by a number of other genome databases including Genome Database for Rosaceae (https://www.rosaceae.org), CottonGen (https://www.cottongen.org), Citrus Genome Database (https://www.citrusgenomedb.org), and Cool Season Food Legume Database (https://www.coolseasonfoodlegume.org).
Figure 2.
Synteny viewer in CuGenDB. (A) Synteny blocks displayed in circos plot. Blue arc indicates the query chromosome and red arcs indicate chromosomes of a compared genome. Dark grey lines between blue and red arcs indicate synteny blocks identified between the two genomes. The line will be changed to red when mouse over. (B) View of the selected synteny block. The query and compared chromosomes of the selected synteny blocks are shown in orange and blue, respectively. The yellow and black lines within each chromosome indicate homologous gene pairs, which are connected by grey lines.
Differential gene expression analysis
In addition to loading and storing gene expression profile data derived from RNA-Seq datasets, the ‘RNA-Seq’ module also provides statistical analysis functions for differentially expressed gene (DEG) identification, and tools to visualize expression profiles. Currently, two types of comparisons can be performed to identify DEGs, including pairwise comparisons of two different samples, and time series comparisons of samples from different stages or under different conditions. edgeR (35) and DESeq (36), the two most popular tools for differential expression analysis of RNA-Seq data, are provided in the ‘RNA-Seq’ module. Users can also change the cutoff of the adjusted P-value and the expression fold change to determine DEGs. The result page of differential expression analysis includes the project description, parameters used for statistical analysis, a list of the top 100 most significant DEG (ordered by adjusted P-values), and links to download files containing expression analysis results for all genes or all identified DEGs. An important feature of the ‘RNA-Seq’ modules is that the identified DEGs can be easily transferred to other modules for downstream functional analysis. The result page of DEGs contains links to tools of GO and pathway enrichment analyses, gene functional classification, and batch query. The identified DEGs will be automatically loaded into these tools as the input.
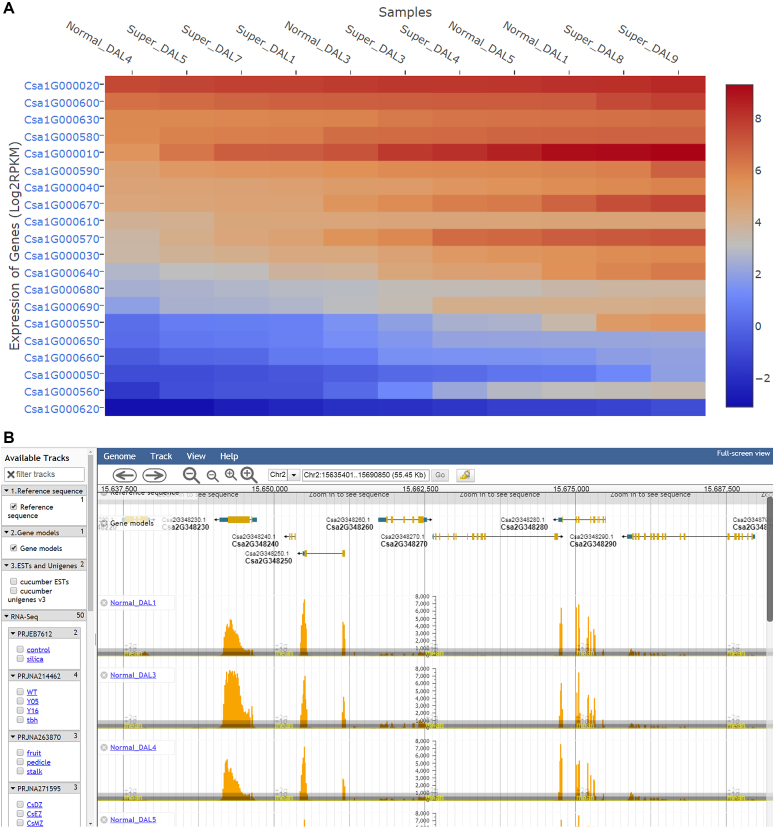
In addition to viewing the expression profiles of individual genes on the gene feature page (Figure 1), the ‘RNA-Seq’ module provides two additional visualiztion tools of gene expression profiles, a heatmap function developed by using the plotly JavaScript library (http://plot.ly) to display expression profiles of multiple genes (Figure 3A), and the JBrowse (33) to display single-base resolution expression abundances under different conditions (Figure 3B).
Figure 3.
Gene expression analysis with the ‘RNA-Seq’ module in CuGenDB. (A) Heatmap showing expression profiles of multiple genes. (B) Single-base resolution expression abundance view in JBrowse.
CONCLUSIONS AND FUTURE DIRECTIONS
We have developed Cucurbit Genomics Database (CuGenDB), which serves as a central portal for genomics, transcriptomics, and genetics of cucurbit species. CuGenDB stores sequences of genomes, mRNAs, proteins, ESTs and unigenes, and comprehensive functional annotations of genes and unigenes, as well as genome synteny blocks, homologous gene pairs, gene expression profiles, biochemical pathways, and genetic datasets of cucurbit species. The database also provides various query, visualization and analysis tools including BLAST, batch query, genome browser, pathway database (CucurbitCyc), GO term and pathway enrichment analysis, genome synteny viewer and differential gene expression analysis. It is worth mentioning that two newly developed modules in CuGenDB, ‘SyntenyViewer’ and ‘RNA-Seq’, have been packed as Tripal extension modules that have been adopted in other genomics databases developed using the Tripal system.
CuGenDB will be continuously updated when new genome, RNA-Seq and genetic datasets of cucurbit species become available. We will continue to improve the functionality of the ‘SyntenyViewer’ and ‘RNA-Seq’ modules, and develop novel related data mining and analysis tools which can be used by the Tripal community. We are currently in the process of developing a module for analyses of small RNA (sRNA) datasets, which will be made available in both CuGenDB and Tripal in the near future. Currently, under the CucCAP project (https://cuccap.org), the cucurbit community has been genotyping >1000 accessions for each of the four major cucurbit crops, cucumber (37), watermelon, melon and squash, using the genotyping-by-sequencing (GBS) technology, among which a core collection of ∼400 accessions for each crop is being established and under deep genome resequencing. A large number of molecular markers will be derived and large-scale phenotypic data generated under CucCAP, together with numerous ongoing efforts from other research groups around the world. Therefore, a breeder-friendly database for phenotypic, genotypic and QTL information is an urgent need for cucurbit crops. Therefore, we will implement tools and query interfaces in CuGenDB to analyze and integrate genotypic and phenotypic data for cucurbit crops using the Breeding Information Management System (BIMS; https://www.rosaceae.org/bims).
ACKNOWLEDGEMENTS
We thank the Tripal community for developing, maintaining and updating the Tripal modules.
FUNDING
USDA National Institute of Food and Agriculture Specialty Crop Research Initiative [2015-51181-24285]; US-Israel Binational Agricultural Research and Development Fund [IS-3333-02, IS-3877-06CR and IS-4223-09C]; USDA Agricultural Research Service, and by SNC Laboratoire ASL, de Ruiter Seeds B.V., Enza Zaden B.V., Gautier Semences S.A., Nunhems B.V., Rijk Zwaan B.V., Sakata Seed Inc, Semillas Fitó S.A., Seminis Vegetable Seeds Inc, Syngenta Seeds B.V., Takii and Company Ltd, Vilmorin and Cie S.A. and Zeraim Gedera Ltd, all of them as part of the support to the International Cucurbit Genomics Initiative (ICuGI). Funding for open access charge: USDA National Institute of Food and Agriculture.
Conflict of interest statement. None declared.
REFERENCES
- 1. McCreight J.D. Grumet R, Katzir N, Garcia-Mas. Cultivation and uses of cucurbit crops. Genetics and Genomics of Cucurbitaceae. Plant Genetics and Genomics: Crops and Models. 2017; Springer International Publishing; 1–12. [Google Scholar]
- 2. Lee J.-M., Kubota C., Tsao S.J., Bie Z., Echevarria P.H., Morra L., Oda M.. Current status of vegetable grafting: diffusion, grafting techniques, automation. Sci. Hortic. 2010; 127:93–105. [Google Scholar]
- 3. Pech J.C., Bouzayen M., Latché A.. Climacteric fruit ripening: ethylene-dependent and independent regulation of ripening pathways in melon fruit. Plant Sci. 2008; 175:114–120. [Google Scholar]
- 4. Bhowmick B.K., Jha S.. Dynamics of sex expression and chromosome diversity in Cucurbitaceae: a story in the making. J. Genet. 2015; 94:793–808. [DOI] [PubMed] [Google Scholar]
- 5. Sui X., Nie J., Li X., Scanlon M.J., Zhang C., Zheng Y., Ma S., Shan N., Fei Z., Turgeon R. et al. Transcriptomic and functional analysis of cucumber (Cucumis sativus L.) fruit phloem during early development. Plant J. 2018; doi:10.1111/tpj.14084. [DOI] [PubMed] [Google Scholar]
- 6. Lough T.J., Lucas W.J.. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 2006; 57:203–232. [DOI] [PubMed] [Google Scholar]
- 7. Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., Lucas W.J., Wang X., Xie B., Ni P. et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009; 41:1275–1281. [DOI] [PubMed] [Google Scholar]
- 8. Garcia-Mas J., Benjak A., Sanseverino W., Bourgeois M., Mir G., Gonzalez V.M., Henaff E., Camara F., Cozzuto L., Lowy E. et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. U.S.A. 2012; 109:11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang L., Koo D.H., Li Y., Zhang X., Luan F., Havey M.J., Jiang J., Weng Y.. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012; 71:895–906. [DOI] [PubMed] [Google Scholar]
- 10. Guo S., Zhang J., Sun H., Salse J., Lucas W.J., Zhang H., Zheng Y., Mao L., Ren Y., Wang Z. et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2012; 45:51–58. [DOI] [PubMed] [Google Scholar]
- 11. Qi J., Liu X., Shen D., Miao H., Xie B., Li X., Zeng P., Wang S., Shang Y., Gu X. et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 2013; 45:1510–1515. [DOI] [PubMed] [Google Scholar]
- 12. Sun H., Wu S., Zhang G., Jiao C., Guo S., Ren Y., Zhang J., Zhang H., Gong G., Jia Z. et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant. 2017; 10:1293–1306. [DOI] [PubMed] [Google Scholar]
- 13. Wu S., Shamimuzzaman M., Sun H., Salse J., Sui X., Wilder A., Wu Z., Levi A., Xu Y., Ling K.-S. et al. The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 2017; 92:963–975. [DOI] [PubMed] [Google Scholar]
- 14. Montero-Pau J., Blanca J., Bombarely A., Ziarsolo P., Esteras C., Martí-Gómez C., Ferriol M., Gómez P., Jamilena M., Mueller L. et al. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018; 16:1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yano R., Nonaka S., Ezura H.. Melonet-DB, a grand RNA-Seq gene expression atlas in melon (Cucumis melo L.). Plant Cell Physiol. 2018; 59:e4. [DOI] [PubMed] [Google Scholar]
- 16. Sanderson L.A., Ficklin S.P., Cheng C.H., Jung S., Feltus F.A., Bett K.E., Main D.. Tripal v1.1: a standards-based toolkit for construction of online genetic and genomic databases. Database. 2013; 2013:bat075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jung S., Lee T., Ficklin S., Yu J., Cheng C.-H., Main D.. Chado use case: storing genomic, genetic and breeding data of Rosaceae and Gossypium crops in Chado. Database. 2016; 2016:baw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jung S., Ficklin S.P., Lee T., Cheng C.-H., Blenda A., Zheng P., Yu J., Bombarely A., Cho I., Ru S. et al. The Genome Database for Rosaceae (GDR): year 10 update. Nucleic Acids Res. 2014; 42:D1237–D1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu J., Jung S., Cheng C.-H., Ficklin S.P., Lee T., Zheng P., Jones D., Percy R.G., Main D.. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014; 42:D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dereeper A., Bocs S., Rouard M., Guignon V., Ravel S., Tranchant-Dubreuil C., Poncet V., Garsmeur O., Lashermes P., Droc G.. The coffee genome hub: a resource for coffee genomes. Nucleic Acids Res. 2015; 43:D1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitchell A., Chang H.-Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S. et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2014; 43:D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Conesa A., Götz S.. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008; 2008:619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T., Jin H., Marler B., Guo H. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng Y., Zhao L., Gao J., Fei Z.. iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics. 2011; 12:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu T.D., Watanabe C.K.. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005; 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- 26. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O.. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Youens-Clark K., Faga B., Yap I. V, Stein L., Ware D.. CMap 1.01: a comparative mapping application for the Internet. Bioinformatics. 2009; 25:3040–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karp P.D., Paley S., Romero P.. The Pathway Tools software. Bioinformatics. 2002; 18(Suppl. 1):S225–S232. [DOI] [PubMed] [Google Scholar]
- 32. Zheng Y., Jiao C., Sun H., Rosli H.G., Pombo M.A., Zhang P., Banf M., Dai X., Martin G.B., Giovannoni J.J. et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant. 2016; 9:1667–1670. [DOI] [PubMed] [Google Scholar]
- 33. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L. et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G.. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004; 20:3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X., Bao K., Reddy U.K., Bai Y., Hammar S.A., Jiao C., Wehner T.C., Ramírez-Madera A.O., Weng Y., Grumet R. et al. The USDA cucumber (Cucumis sativus L.) collection: genetic diversity, population structure, genome-wide association studies and core collection development. Hort. Res. 2018; 5:64. [DOI] [PMC free article] [PubMed] [Google Scholar]