Abstract
Flatworms (Platyhelminthes) are a basally branching phylum that harbours a wealth of fascinating biology, including planarians with their astonishing regenerative abilities and the parasitic tape worms and blood flukes that exert a massive impact on human health. PlanMine (http://planmine.mpi-cbg.de/) has the mission objective of providing both a mineable sequence repository for planarians and also a resource for the comparative analysis of flatworm biology. While the original PlanMine release was entirely based on transcriptomes, the current release transitions to a more genomic perspective. Building on the recent availability of a high quality genome assembly of the planarian model species Schmidtea mediterranea, we provide a gene prediction set that now assign existing transcripts to defined genomic coordinates. The addition of recent single cell and bulk RNA-seq datasets greatly expands the available gene expression information. Further, we add transcriptomes from a broad range of other flatworms and provide a phylogeny-aware interface that makes evolutionary species comparisons accessible to non-experts. At its core, PlanMine continues to utilize the powerful InterMine framework and consistent data annotations to enable meaningful inter-species comparisons. Overall, PlanMine 3.0 thus provides a host of new features that makes the fascinating biology of flatworms accessible to the wider research community.
INTRODUCTION
Planarians are members of the phylum Platyhelminthes (flatworms) (1). Many, but not all, species have the fascinating ability to regenerate complete and perfectly proportioned animals from random tissue pieces (2–4). Regeneration relies on abundant pluripotent adult stem cells that mediate the continuous turn-over of all cell types and make planarians a powerful model system for multiple pertinent questions in current life sciences research (5). Moreover, the close phylogenetic neighbourhood of planarians harbour more interesting animals. Chief examples here are the parasitic tape worms and blood flukes that exert a massive impact on human health and have evolved a broad diversity of life cycles (6). The great taxonomic diversity of flatworms raises additional layers of intriguing questions, including the evolution of parasitic life styles from free-living relatives (7) or why some planarians regenerate while others cannot (8–10). Systematic and mechanistic inter-species comparisons amongst flatworms thus have the potential to provide insights into questions that cannot be addressed in individual model systems.
PlanMine’s mission is to catalyse the comparative analysis of flatworm biology. This entails a dual purpose both as a model system resource for the planarian model species, but also as interactively mineable sequence repository of flatworm data. The PlanMine database and web application was first released in 2015 (11) and mainly focused on transcriptomes as the major type of sequence information available at the time. The current release transitions to a more genomic perspective on basis of the recently released high quality genome assembly of the planarian model species Schmidtea mediterranea (12). We provide a gene prediction set that now anchors transcript sequences within the genomic landscape. The addition of recent single cell sequencing data and multiple RNA-seq datasets generated by the planarian research community greatly improve the breadth and depth of provided gene expression information. Finally, we add transcriptomes of a broad range of other flatworms and provide a phylogeny-aware interface that facilitates evolutionary species comparisons. At its core, PlanMine continues to rely on the powerful InterMine framework (13) and uniform core annotation workflows continue to ensure the necessary data consistency for inter-species comparisons. Overall, PlanMine 3.0 thus provides a host of new functionalities to the community, but as an organic addition to the successful and widely utilized PlanMine platform. A concise discussion of the new features is given below.
Schmidtea mediterranea gene models
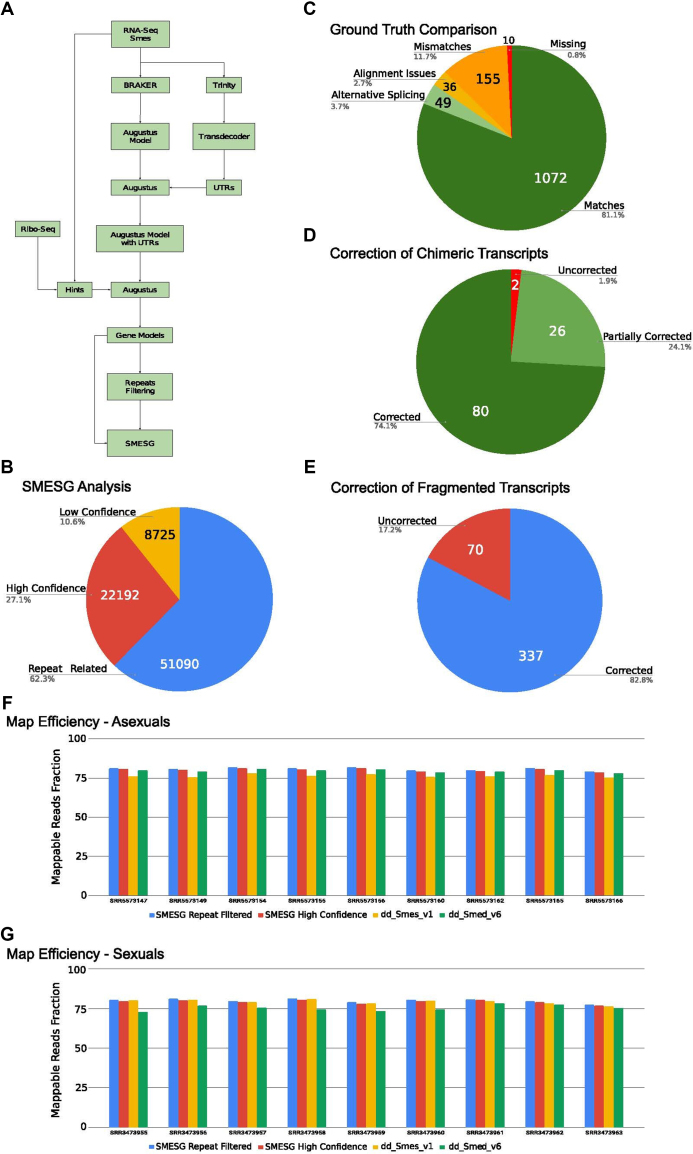
Though tremendously useful, de novo assembled transcriptomes cannot provide information regarding the genomic context of genes and thus the regulatory mechanisms that govern gene expression. The recently released high quality S. mediterranea genome assembly (12) offered an opportunity to overcome this problem via de novo gene annotations. We used a customized BRAKER/Augustus (14,15) pipeline for predicting genes in the S. mediterranea genome assembly (Figure 1A; see online user guide for further information). Briefly, we used published RNA-seq data of the sexual S. mediterranea strain as input for BRAKER and subsequent Augustus initial training. To improve the UTR representations of the resulting gene models, we first generated a new transcriptome assembly with Ribo-seq data to better represent the 5′ end of transcripts and used TransDecoder (16) to extract 5′ and 3′ UTRs of complete transcripts as additional training input for Augustus. Using the combined Ribo-seq and RNA-seq datasets as hints for Augustus gene predictions, our pipeline predicted an initial set of 82 007 genes and 132 355 mRNAs (Figure 1B) that are available as browsable tracks in PlanMine v3.0 and the associated UCSC genome browser instance (SMESG–full). The raw predictions were refined by filtering for models associated with the highly abundant repeat elements (12) with a custom Repeat Masker (RMReference) library and for minimal expression levels using lowly expressed genes to define the cut-off (e.g. Smed-wnt1 and Smed-ovo). The resulting high confidence (HC) prediction set comprises 22 192 gene models and 31 102 transcript models (Figure 1B), which is broadly consistent with the number of protein-coding genes in metazoan genomes (17). The HC predictions (SMESG-HC), the combined high and low confidence predictions (SMESG- repeat filtered) and the unfiltered SMESG-full dataset are all available for download under the PlanMine ‘data sources’ tab and can be viewed as tracks in the JBrowse window embedded in gene pages (see below).
Figure 1.
(A) Workflow for the SMESG gene predictions, see text and user guide for explanations. (B) Pie chart sub-categorization of the 82 007 gene models in the SMESG dataset. Repeat related: Models that overlap >25% with a genomic repeat annotation. Low confidence: Models with <25% repeat overlap and <0.1 rpkm expression level. HC: Models with <25% repeat overlap and >0.1 rpkm expression in the above RNA-seq dataset. (C) Ground truth comparison of SMESG predictions versus a set of 1322 HC transcripts (see user guide for details). ‘Matches’: Base pair level identity between SMESG prediction and transcript; deviations include ‘Alternative splicing’ as likely biological cause, ‘Mismatches’ with length deviation between transcript and gene model and ‘Missing’ genes without gene prediction. ‘Alignment issues’ group transcripts that could not be assayed due to technical GMAP alignment failures. (D) Correction of transcriptome chimeras by the SMESG gene models. Pie chart summary of the fraction of 108 chimeras in the dd_Smed_v6 reference transcriptome (see user guide for details) in which both (Corrected), at least one (Partially corrected) or none of the two ORFs (Uncorrected) were matched by SMESG models. (E) Correction of 407 genes with fragmented transcript representation in the reference transcriptome dd_Smed_v6 (see user guide for details). ‘Corrected’: genes with accurate full-length representation in SMESG; ‘Uncorrected’: genes with incomplete SMESG representation. (F and G) RNA-seq mapping reference comparison between indicated SMESG subsets and current reference transcriptomes. (F) RNA-seq data from the asexual Schmidtea mediterranea strain (20); (G) RNA-seq datasets from the sexual S. mediterranea strain (21). Mapping references are encoded by bar colour, bar height encodes the fraction of reads mappable against the respective reference. SRR accession number of individual RNA-seq datasets are indicated below the bars.
Just like de novo assembled transcriptomes, de novo predicted gene models are subject to various potential technical artifacts. To evaluate the quality of the gene models versus the current S. mediterranea reference transcriptome assembly, we used a set of 1300 HC S. mediterranea transcripts as ‘ground truth’ (see user guide for further information). Using the ParsEval tool (18) to compare the mapped (GMAP; 19) HC transcripts to the HC gene predictions, we found that 84% of the HC transcripts either perfectly matched a gene prediction or a predicted splice isoform (Figure 1C). About 11.7% of HC transcripts were categorized as mismatches, often due to apparent truncations in the 5′ or 3′ UTRs in the corresponding gene models. Only 10 HC transcripts (0.8%) did not have a corresponding gene model due to a prediction failure. Overall, the ground truth comparison quantitatively confirms the high degree of precision of our gene predictions, which may represent an underestimate due to the unknown contribution of transcript errors especially on the ‘mismatch’ category.
We also carried out reverse comparisons to estimate the extent by which the gene predictions remedy known shortcomings of de novo assembled transcriptomes. Chimeric transcripts are one such problem that we addressed by first establishing filtering criteria to identify representative examples in the current reference transcriptome (see user guide for further information). Out of a total of 108 chimeric transcripts identified by our pipeline, the gene predictions split >95% of the fused open reading frames (ORFs) into independent genes (Figure 1D). Further, in ∼24% or ∼74% of cases, one or both ORFs were represented by gene predictions. Therefore, the gene predictions largely overcome the problem of chimeric transcripts. A further common challenge in de novo assembled transcriptomes are fragmented transcripts. Out of 407 representative examples identified via a BLAST mapping strategy (see user guide for further information), the gene predictions correctly represent 82.8% in a single gene model (Figure 1E). A further 17.2% fail to correct the split or suffer from a mismatch elsewhere. The implication of reduced gene fragmentation is further supported by an ORF length comparison between the reference transcriptome and the HC gene predictions (see user guide for further information). As orthogonal assessment of transcript quality, we also compared the performance of the gene predictions versus existing reference transcriptomes as mapping reference for RNA-seq datasets (20, 21). Figure 1F and G show that the gene predictions consistently yield equal or higher fractions of mappable reads both for datasets from asexual or sexual strains of S. mediterranea, thus providing additional evidence that on average, the transcript representation in the predictions is en par or better than the current reference transcriptomes. This analysis further broadly confirms the equal utility of the gene predictions for work with both the asexual and sexual strains of S. mediterranea, even though the predictions were generated on basis of the sexual strain genome.
Overall, the SMESG gene predictions are therefore of comparable quality to the de novo assembled reference transcriptomes in current use by the community. We feel that the unique provision of genomic context within the S. mediterranea genome assembly justify a transition to the gene models and associated gene IDs at this point in time and we encourage the S. mediterranea research community to evaluate their utility as potential future community standard (see below).
S. mediterranea gene pages
Although we eventually intend genome-based gene annotations as the main S. mediterranea data type in PlanMine, the 3.0 release continues to provide de novo assembled transcriptomes in parallel. This allows the critical evaluation and user-driven improvement of the current gene models (see below), but also ensures continuity with the established transcripts and transcript IDs and the comparative analysis of the many flatworm species for which only transcriptomes are available. This means that for S. mediterranea, the only species with genome representation in the current PlanMine release, each gene model is represented by a gene page and associated transcripts by individual transcript pages. All other flatworm species in PlanMine continue to be represented by transcript pages only (see below).
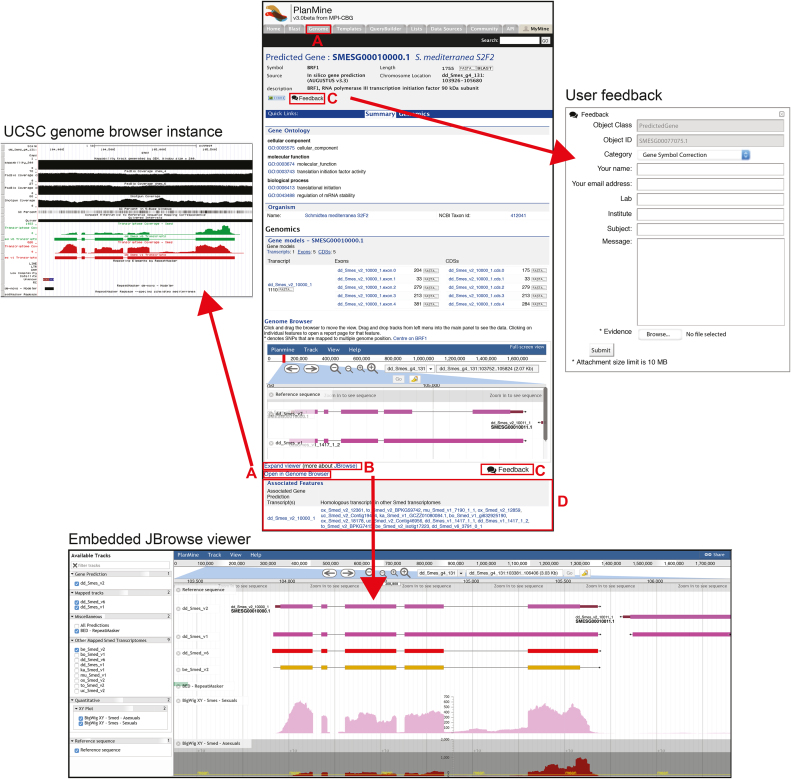
The chief objective of the gene page (Figure 2) is the provision of an overview of the genomic context of the gene model and of homologous sequences in PlanMine. Gene pages include the following elements: The gene ID (see user guide) at the top of the page (Figure 2) constitutes a unique identifier for each gene model. Gene models are designated with an alphabetical species prefix code followed by a ‘G’ to indicate a gene identifier and then a zero-padded nine digit number (e.g. SMESG000020421.1) The suffix ‘.1’ indicates the gene prediction release number. The associated predicted transcript models take a similar form with the ‘G’ replaced by a ‘T’ and the last three digits representing the transcript isoform (e.g. SMEST020421001.1 as the first transcript of the SMESG000020421.1 locus in the .1 prediction release). The ‘SMES’ in the identifier designates the S.mediterraneasexual strain genome as origin of the predictions. Internal strategies are in place to update, add or retire gene IDs in order to facilitate the anticipated rapid convergence on an accurate reference gene set for S. mediterranea. For SMES gene models or dd_Smed_v6 transcripts that match published GenBank mRNA sequences, we link to the corresponding GenBank page and display the name (e.g. annotation) of the deposited sequence. Further, we additionally list putative gene symbols derived on basis of orthology for all SMES gene models and dd_Smed_v6 transcripts (see online user guide for further information). However, due to the various pitfalls associated with automatized orthology establishment of the often highly divergent planarian gene sequences, the gene symbols in the current PlanMine release should be considered only as indicator of gene function and we encourage the use of the gene IDs to designate gene identity.
Figure 2.
Overview of gene information page containing (A) links to our UCSC Genome Browser instance hosting the Schmidtea mediterranea genome assembly, (B) a JBrowse view giving a genomic perspective of the gene models and mapped transcript information, (C) opportunities for community feedback with regard to the included gene annotations, (D) a table containing homologous transcripts from community contributed S. mediterranea transcriptomes that are associated with our gene predictions.
To visualize the genomic context of the gene, a JBrowse (22) based Genome Browser view is embedded in each gene page (Figure 2B). The default view displays the genomic location of the SMES gene model with the predicted transcripts, along with overlapping contigs/transcripts from the previous sexual and asexual S. mediterranea transcriptome assemblies in order to give those too a genomic dimension. The expanded JBrowse genome browser view (Figure 2B) allows the user-configurable activation of additional display tracks, including further S. mediterranea transcriptome assemblies and RNA-seq read mapping data. While the JBrowse window on the gene page is primarily meant to view and evaluate the genomic context of gene models, the UCSC genome browser instance that we maintain in parallel is meant for genome mining (Figure 2A). For this purpose, it provides many more annotation tracks and associated tools (e.g. BLAT). A further important feature of the gene page is the associated features section (Figure 2D) that lists all S. mediterranea transcripts that intersect the map coordinates of the gene model (see user guide for further information). Effectively, this list amounts to a consolidation of transcripts from the various previous S. mediterranea transcriptome assemblies under a single gene ID. Finally, the current PlanMine release expands and encourages user feedback options. Prominently placed ‘Feedback’ buttons in the gene ID/Symbol and JBrowse sections of the gene page and the JBrowse transcriptome viewer on transcript pages (see below) link to a feedback form (Figure 2C) with pre-configured topics (e.g. gene symbol, gene model or missing gene model). The internal collection and archiving of user feedback has been designed in order to allow facile integration in future releases of PlanMine, towards the goal of reliable and curated gene models and gene symbols.
TRANSCRIPT PAGES
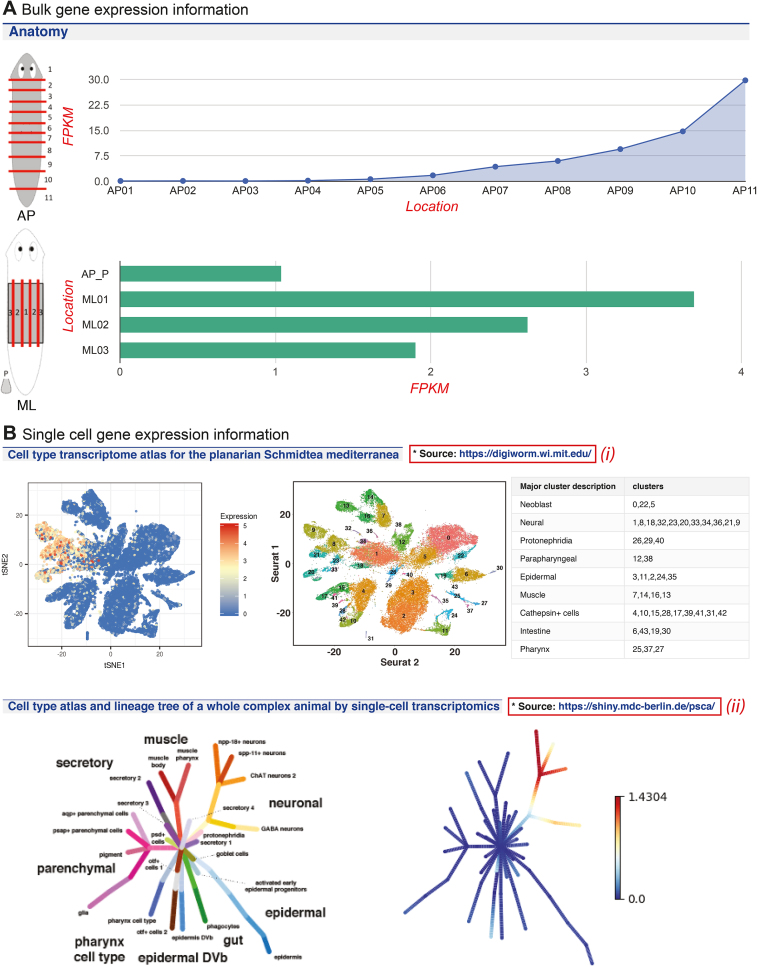
The transcript pages provide a concise summary of transcript structure, homologies and functional indications. In the case of prediction-associated transcripts (SMEST), the transcript page complements the corresponding gene page with information on gene expression dynamics; for de novo transcriptome assemblies, the transcript pages provide the sole source of sequence information. Additions to S. mediterranea transcript pages include links to the associated gene model gene page and a ‘feedback’ button to report sequences that lack a gene model. Further, a large amount of gene expression information that has been released by the community since the previous PlanMine publication has been added to transcript pages, including 12 bulk RNA-seq datasets under different RNAi conditions and a serial section sequencing dataset that documents gene expression along the A/P and M/L axis (Figure 3A). Further, for SMEST and the dd_Smed_v6 reference transcriptome transcripts, we make available two recent large-scale single cell sequencing datasets (Figure 3B). The strategic utility of these data is to complement the bulk gene expression data of the preceding section with information on the specific cell types and cell lineages in which S. mediterranea genes or transcripts are expressed. Specifically, PlanMine displays the tSNE maps of the Reddien lab dataset (23) and the lineage tree information of the Rajewsky lab dataset (24). Link-outs provide facile access to more data display and mining tools provided by the respective labs. Overall, the intuitive visualization of the expression data provides a crucial new dimension of fully mineable transcript annotations and furthers the original PlanMine mission of working with collaborators and contributors to provide one-stop access to disparate datasets.
Figure 3.
Gene expression data from a variety of internal and community sources is integrated into PlanMine including (A) bulk gene expression studies with Smed-wnt11–1/dd_Smed_v6_14391_0_1 as example, (B) single cell gene expression studies with dd_Smed_v6_6859_0_1 as example.
COMPARATIVE ANALYSIS OF FLATWORM GENE SEQUENCES
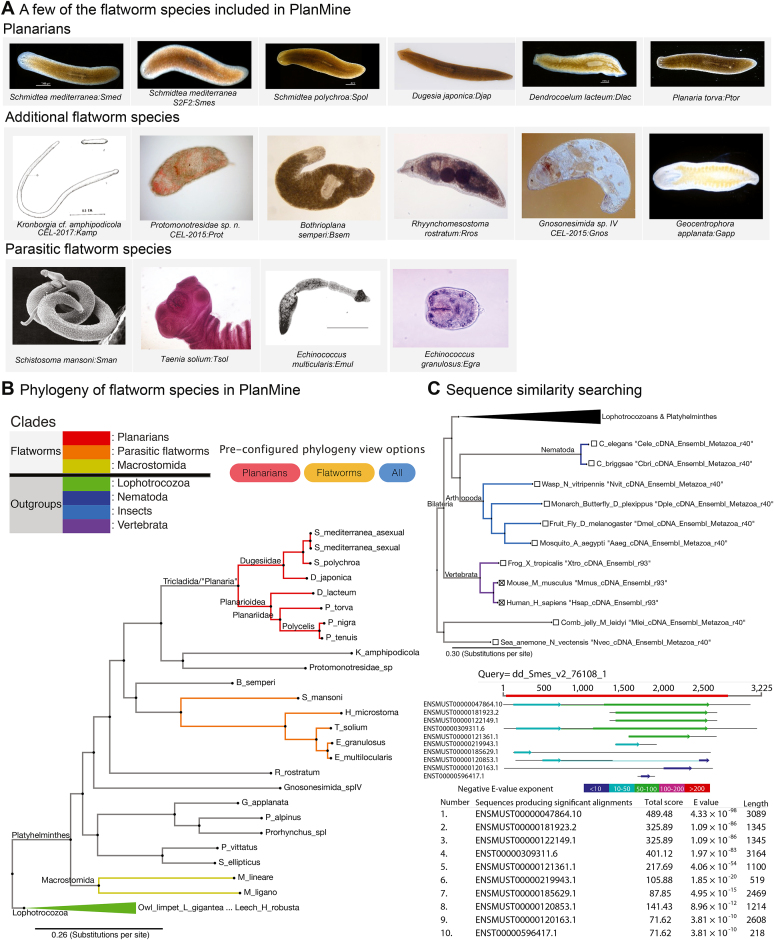
This release of PlanMine further significantly enhances utility towards its second mission objective, the comparative analysis of flatworm gene sequences. First, we include a wide range of additional species and transcriptomes, bringing the total number of represented species from seven (including the S. mediterranea sexual strain) up to 25 flatworm species. This includes the addition of the planarian model species Dugesia japonica along with the addition of five parasitic flatworm transcriptomes (6), and a taxonomically diverse set of 14 other flatworm species (25–27). Several lophotrochozoan outgroup species have also been added in order to facilitate the analysis of flatworm gene evolution, including limpet, oyster and leech transcriptomes (Ensembl release 34). To improve the accessibility of the data to non-experts, all flatworm species are also represented in tabular form using images (Figure 4A) and these link to further background information in the form of a curated species page that include more photos and descriptions of external anatomy, habitat, distribution and reproductive strategy or link-outs to public repositories.
Figure 4.
(A) There are 25 flatworm species included in the latest PlanMine release including planarian species, parasitic flatworms, macrostomids and other flatworm species. (B) Included species are presented in form of a phylogenetic tree to represent their evolutionary relationships. (C) The phylogeny can also be used to easily select appropriate species for sequence similarity searching using BLAST. BLAST options include nematode, insect and vertebrate outgroup transcriptomes from ENSEMBL release 93 and ENSEMBL Metazoa release 40.
Second, we present all species in an intuitive phylogeny-aware manner. Comparative approaches necessitate consideration of the phylogenetic relationship between the different species. To make this crucial information accessible also to non-experts, we present all PlanMine species in the context of our in-house generated multi-gene phylogeny (Figure 4B; see online user guide for further information). Important clades in the phylogeny are coloured so that planarians, parasitic worms and macrostomids can be clearly identified along with key outgroup clades. Further, we include several pre-configured tree views in order to allow users to restrict the information content for their specific purposes (e.g. planarians only, flatworms only or all species). The trees themselves are also interactive thanks to the integration of the Phylo.io JavaScript framework (28), which allows clades to be expanded or collapsed, the order of the leaf nodes changed, branches to be trimmed or even the whole phylogeny re-rooted.
Our phylogeny-aware presentation of sequence resources carries over to the sequence server-based BLAST page. The underlying BLAST database additionally includes 14 outgroup species from nematodes, arthropods, vertebrates, comb jelly and sea anemone, along with the S. mediterranea genome and gene annotations. Importantly, the individual species are again presented in the form of a phylogenetic tree, which allows the intuitive user-configurable selection of appropriate planarian, flatworm or outgroups species to include in BLAST searches. Further, all S. mediterranea transcriptome assemblies remain accessible for those who still wish to use them. Outgroup species are largely taken from ENSEMBL or ENSEMBL Metazoa with metadata indicating which release they correspond to. The Locophotrocozoan outgroup transcriptomes are included in PlanMine as ENSEMBL Metazoa release 34 transcriptomes that have been re-annoated using our standard transcriptome annotation pipeline. Advanced BLAST hit visualization customizations to the underlying SequenceServer (bioRxiv, https://doi.org/10.1101/033142) are available as with the original PlanMine release and BLAST results outlining corresponding high scoring pairs link to corresponding transcript pages in PlanMine or Ensembl/Ensembl Metazoa as appropriate. The phylogeny-aware data presentation in PlanMine allows also non-experts to address such questions as the identification of how gene expression changes correlate with phenotype divergence amongst planarians, gene losses that are specific to parasitic versus free-living flatworms or the evolution of ‘new’ genes without sequence homology outside of the flatworms.
Overall, we are therefore confident that the 3.0 PlanMine update will prove valuable to the flatworm research community and beyond.
FUTURE PLANS
Our near-term focus remains on further polishing of the SMES reference gene set, specifically the inclusion of the small fraction of currently ‘missing’ genes and of non-protein coding genes. New SMESG releases will be announced via the PlanMine website and our gene ID versioning system is designed to permit non-disruptive gene model updates. Further, we will deposit the S. mediterranea genome and reference gene set in a public repository. In the medium term, we aim to add further planarian and other flatworm genomes to PlanMine. This will entail enhancements of our UCSC genome browser instance for inter-species comparisons and for the incorporation and mining of the functional genomics data that is increasingly generated by the flatworm research community. Beyond, we will continue to work with the flatworm research community to incorporate new approaches towards the common objective of mining the rich biology and biodiversity of flatworms.
DATA AVAILABILITY
Supplementary Data are provided via the PlanMine 3.0 online user guide located under http://planmine-test.mpi-cbg.de/planmine-v3/user_guide.html.
ACKNOWLEDGEMENTS
We thank following Services and Facilities of the MPI-CBG Dresden for their support: Bioinformatics & Scientific Computing Facility and Computer Department. We thank Miquel Vila-Farré for phylogeny advice and Christopher Laumer, Gonzalo Giribet, Andreas Hejnol and Eugene Berezikov for the provision of transcriptomes. In addition, we thank Andreas Dahl and the Deep Sequencing Group (SFB 655/BIOTEC) for RNA sequencing support and Heino Andreas and Stephanie von Kannen for expert animal care.
FUNDING
European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program [649024]; Max Planck Society. Funding for open access charge: ERC Grant [64902].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sluys R., Riutort M.. Planarian diversity and phylogeny. Methods Mol. Biol. 2018; 1774:1–56. [DOI] [PubMed] [Google Scholar]
- 2. Vila-Farré M., Rink C.J.. The ecology of freshwater planarians. Methods Mol. Biol. 2018; 1774:173–205. [DOI] [PubMed] [Google Scholar]
- 3. Reddien P.W., Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004; 20:725–757. [DOI] [PubMed] [Google Scholar]
- 4. Saló E., Agata K.. Planarian regeneration: a classic topic claiming new attention. Int. J. Dev. Biol. 2012; 56:3–4. [DOI] [PubMed] [Google Scholar]
- 5. Rink J.C. Stem cells, patterning and regeneration in planarians: self-organization at the organismal scale. Methods Mol. Biol. 2018; 1774:57–172. [DOI] [PubMed] [Google Scholar]
- 6. Tsai I.J., Zarowiecki M., Holroyd N., Garciarrubio A., Sanchez-Flores A., Brooks K.L., Tracey A., Bobes R.J., Fragoso G., Sciutto E. et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013; 496:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collins J.J., Newmark P.A.. It's no fluke: the planarian as a model for understanding schistosomes. PLoS Pathog. 2013; 9:e1003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sikes J.M., Newmark P.A.. Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature. 2013; 500:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu S.-Y., Selck C., Friedrich B., Lutz R., Vila-Farré M., Dahl A., Brandl H., Lakshmanaperumal N., Henry I., Rink J.C.. Reactivating head regrowth in a regeneration-deficient planarian species. Nature. 2013; 500:81–84. [DOI] [PubMed] [Google Scholar]
- 10. Umesono Y., Tasaki J., Nishimura Y., Hrouda M., Kawaguchi E., Yazawa S., Nishimura O., Hosoda K., Inoue T., Agata K.. The molecular logic for planarian regeneration along the anterior-posterior axis. Nature. 2013; 500:73–76. [DOI] [PubMed] [Google Scholar]
- 11. Brandl H., Moon H., Vila-Farré M., Liu S.-Y., Henry I., Rink J.C.. PlanMine - a mineable resource of planarian biology and biodiversity. Nucleic Acids Res. 2016; 44:D764–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grohme M., Schloissnig S., Rozanski A., Pippel M., Young G., Winkler S., Brandl H., Henry I., Dahl A., Powell S.. The genome of S. mediterranea and the evolution of cellular core mechanisms. Nature. 2018; 554:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lyne R., Sullivan J., Butano D., Contrino S., Heimbach J., Hu F., Kalderimis A., Lyne M., Smith R.N., Stepan R. et al. Cross-organism analysis using InterMine. Genesis. 2015; 53:547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanke M., Waack S.. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003; 19(Suppl. 2):ii215–ii225. [DOI] [PubMed] [Google Scholar]
- 15. Hoff K.J., Lange S., Lomsadze A., Borodovsky M., Stanke M.. BRAKER1: Unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016; 32:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013; 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. et al. Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Standage D.S., Brendel V.P.. ParsEval: parallel comparison and analysis of gene structure annotations. BMC Bioinformatics. 2012; 13:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu T.D., Watanabe C.K.. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005; 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- 20. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fincher C.T., Wurtzel O., de Hoog T., Kravarik K.M., Reddien P.W.. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science. 2018; 360:eaaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Plass M., Solana J., Wolf F.A., Ayoub S., Misios A., Glažar P., Obermayer B., Theis F.J., Kocks C., Rajewsky N.. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. 2018; 360:eaaq1723. [DOI] [PubMed] [Google Scholar]
- 23. Martín-Durán J.M., Ryan J.F., Vellutini B.C., Pang K., Hejnol A.. Increased taxon sampling reveals thousands of hidden orthologs in flatworms. Genome Res. 2017; 27:1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grudniewska M., Mouton S., Simanov D., Beltman F., Grelling M., de Mulder K., Arindrarto W., Weissert P.M., van der Elst S., Berezikov E.. Transcriptional signatures of somatic neoblasts and germline cells in Macrostomum lignano. Elife. 2016; 5:e20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laumer C.E., Hejnol A., Giribet G.. Nuclear genomic signals of the ‘microturbellarian’ roots of platyhelminth evolutionary innovation. Elife. 2015; 4:e05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson O., Dylus D., Dessimoz C.. Phylo.io: interactive viewing and comparison of large phylogenetic trees on the web. Mol. Biol. Evol. 2016; 33:2163–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scimone M.L., Cote L.E., Reddien P.W.. Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature. 2017; 551:623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rouhana L., Tasaki J., Saberi A., Newmark P.A.. Genetic dissection of the planarian reproductive system through characterization of Schmidtea mediterranea CPEB homologs. Dev. Biol. 2017; 426:43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary Data are provided via the PlanMine 3.0 online user guide located under http://planmine-test.mpi-cbg.de/planmine-v3/user_guide.html.