Abstract
Protein post-translational modifications (PTMs), including phosphorylation, ubiquitination, methylation, acetylation, glycosylation et al, are very important biological processes. PTM changes in some critical genes, which may be induced by base-pair substitution, are shown to affect the risk of diseases. Recently, large-scale exome-wide association studies found that missense single nucleotide polymorphisms (SNPs) play an important role in the susceptibility for complex diseases or traits. One of the functional mechanisms of missense SNPs is that they may affect PTMs and leads to a protein dysfunction and its downstream signaling pathway disorder. Here, we constructed a database named AWESOME (A Website Exhibits SNP On Modification Event, http://www.awesome-hust.com), which is an interactive web-based analysis tool that systematically evaluates the role of SNPs on nearly all kinds of PTMs based on 20 available tools. We also provided a well-designed scoring system to compare the performance of different PTM prediction tools and help users to get a better interpretation of results. Users can search SNPs, genes or position of interest, filter with specific modifications or prediction methods, to get a comprehensive PTM change induced by SNPs. In summary, our database provides a convenient way to detect PTM-related SNPs, which may potentially be pathogenic factors or therapeutic targets.
INTRODUCTION
Germline genetic variants, mostly single nucleotide polymorphisms (SNPs), have been shown to significantly associate with complex diseases or traits, such as cancer, type 2 diabetes, cardiovascular diseases, heights et al. These susceptibility genetic polymorphisms could be divided into two groups: regulatory variants located in noncoding regions and missense or synonymous variants located in coding regions (1). The regulatory variants have been identified by genome-wide association studies (GWAS) and many tools have been built for the annotation of these variants (2,3). Recently, more and more large-scale exome-wide association studies showed the coding variants have also been significantly correlated with complex diseases or traits, especially for these low-frequency or rare missense variants with relatively high effect size (odds ratio > 1.5) (4–8). Systematic investigation of the functional mechanism underlying these missense variants will be the next challenge for the researchers. It is hypothesized that these missense variants may alter the amino acid sequences, and thus significantly influence some critical biological processes by inducing the protein structure disorders (9,10), or disrupting protein-macromolecules interactions (11), or affecting post-translational modifications (PTMs) (12,13). However, useful tools for functional annotation of these coding variants are still lacking, especially for variants potentially affect PTMs.
PTM refers to the covalent and enzymatic modification of proteins during or after biosynthesis (14). Such modifications come in a wide variety of types, such as phosphorylation, glycosylation, ubiquitination, methylation, acetylation etc. They play a crucial role in many biological processes including regulation protein folding (15), cellular differentiation (16), protein degradation (17), signalling and regulatory processes (18,19), regulation of gene expression (20,21), interaction with ligands or other proteins (22–24), and protein functional state (25,26). Alterations of these PTM target sites were found to be directly involved in the development of diseases. For example, the G553E mutation (rs118005095) was identified to completely abrogate PTMs of huntingtin gene (HTT) and induce cellular toxicity of the protein (27). The missense mutation of Lys27Met (K27M) in the gene encoding histones H3.3 (H3F3A) reduces the overall methylation level of H3K27me3 by inhibiting the enzymatic activity of polycomb repressive complex 2, and increases the risk of diffuse intrinsic pontine gliomas (28). Therefore, identifying and understanding variants that affect PTMs is critical in the study of cell biology, disease treatment and prevention.
Although there are many excellent SNP interpretation tools, such as VarCards (29), ANNOVAR (2) and Ensembl Variant Effect Predictor (VEP) (3), none of which includes a PTM analysis for exonic variants. The PTM-related databases either only focus on annotation of experimental identified PTM sites (PhosphoSitePlus, HPRD, PTMfunc or ActiveDriverDB) (24,30–32), or only predict a single type of undiscovered PTM sites (such as NetPhos, CKSAAP or MePred-RF) (33–35). None of these above consider the effect of SNPs on PTM systematically, although these SNPs may be pathogenic factors or therapeutic targets. Even though there is a database integrated SNP and PTM, PhosSNP (36), it only focus on phosphorylation based on GPS (37). Therefore, a resource for systematically annotating SNPs on most common types of PTM is needed to help researchers to easily discover the potential function of exonic variants.
Here, we constructed a comprehensive platform to collect and integrate SNPs and multiple PTM information. A total of 1,043,608 germline missense variants from the dbSNP was used and each SNP was matched with its protein sequence. We then utilized 24 published database or tools, including four databases with experimental PTM data (PhosphoSitePlus, HPRD, dbPTM and Phospho.ELM) (30,31,38,39), which covers nearly all types of PTM and 20 PTM prediction tools (ReKINect, GPS, PPSP, Musite, MusiteDeep, NetPhorest, NetPhos, PhosPred-RF, hCKSAAP_UbSite, UbiProber, UbiSite, UbPred, GPS-MSP, MePred-RF, NetOGlyc, NetNGlyc, YinOYang, NetAcet, GPS-PAIL, GPS-SUMO) (33–35,37,40–55), which covers six common types of PTMs (phosphorylation, ubiquitination, methylation, glycosylation, acetylation and sumoylation) to predict whether a SNP could affect protein PTM. To help users interpret results from different prediction tools, we have developed a well-designed scoring system to compare the performance of various bioinformatics tools, thus allowing the users to have a better and clear overview of prediction results.
DATA COLLECTION AND PROCESSING
Data collection
A total of 1,043,608 missense SNPs was downloaded from dbSNP (138) in which both common and rare variants located in protein coding regions across human genome were obtained (Figure 1). The SNP information, such as allele change, amino acid change, chromosome location (hg19/hg38), gene symbol, protein sequence, minor allele frequency, polyphen2, and SIFT score were annotated via VEP (3). The mapping of protein sequences and missense SNPs was achieved by canonical Ensembl Protein ID (ENSP) and HGVSp. Genomic coordinates of SNPs were mapped to protein amino acid substitutions using the VEP (3) and ‘biomaRt’ package in R. Reference nucleotide sequences (RefSeq) were referred to National Center for Biotechnology Information (NCBI) nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/). Cancer gene list used for result interpretation was downloaded from Catalogue of Somatic Mutations in Cancer (COSMIC).
Figure 1.
Overview and workflow of AWESOME. The database integrates genomic information and PTM database/tools to annotate missense SNPs that potentially affect PTM. We downloaded all missense SNPs from dbSNP (138), then mapped these SNPs to canonical protein sequence via VEP and ‘biomaRt’ package in R. Four experimental PTM database were applied to annotate PTM-related SNPs. Twenty bioinformatics tools were applied to predict PTM-related SNPs. All of the results were rearranged and scored, and then presented at the website.
PTM annotation of SNPs
The PTM changes affected by SNPs were annotated by experimental data and prediction tools. The experimental data of protein PTM sites were obtained from PhosphoSitePlus (30), HPRD (31), dbPTM (38) and Phospho.ELM (39). It contains not only the six common types of PTM (phosphorylation, ubiquitination, methylation, glycosylation, acetylation and sumoylation), but also uncommon types, such as sulfation, carboxylation, hydroxylation etc.
We used multiple tools to predict whether a SNP could affect the six common types of PTM. Of which the tools used for each PTM type are listed below.
The potential change of phosphorylation affected by SNPs was predicted by 8 tools: ReKINect (40), PPSP (41), NetPhos (33), GPS (37), Musite (42), MusiteDeep (43), PhosPred-RF (46) and NetPhorest (44,45). These prediction tools were based on different methods. The ReKINect (40) is a computational framework that predicts the functionality of mutation. The PPSP (41), NetPhos (33), NetPhorest (44,45) and GPS (37) are developed by using Bayesian decision theory, ensembles of neural networks, neural networks and group-based phosphorylation scoring method, respectively. The Musite (42), MusiteDeep (43) and PhosPred-RF (46) are machine learning-based phosphorylation site prediction tools. For ReKINect, we inputted ±15 amino acids (aa) around the SNP and run the script locally. After the reference and mutant sequences were submitted, the ‘destruction of phosphorylation site’ was seen for PTM-related SNPs. For GPS, we inputted ±25 aa around the SNP and selected all kinases with ‘high threshold’ to predict kinase-specific phosphorylation sites on local software. For Musite and MusiteDeep, we performed a human general phosphorylation site prediction using provided models with ±40 aa (Musite) and ±17 aa (MusiteDeep) around the SNP as input. For the other four tools, we inputted ±31 aa (NetPhos), ±5 aa (NetPhorest and PPSP) and ±12 aa (PhosPred-RF) around the SNP, respectively, with default setting and computed in local server or at websites.
The potential change of ubiquitination affected by SNPs was predicted by four tools, which cover all bioinformatics methods available. The UbPred (49) is a random forest-based predictor. The UbiProber (47), UbiSite (48) and hCKSAAP_UbSite (34) use support vector machine to make predictions. For UbiSite, we inputted ±6 aa around the SNP with specificity level as ‘low threshold (85%)’. For UbiProber, we used ‘H.sapiens model’ and set specificity as zero to get scores of all sites with ±13 aa around the SNP input. For the other two tools, we inputted ±17 aa (hCKSAAP_UbSite) and ±40 aa (UbPred) around the SNP, respectively, with default settings.
The potential change of glycosylation affected by SNPs was predicted by three tools, which cover three most common types of glycosylation in humans: N-linked glycosylation and O-linked glycosylation, with the later includes two types of sugar: O-N-acetylgalactosamine (O-GalNAc) and O-N-acetylglucosamine (O-GlcNAc). The NetNGlyc server predicts N-glycosylation sites in human proteins using artificial neural networks that examine the sequence context of Asn-Xaa-Ser/Thr sequons. The NetOGlyc (51) server produces neural network predictions of mucin type GalNAc O-glycosylation sites in mammalian proteins. The YinOYang (52) predicts O-ß-GlcNAc attachment sites by using neural network as well. We inputted ±50 aa around the SNP (YinOYang and NetNGlyc) and full length of protein (NetOGlyc) with default settings for these three tools, respectively.
The potential change of methylation affected by SNPs was predicted by two tools. The GPS-MSP (50) is a methyl-group specific predictor for the prediction of protein methylation modifications. We performed all methylation (R.mono, R.di, K.mono, K.di, K.tri) predictions with the lowest threshold for ±25 aa around the SNP. The MePred-RF (35) is based on machine learning and can identify potential methylation sites within the input proteins. We inputted ±5 aa around the SNP with default settings.
The potential change of acetylation affected by SNPs was predicted by two tools, which cover two types of acetylation modification widely occurred in proteins: N-terminal acetylation and lysine acetylation. The NetAcet (53) was used to predict N-terminal acetylation and the GPS-PAIL (54) was carried out to predict lysine acetylation. We inputted an N-terminal 20 aa (NetAcet) and ±20 aa (GPS-PAIL) around the SNP, respectively, with default settings.
The potential change of sumoylation affected by SNPs was predicted by GPS-PAIL (54). We used three different thresholds (‘low’, ‘medium’ and ‘high’) with ±20 aa around the SNP as input options.
For all the above PTM prediction tools, the reference and mutant sequences were submitted separately to predict the gain or loss of PTM sites. All the predicted PTM sites were marked with protein ID and mutation position, of which the results were then rearranged and all of the modification sites were matched to corresponding missense SNPs.
PTM prediction performance evaluation by a score system
To evaluate the prediction performance of different methods and estimate specificity under various thresholds, we performed validation tests. For each type of PTM, we chose a list of genes that were well-studied previously as a positive control (Supplementary Data 1). Receiver operating characteristic (ROC) curves were plotted and the Area Under the Curves (AUC) were calculated by taking different thresholds for each PTM prediction tool. The minimal distance between ROC curve and point (0,1) was used to determinate the optimal cut-off point. For some methods without comparable scores, like ReKINect (40), PPSP (41) and GPS (37), we also reported the specificity and sensitivity value by using a default threshold. We then presented a cumulative scoring system for three types of PTM with more than one prediction method: phosphorylation, ubiquitination and methylation.
The scores were calculated as below: First, we defined an optimal cut-off value for each method as mentioned above. Second, we calculated the specificity under the cut-off value as a score. If a SNP is predicted to gain a PTM site, it will be added to the score. If lost, the score will be deducted. If it isn’t a PTM-related SNP, a score of zero will be given. Finally, we summed all the values given from each method to get a final score.
Website building
The current version of AWESOME has been developed using MongoDB 3.6.5 (https://www.mongodb.com/) and runs on a Linux-based Nginx Web server. NodeJS 8.10.0 (https://nodejs.org/en/) is used for server-side scripting. We designed and built the interactive interface using ReactJS (https://reactjs.org/), a modern JavaScript library for building user interfaces. We recommend using a modern web browser such as Google Chrome (preferred), Firefox or Safari to achieve the best display effect.
DATABASE CONTENT AND USAGE
Data summary
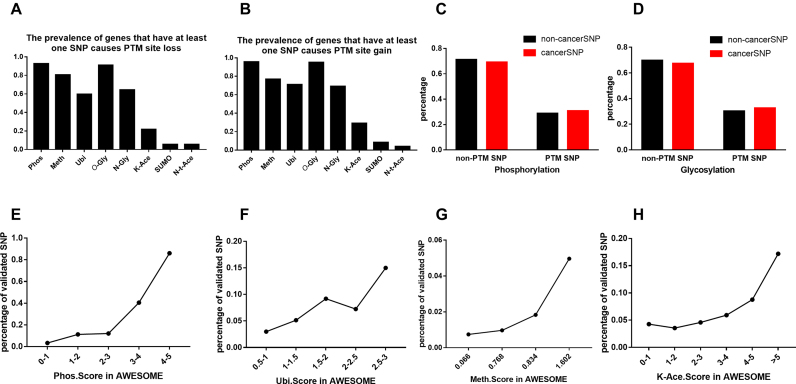
The current version of AWESOME consists of six types of PTM from prediction tools and nearly all types of PTM from experimental data. A total of 481,557 missense SNPs in 17,578 genes were predicted to alter at least one type of PTM (Figure 2A and B). These PTM-related SNPs were nearly 50% of all missense SNPs (36% SNPs affected phosphorylation, 16% SNPs affected ubiquitination, 21% SNPs affected methylation, 37% SNPs affected glycosylation, 11% SNPs affected acetylation, respectively). We then tested whether these PTM-related SNPs were enriched in some key genes involved in the development of diseases. We found that SNPs in cancer-related genes (from the COSMIC database) are more likely to affect their protein phosphorylation (chi-test P < 0.0001) and glycosylation (chi-test P < 0.0001) (Figure 2C and D). It reflected that protein phosphorylation alterations may play an important role in the development of cancer, which is consistent with previous studies (56).
Figure 2.
Statistical results of PTM-related SNPs in AWESOME. (A, B) The prevalence of genes that have at least one SNP that causes PTM loss/gain for at least one type of modification. (C, D) The percentage of predicted phosphorylation/glycosylation-related cancer associated SNPs and non-cancer SNPs. (E, H) The percentage of experimental data validated PTM-related SNPs in different prediction score ranges.
Another important feature of AWESOME is that the scoring system that we built has an effective way to estimate the prediction results. PTM-related SNPs with the validated experimental PTM data have a higher score. With the increase of the score, more experimental validated PTM-related SNPs are shown (Figure 2E–H). About 80% of SNPs with phosphorylation score >4 are based on experimental validated PTM sites. Other PTM predictions have similar performance.
Web design and interface
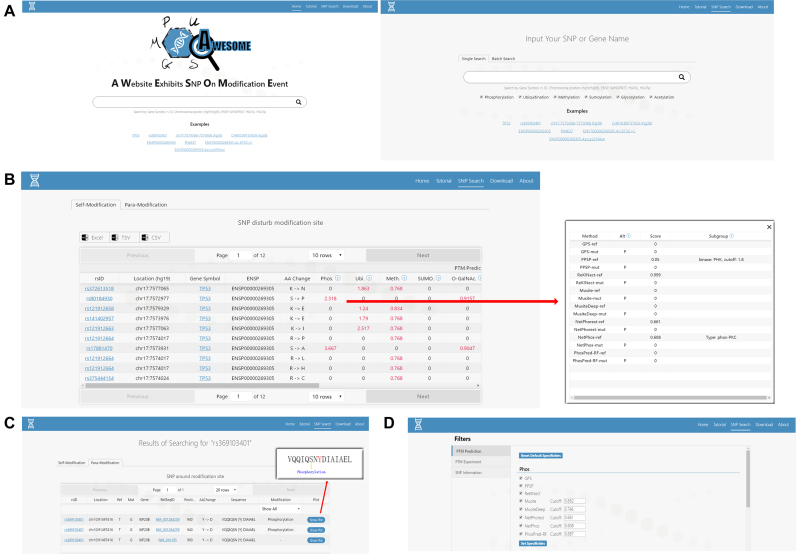
The AWESOME website (http://www.awesome-hust.com) features a user-friendly query interface and a set of custom filter function that provides a comprehensive overview of PTM change related SNPs. Users can input gene symbol, SNP rsID, chromosome position (hg19/hg38), Ensembl Protein ID, SWISSPROT ID, HGVSc ID and HGVSp ID to retrieve the results. We have shown examples for each input format available under the query box. A batch search option will be available on the SNP search page to help users search and download multiple variants data quickly (Figure 3A).
Figure 3.
Examples of some key elements of AWESOME’s user interface. (A) the single search page and the batch search page; (B) The ‘SNP Search’ page presents ‘Self-Modification’ results, including SNP basic information and PTM annotation results with an extended box displays detailed results; (C) The ‘SNP Search’ page presents ‘Para-Modification’ results, including SNP basic information and PTM annotation results for SNPs near a PTM sites; (D) The filter page for setting custom options.
Once the query is submitted and processed, the AWESOME will provide two tabular summary results for all queried SNPs (Figure 3B). The ‘Self-Modification’ part shows result for SNPs that locate just at the PTM site and may directly make gene gain or loss a PTM. It also contains SNP basic information, including SNP rsID, chromosome position, gene symbol, Ensmbl Protein ID, amino acid change. The PTM results are divided into two major columns named ‘PTM Prediction’ and ‘PTM Experiment’ for results based on prediction and experimental PTM data, respectively. For each type of PTM, a score is given to show whether the SNP could lead to a loss of PTM (positive value), a gain of PTM (negative value) or no change (zero). Users can obtain details for the PTM results (from each tool) by clicking on the score (Figure 3B). Furthermore, users can sort specific column in alphabetical order or value order by clicking on the column name. The ‘Para-Modification’ annotates SNPs that locates upstream or downstream (±7 amino acids) of the experimental validated PTM sites. Those SNPs may affect the PTM level. A schematic diagram for the position of SNP site and PTM site can be viewed in the last column of PTM result (Figure 3C).
At the bottom of the result page, a filter box with 3 categories is available (Figure 3D). Users can (i) filter with the type of PTM or prediction method with custom threshold; (ii) filter with four experimental databases and can select one or more databases to get custom results; (iii) filter with SNP information including chromosome location (hg38), PolyPhen score, SIFT score, HGVSc ID, HGVSp ID and the minor allele frequency for specific populations. Once the options in SNP information category are checked, new columns with additional data will be added to the end results immediately.
DISCUSSION
Protein post-translational modifications, including phosphorylation, ubiquitination, methylation, acetylation, glycosylation, are involved in the development of many diseases. Therefore, understanding the PTMs affected by SNPs will help researchers to explore the function of susceptibility coding variants identified by large-scale association studies. In our project, we provide a systematic annotation for the potential protein PTM affected by germline coding variants. All the properties are transparently mapped onto the present version profile via an easy-to-use and interactive user interface.
Compared with other annotation tools for coding variants, the AWESOME has the following advantages. (i) AWESOME integrates nearly all kinds of common post-translational modifications. (ii) AWESOME integrates 1,043,608 coding SNPs that cover both common and rare variants. (iii) AWESOME annotates with multiple tools based on both prediction and experimental data. (iv) AWESOME makes a well-designed scoring system, which helps users to find the most possible PTM-related SNPs. (v) AWESOME has a user-friendly interface that supports query by gene, rsID or chromosome location etc. and a batch query and download.
CONCLUSION
The AWESOME provides useful information on PTM-related variants to help researchers interpret the disease-related coding variants by PTM function. It will be continually updated whenever new PTM prediction tools or experimental data are released in public.
Supplementary Material
ACKNOWLEDGEMENTS
We thank for Weilin Nie for the help with web design and building. We are indebted to Pau Creixell, Xavier Robin and Rune Linding who provided original script of ReKINect. We are grateful to Shiyao Yu, Shi Chen and Xuhang Ying for improving script and solving programming issues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Plan Program [2016YFC1302702 to X.M.]; National Program for Support of Top-notch Young Professionals, National Natural Science Foundation of China [81171878, 81222038 to X.M.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gallagher M.D., Chen-Plotkin A.S.. The post-GWAS era: from association to function. Am. J. Hum. Genet. 2018; 102:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang K., Li M., Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F.. The ensembl variant effect predictor. Genome Biol. 2016; 17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu J., Beaty T.H., Scott A.F., Hetmanski J., Parker M.M., Wilson J.E., Marazita M.L., Mangold E., Albacha-Hejazi H., Murray J.C. et al. . Whole exome association of rare deletions in multiplex oral cleft families. Genet. Epidemiol. 2017; 41:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao Y., Yun D., Zou X., Jiang T., Li G., Hu L., Chen J., Xu J., Mao Y., Chen H. et al. . Whole exome-wide association study identifies a missense variant in SLC2A4RG associated with glioblastoma risk. Am. J. Cancer Res. 2017; 7:1937–1947. [PMC free article] [PubMed] [Google Scholar]
- 6. Li J., Chang J., Tian J., Ke J., Zhu Y., Yang Y., Gong Y., Zou D., Peng X., Yang N. et al. . A rare variant P507L in TPP1 interrupts TPP1-TIN2 interaction, influences telomere length, and confers colorectal cancer risk in Chinese population. Cancer Epidemiol. Biomarkers Prev. 2018; 9:1029–1035. [DOI] [PubMed] [Google Scholar]
- 7. Chang J., Zhong R., Tian J., Li J., Zhai K., Ke J., Lou J., Chen W., Zhu B., Shen N. et al. . Exome-wide analyses identify low-frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat. Genet. 2018; 50:338–343. [DOI] [PubMed] [Google Scholar]
- 8. Chang J., Tian J., Yang Y., Zhong R., Li J., Zhai K., Ke J., Lou J., Chen W., Zhu B. et al. . A rare missense variant in TCF7L2 associates with colorectal cancer risk by interacting with a GWAS-identified regulatory variant in the MYC enhancer. Cancer Res. 2018; 17:5164–5172. [DOI] [PubMed] [Google Scholar]
- 9. Iacovache I., De Carlo S., Cirauqui N., Dal Peraro M., van der Goot F.G., Zuber B.. Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process. Nat. Commun. 2016; 7:12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suri M., Evers J.M.G., Laskowski R.A., O’Brien S., Baker K., Clayton-Smith J., Dabir T., Josifova D., Joss S., Kerr B. et al. . Protein structure and phenotypic analysis of pathogenic and population missense variants in STXBP1. Mol. Genet. Genomic Med. 2017; 5:495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nishi H., Nakata J., Kinoshita K.. Distribution of single-nucleotide variants on protein-protein interaction sites and its relationship with minor allele frequency. Protein Sci. 2016; 25:316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hendriks W.J., Pulido R.. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim. Biophys. Acta. 2013; 1832:1673–1696. [DOI] [PubMed] [Google Scholar]
- 13. Alodaib A., Sobreira N., Gold W.A., Riley L.G., Van Bergen N.J., Wilson M.J., Bennetts B., Thorburn D.R., Boehm C., Christodoulou J.. Whole-exome sequencing identifies novel variants in PNPT1 causing oxidative phosphorylation defects and severe multisystem disease. Eur. J. Hum. Genet. 2016; 25:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Knorre D.G., Kudryashova N.V., Godovikova T.S.. Chemical and functional aspects of posttranslational modification of proteins. Acta Naturae. 2009; 1:29–51. [PMC free article] [PubMed] [Google Scholar]
- 15. Bah A., Vernon R.M., Siddiqui Z., Krzeminski M., Muhandiram R., Zhao C., Sonenberg N., Kay L.E., Forman-Kay J.D.. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015; 519:106–109. [DOI] [PubMed] [Google Scholar]
- 16. Henley J.M., Craig T.J., Wilkinson K.A.. Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 2014; 94:1249–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Varshavsky A. The ubiquitin system, autophagy, and regulated protein degradation. Annu. Rev. Biochem. 2017; 86:123–128. [DOI] [PubMed] [Google Scholar]
- 18. Song Y., Brady S.T.. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol. 2015; 25:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mowen K.A., David M.. Unconventional post-translational modifications in immunological signaling. Nat. Immunol. 2014; 15:512–520. [DOI] [PubMed] [Google Scholar]
- 20. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 21. Wu S.C., Kallin E.M., Zhang Y.. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res. 2010; 20:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishi H., Hashimoto K., Panchenko A.R.. Phosphorylation in protein-protein binding: effect on stability and function. Structure. 2011; 19:1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zanzoni A., Carbajo D., Diella F., Gherardini P.F., Tramontano A., Helmer-Citterich M., Via A.. Phospho3D 2.0: an enhanced database of three-dimensional structures of phosphorylation sites. Nucleic Acids Res. 2011; 39:D268–D271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beltrao P., Albanese V., Kenner L.R., Swaney D.L., Burlingame A., Villen J., Lim W.A., Fraser J.S., Frydman J., Krogan N.J.. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012; 150:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghioni P., D’Alessandra Y., Mansueto G., Jaffray E., Hay R.T., La Mantia G., Guerrini L.. The protein stability and transcriptional activity of p63alpha are regulated by SUMO-1 conjugation. Cell Cycle. 2005; 4:183–190. [DOI] [PubMed] [Google Scholar]
- 26. Kudlow J.E. Post-translational modification by O-GlcNAc: another way to change protein function. J. Cell. Biochem. 2006; 98:1062–1075. [DOI] [PubMed] [Google Scholar]
- 27. Martin D.D.O., Kay C., Collins J.A., Nguyen Y.T., Slama R.A., Hayden M.R.. A human huntingtin SNP alters post-translational modification and pathogenic proteolysis of the protein causing Huntington disease. Sci. Rep. 2018; 8:8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan K.M., Fang D., Gan H., Hashizume R., Yu C., Schroeder M., Gupta N., Mueller S., James C.D., Jenkins R. et al. . The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013; 27:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J., Shi L., Zhang K., Zhang Y., Hu S., Zhao T., Teng H., Li X., Jiang Y., Ji L. et al. . VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018; 46:D1039–D1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E.. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015; 43:D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. et al. . Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009; 37:D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krassowski M., Paczkowska M., Cullion K., Huang T., Dzneladze I., Ouellette B.F.F., Yamada J.T., Fradet-Turcotte A., Reimand J.. ActiveDriverDB: human disease mutations and genome variation in post-translational modification sites of proteins. Nucleic Acids Res. 2018; 46:D901–D910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blom N., Gammeltoft S., Brunak S.. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999; 294:1351–1362. [DOI] [PubMed] [Google Scholar]
- 34. Chen Z., Zhou Y., Song J., Zhang Z.. hCKSAAP_UbSite: improved prediction of human ubiquitination sites by exploiting amino acid pattern and properties. Biochim. Biophys. Acta. 2013; 1834:1461–1467. [DOI] [PubMed] [Google Scholar]
- 35. Wei L., Xing P., Shi G., Ji Z.L., Zou Q.. Fast prediction of protein methylation sites using a sequence-based feature selection technique. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017; doi:10.1109/TCBB.2017.2670558. [DOI] [PubMed] [Google Scholar]
- 36. Ren J., Jiang C., Gao X., Liu Z., Yuan Z., Jin C., Wen L., Zhang Z., Xue Y., Yao X.. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol. Cell. Proteomics. 2010; 9:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X.. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics. 2008; 7:1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang K.Y., Su M.G., Kao H.J., Hsieh Y.C., Jhong J.H., Cheng K.H., Huang H.D., Lee T.Y.. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 2016; 44:D435–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dinkel H., Chica C., Via A., Gould C.M., Jensen L.J., Gibson T.J., Diella F.. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 2011; 39:D261–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Creixell P., Schoof E.M., Simpson C.D., Longden J., Miller C.J., Lou H.J., Perryman L., Cox T.R., Zivanovic N., Palmeri A. et al. . Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell. 2015; 163:202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xue Y., Li A., Wang L., Feng H., Yao X.. PPSP: prediction of PK-specific phosphorylation site with Bayesian decision theory. BMC Bioinformatics. 2006; 7:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao J., Thelen J.J., Dunker A.K., Xu D.. Musite, a tool for global prediction of general and kinase-specific phosphorylation sites. Mol. Cell. Proteomics. 2010; 9:2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang D., Zeng S., Xu C., Qiu W., Liang Y., Joshi T., Xu D.. MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction. Bioinformatics. 2017; 33:3909–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller M.L., Jensen L.J., Diella F., Jorgensen C., Tinti M., Li L., Hsiung M., Parker S.A., Bordeaux J., Sicheritz-Ponten T. et al. . Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 2008; 1:ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Linding R., Jensen L.J., Ostheimer G.J., van Vugt M.A., Jorgensen C., Miron I.M., Diella F., Colwill K., Taylor L., Elder K. et al. . Systematic discovery of in vivo phosphorylation networks. Cell. 2007; 129:1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wei L., Xing P., Tang J., Zou Q.. PhosPred-RF: a novel sequence-based predictor for phosphorylation sites using sequential information only. IEEE Trans. Nanobiosci. 2017; 16:240–247. [DOI] [PubMed] [Google Scholar]
- 47. Chen X., Qiu J.D., Shi S.P., Suo S.B., Huang S.Y., Liang R.P.. Incorporating key position and amino acid residue features to identify general and species-specific Ubiquitin conjugation sites. Bioinformatics. 2013; 29:1614–1622. [DOI] [PubMed] [Google Scholar]
- 48. Huang C.H., Su M.G., Kao H.J., Jhong J.H., Weng S.L., Lee T.Y.. UbiSite: incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines. BMC Syst. Biol. 2016; 10(Suppl. 1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Radivojac P., Vacic V., Haynes C., Cocklin R.R., Mohan A., Heyen J.W., Goebl M.G., Iakoucheva L.M.. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010; 78:365–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deng W., Wang Y., Ma L., Zhang Y., Ullah S., Xue Y.. Computational prediction of methylation types of covalently modified lysine and arginine residues in proteins. Brief. Bioinform. 2017; 18:647–658. [DOI] [PubMed] [Google Scholar]
- 51. Steentoft C., Vakhrushev S.Y., Joshi H.J., Kong Y., Vester-Christensen M.B., Schjoldager K.T., Lavrsen K., Dabelsteen S., Pedersen N.B., Marcos-Silva L. et al. . Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013; 32:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gupta R., Brunak S.. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002; 310–322. [PubMed] [Google Scholar]
- 53. Kiemer L., Bendtsen J.D., Blom N.. NetAcet: prediction of N-terminal acetylation sites. Bioinformatics. 2005; 21:1269–1270. [DOI] [PubMed] [Google Scholar]
- 54. Deng W., Wang C., Zhang Y., Xu Y., Zhang S., Liu Z., Xue Y.. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci. Rep. 2016; 6:39787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., Ren J.. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014; 42:W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krueger K.E., Srivastava S.. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol. Cell. Proteomics. 2006; 5:1799–1810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.