Abstract
Extensive use of next-generation sequencing (NGS) for pathogen profiling has the potential to transform our understanding of how genomic plasticity contributes to phenotypic versatility. However, the storage of large amounts of NGS data and visualization tools need to evolve to offer the scientific community fast and convenient access to these data. We introduce BACTOME as a database system that links aligned DNA- and RNA-sequencing reads of clinical Pseudomonas aeruginosa isolates with clinically relevant pathogen phenotypes. The database allows data extraction for any single isolate, gene or phenotype as well as data filtering and phenotypic grouping for specific research questions. With the integration of statistical tools we illustrate the usefulness of a relational database structure for the identification of phenotype–genotype correlations as an essential part of the discovery pipeline in genomic research. Furthermore, the database provides a compilation of DNA sequences and gene expression values of a plethora of clinical isolates to give a consensus DNA sequence and consensus gene expression signature. Deviations from the consensus thereby describe the genomic landscape and the transcriptional plasticity of the species P. aeruginosa. The database is available at https://bactome.helmholtz-hzi.de.
INTRODUCTION
The study of how the genomic structure of the important opportunistic pathogen Pseudomonas aeruginosa contributes to its phenotypic versatility is an intense focus of interest (1–4). The P. aeruginosa research community was among the first to address the issue of genome-wide data storage in a format suitable for facilitating downstream comparative analyses and distribution of genome-wide information to the wider research community (5). In 2000 the first P. aeruginosa genome was sequenced (6) and since 2005, the Pseudomonas Genome Database team and participating community members have maintained a website that provides an updated P. aeruginosa PAO1 gene annotation, and facilitates whole genome comparative analyses between the type strain PAO1 and other Pseudomonas strains (7).
Here, we created a centralized DNA-sequence and RNA-expression database, which stores, displays and links genomic, transcriptomic as well as phenotypic information of a plethora of clinical P. aeruginosa isolates. We illustrate the usefulness of a relational database structure that enables data filtering for the identification of associations between sequence- and gene expression-variations and a particular phenotype and demonstrate that linking phenotypic information with genomic and transcriptomic data constitutes a good source for discovery studies in the era of big data. The BACTOME database can furthermore serve as a multipurpose tool to facilitate the work of researchers who use sequence-based technologies to study the evolution, phylogeny, diversity and adaptation strategies of P. aeruginosa isolates.
DATABASE CONTENT
Genomic data to explore sequence variability across heterogeneous lineages
Sequencing of a plethora of genomes of one bacterial species gives detailed information on the overall genomic composition and the intra-species sequence diversity (8–10). The P. aeruginosa BACTOME database as of March 2018 was created based on 99 diverse clinical P. aeruginosa isolates of which all were fully genome sequenced. For 96 of them also the transcriptomes were recorded. BACTOME harbors integrated pipelines and up-dates the retrievable information so that genomic and transcriptomic sequences of additional P. aeruginosa isolates can be added.
SNP extraction—analyzing sequence diversity
In order to learn more about evolutionary processes, the identification of the genetic loci that underlie the selection of favorable phenotypes is critical (11,12). Diversity in sequences may reflect phylogenetic relatedness of the clinical isolates, but also genes under positive or negative selection are hot spots of sequence variations (13,14). One focus of the database is therefore to provide lists of single nucleotide polymorphisms (SNPs) as compared to the UCBPP-PA14 reference genome (15) for any gene or isolate of interest. The retrieval can be restricted to defined groups of SNPs within selected genes, e.g. those leading to amino acid exchanges or affecting predicted regulator binding sites. For the latter, information is integrated from Prodoric (16), ChIP-seq data (17) or on native RNA folding predictions (RNAscan, ViennaRNA Package) (18). There is a customizable display for parameter settings such as SNP quality score (SAMtools (19) derived), minimal read coverage and length of included up- and downstream flanking regions of the defined genes.
Pangenome—the P. aeruginosa core and accessory genome
The repertoire of genes in a particular species in a dataset of n genomes is referred to as the pangenome, where n is the number of included genomes (20). To date, the database comprises 18 319 (pangenome) genes from 101 genomes (99 clinical isolates, and reference strains PAO1 and UCBPP PA14). We used a reciprocal Blast search algorithm and found 3814 genes which were present in all P. aeruginosa isolates (core genome) with >90% homology. All other genes (8966) were assigned as accessory (accessory genome), 5539 of them are only present in single isolates (singletons). Information on whether a gene of interest belongs to the core or accessory genome can be assessed through the database, including information on the degree of nucleotide identity across the clinical P. aeruginosa isolates as compared to the UCBPP-PA14 gene as a reference.
Consensus sequence—a sequence alignment as a reference
In addition to the overall genomic composition of the P. aeruginosa isolates, BACTOME provides information on the degree of sequence variation within the P. aeruginosa core genes on the single nucleotide level. This facilitates the identification of e.g. sequence motifs as predictive features of protein function. The functions of these sequences should be largely conserved across many clinical isolates but they might also be targets of evolutionary processes (21).
A novel algorithm was developed to create a position-wise sequence alignment. The most frequently occurring nucleotide in each position was taken as the consensus. A color-coded diversity display enables a quick estimation for sequence conversation at each nucleotide position (Figure 1). Besides the differentiation between e.g. phylogeny-derived and potential patho-adaptive variations, the created consensus sequence can be used as template to facilitate the design of polymerase chain reaction primers or probes for diagnostic or surveillance purposes. Of note, currently only core genes which are identical in length in all of the genomes are included in the database.
Figure 1.
Consensus sequence and nucleotide diversity among clinical isolates.
Transcriptome data to explore gene expression variability across heterogeneous linages
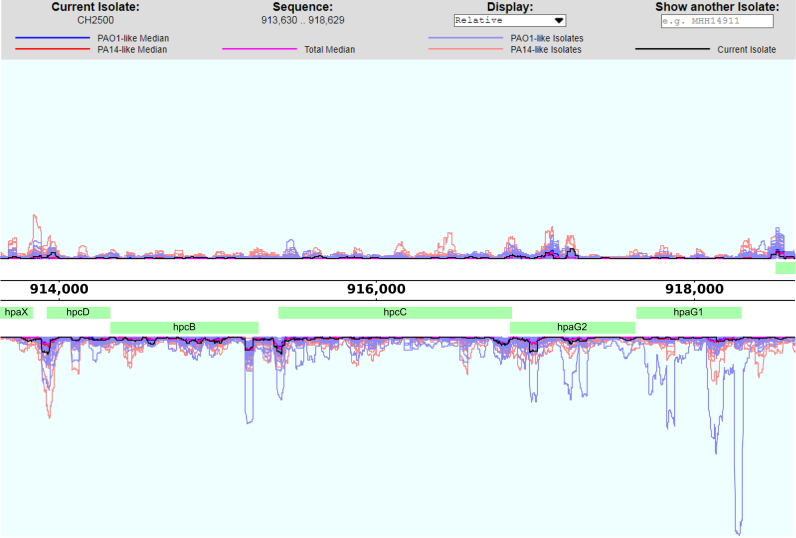
Bacterial genomes are shaped by evolutionary processes and encode optimized gene expression-based systems that guarantee phenotypic plasticity. Thus, more insights into the variation of transcription patterns across heterogeneous lineages should aid the identification of causative patho-adaptive mutations that shape the transcriptional profile. In addition to sequence information, BACTOME comprises gene expression data of 96 clinical P. aeruginosa isolates to gain profound insights into the variation of transcription patterns and genomic loci that contribute to variation in mRNA expression levels. All transcriptomes were recorded under the same culture conditions (growth in LB medium to an OD600 of 2). The TRANSCRIPTOME section in BACTOME includes an interactive genome browser to visualize and navigate the gene expression variations across the P. aeruginosa UCBPP-PA14 reference genome for all isolates (menu item ‘Relative Expression Distribution’, Figure 2). A compilation of all RNA sequencing reads for all PA14-like as well as all PAO1-like isolates is included. The depiction of the transcriptional profile of an individual isolate in the context of the transcriptional profiles of other clinical isolates allows for a more informative interpretation of differentially regulated genes. For instance, a differential expression of a gene that is otherwise very stably expressed across the clinical isolates will take on new significance. The web-based navigable depiction of the P. aeruginosa transcriptional landscape additionally displays corresponding protein sequences and operon structures as well as predicted transcription factor binding sites.
Figure 2.
Interactive genome browser to visualize and navigate the gene expression variations across clinical isolates.
Besides the graphical genome-wide transcription pattern display, BACTOME also allows the extraction of individual fold change information throughout the genome for any gene of interest (menu item ‘Gene Expression Extraction’).
Phenotypic data to explore variability across heterogeneous linages
A major challenge is to understand the causal relationship between genotypes and phenotypes (15). Data-driven genomics projects move from analyzing the genomic sequence to predicting gene functions and bacterial behavior. However, phenotype–genotype correlation studies stringently require that phenotypes are consistently annotated for sequenced strains (22).
We determined and systematically categorized the expression of clinically relevant phenotypes of the P. aeruginosa isolates and stored the respective data in the database. These included phenotypes such as virulence as determined by the use of a Galleria mellonella model, biofilm formation capabilities, colony morphologies and antibiotic resistance to five antipseudomonal antibiotics (ciprofloxacin (CIP), meropenem, ceftazidime (CAZ), tobramycin and colistin). The PHENOTYPE section of BACTOME not only depict the distribution of all different phenotypic traits across all isolates, but also allows to view single isolate information either by isolate ID or selection within an interactive phylogenetic tree.
Linking sequence and expression information to phenotypes
With high-dimensional genotype and phenotype data, there is a need for infrastructure capable of handling both types of data and at the same time associating them in a meaningful fashion. Besides individual information, BACTOME provides a filter for groups of isolates with specific phenotypes or phenotype combinations. This enables the categorization of the isolates according to their phenotype and to define groups of isolates exhibiting a positive or negative phenotype (e.g. CIP-resistant and -sensitive isolates, respectively).
Phenotypes to genotypes
Groups of isolates that exhibit a well-defined phenotype can be further analyzed by mutation enrichment comparison studies to retrieve lists of mutations that are significantly enriched in one of the two groups from the database. The comparison tool uses Fisher’s exact test to search for significant accumulations of mutations in groups of isolates. Comparison options include a nucleotide-specific as well as a gene-wise search to include the detection of whole gene mutational hot spots. Additionally, a third comparison mode allows for the detection of intragenic stop sites. The user may custom adjust the SNP quality threshold and minimum read coverage at the included positions, and select if insertions or deletions shall be included. Furthermore, SNPs which occur at the same nucleotide position but lead to different effects in different clinical isolates (e.g. represent synonymous mutations in some isolates and non-synonymous mutations in others) may be combined in the analysis to identify general, position-specific mutational hot spots. For an estimation of relevant P-value cut-offs of the obtained results, the analysis may be repeated with randomly permutated datasets for comparison.
As an example, when the group comparison is done with all CIP susceptible (n = 40) versus all non-susceptible (n = 56) isolates, which are currently in the BACTOME database, the top two hits of mutations enriched in the non-susceptible group are SNPs in the gyrA and parC genes. Both mutations are causing well-known amino acid exchanges (T83I in the DNA gyrase gyrA and the S87L in the topoisomerase IV parC) which result in antibiotic target mutations leading to CIP resistance (23).
Phenotypes to expression profiles
As mentioned above, BACTOME allows for the categorization of P. aeruginosa isolates according to their phenotype, e.g. antibiotic resistance. Lists of isolates that exhibit a well-defined resistance phenotype can be retrieved and further used for the identification of gene expression variations among the two phenotypically different isolate groups using a group comparison tool which is based on t-tests. When the group comparison is done exemplarily with all CAZ susceptible (n = 41) versus all non-susceptible (n = 55) isolates, upregulation of the beta-lactamase ampC appears as the top hit of gene expression variation distinguishing both groups. The CAZ-degrading function of AmpC is well known (24).
CONCLUSION
We presented a novel database system to store and query genomic and transcriptomic data of the human opportunistic pathogen P. aeruginosa. The genomic sequence data of a plethora of clinical isolates are organized into a phylogenetic tree that can be navigated to facilitate analysis on sequence diversity, evolution and the design of primers/probes. BACTOME also includes associated metadata (e.g. information on the bacterial resistance phenotype) in a searchable framework. With this, BACTOME provides the infrastructure needed to mine big data sets generated by next-generation sequencing efforts and serves the evaluation of the correlation of DNA- as well as RNA-sequence variations with bacterial phenotypes as a systematic strategy for large-scale analysis of genotype–phenotype correlations.
FUNDING
German Federal Ministry of Education and Research [1616038C to S.H.]; German Research Foundation [DFG SFB 900]; Helmholtz Association [iMED]; EU (Starter and Consolidator Grant of the European Research council) [RESISTOME 260276, COMBAT 724290]. Funding for open access charge: ERC Grant [COMBAT].
Conflict of interest statement. None declared.
REFERENCES
- 1. Klockgether J., Cramer N., Wiehlmann L., Davenport C.F., Tümmler B.. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011; 2:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dötsch A., Klawonn F., Jarek M., Scharfe M., Blöcker H., Häussler S.. Evolutionary conservation of essential and highly expressed genes in Pseudomonas aeruginosa. BMC Genomics. 2010; 11:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mathee K., Narasimhan G., Valdes C., Qiu X., Matewish J.M., Koehrsen M., Rokas A., Yandava C.N., Engels R., Zeng E. et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3100–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turner K.H., Wessel A.K., Palmer G.C., Murray J.L., Whiteley M.. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:4110–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Winsor G.L., Lo R., Ho Sui S.J., Ung K.S., Huang S., Cheng D., Ching W.K., Hancock R.E., Brinkman F.S.. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 2005; 33:D338–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M. et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000; 406:959–964. [DOI] [PubMed] [Google Scholar]
- 7. Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S.. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016; 44:D646–D653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Silby M.W., Winstanley C., Godfrey S.A., Levy S.B., Jackson R.W.. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 2011; 35:652–680. [DOI] [PubMed] [Google Scholar]
- 9. Wiehlmann L., Wagner G., Cramer N., Siebert B., Gudowius P., Morales G., Köhler T., van Delden C., Weinel C., Slickers P. et al. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:8101–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marvig R.L., Sommer L.M., Jelsbak L., Molin S., Johansen H.K.. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol. 2015; 10:599–611. [DOI] [PubMed] [Google Scholar]
- 11. Freschi L., Jeukens J., Kukavica-Ibrulj I., Boyle B., Dupont M.J., Laroche J., Larose S., Maaroufi H., Fothergill J.L., Moore M. et al. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front. Microbiol. 2015; 6:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deraspe M., Raymond F., Boisvert S., Culley A., Roy P.H., Laviolette F., Corbeil J.. Phenetic comparison of prokaryotic genomes using k-mers. Mol. Biol. Evol. 2017; 34:2716–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith E.E., Buckley D.G., Wu Z., Saenphimmachak C., Hoffman L.R., D’Argenio D.A., Miller S.I., Ramsey B.W., Speert D.P., Moskowitz S.M. et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marvig R.L., Sommer L.M., Molin S., Johansen H.K.. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 2015; 47:57–64. [DOI] [PubMed] [Google Scholar]
- 15. Lee D.G., Urbach J.M., Wu G., Liberati N.T., Feinbaum R.L., Miyata S., Diggins L.T., He J., Saucier M., Déziel E. et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006; 7:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grote A., Klein J., Retter I., Haddad I., Behling S., Bunk B., Biegler I., Yarmolinetz S., Jahn D., Münch R.. PRODORIC (release 2009): a database and tool platform for the analysis of gene regulation in prokaryotes. Nucleic Acids Res. 2009; 37:D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schulz S., Eckweiler D., Bielecka A., Nicolai T., Franke R., Dötsch A., Hornischer K., Bruchmann S., Düvel J., Häussler S.. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog. 2015; 11:e1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011; 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R.. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005; 15:589–594. [DOI] [PubMed] [Google Scholar]
- 21. Cheatle Jarvela A.M., Hinman V.F.. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo. 2015; 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brbic M., Piskorec M., Vidulin V., Krisko A., Smuc T., Supek F.. The landscape of microbial phenotypic traits and associated genes. Nucleic Acids Res. 2016; 44:10074–10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bruchmann S., Dötsch A., Nouri B., Chaberny I.F., Häussler S.. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother. 2013; 57:1361–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cabot G., Ocampo-Sosa A.A., Tubau F., Macia M.D., Rodriguez C., Moya B., Zamorano L., Suárez C., Peña C., Martínez-Martínez L. et al. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother. 2011; 55:1906–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]