Abstract
High functional heterogeneity of cancer cells poses a major challenge for cancer research. Single-cell sequencing technology provides an unprecedented opportunity to decipher diverse functional states of cancer cells at single-cell resolution, and cancer scRNA-seq datasets have been largely accumulated. This emphasizes the urgent need to build a dedicated resource to decode the functional states of cancer single cells. Here, we developed CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/ or http://202.97.205.69/CancerSEA/), the first dedicated database that aims to comprehensively explore distinct functional states of cancer cells at the single-cell level. CancerSEA portrays a cancer single-cell functional state atlas, involving 14 functional states (including stemness, invasion, metastasis, proliferation, EMT, angiogenesis, apoptosis, cell cycle, differentiation, DNA damage, DNA repair, hypoxia, inflammation and quiescence) of 41 900 cancer single cells from 25 cancer types. It allows querying which functional states are associated with the gene (or gene list) of interest in different cancers. CancerSEA also provides functional state-associated PCG/lncRNA repertoires across all cancers, in specific cancers, and in individual cancer single-cell datasets. In summary, CancerSEA provides a user-friendly interface for comprehensively searching, browsing, visualizing and downloading functional state activity profiles of tens of thousands of cancer single cells and the corresponding PCGs/lncRNAs expression profiles.
INTRODUCTION
Human cancer is a highly diverse and complex disease composed of cancer cells with distinct genetic, epigenetic and transcriptional status, forming heterogeneous functional populations of cancer cells, which poses a major obstacle to cancer diagnosis and treatment (1–4). Some cancer cells have high cell proliferation activity, some have tumor aggressiveness and metastasis capacity, some show stem-cell-like properties, while some exhibit ‘lazy’ state of quiescence (5–7). These functionally heterogeneous cancer cells act cooperatively or competitively during the entire tumor evolution, leading to distinct tumor phenotypes (8–10). Therefore, it is essential to comprehensively and adequately decode the functional states of cancer cells.
Single-cell sequencing-based technologies open up new avenues for exploring complex ecosystems, especially cancers, revolutionizing whole-organism science (11). It provides an unprecedented opportunity to decipher the functional states of cancer cells at single cell resolution, thus, allowing researchers to accurately and unbiasedly explore the functional heterogeneity of cancer cells, and deepening the understanding of cancer cell as a functional unit to perform specific biological functions in the initiation and progression of cancer. In 2014, a pioneering study of glioblastoma used single-cell RNA sequencing (scRNA-seq) to uncover previously unexpected heterogeneity in cancer-related functional states, such as stemness, proliferation, and hypoxia (5). Profiling 4347 single cells from six human oligodendrogliomas by scRNA-seq, Tirosh et al. found that these single cells exhibited widespread heterogeneity in stemness and differentiation, and revealed that a few cancer cells with high stemness may act as cancer stem cells to fuel the growth of cancer (12). And a study about chronic myeloid leukemia revealed that cells with different activities of quiescence, proliferation, and stemness have different sensitivity to tyrosine kinase inhibitor (TKI) treatments, leading to frequent relapse for this disease (6).
The rapid development of scRNA-seq leads to the accelerated accumulation of a large amount of scRNA-seq datasets, and recently several related databases have been developed. For example, SCPortalen collected and annotated scRNA-seq datasets in human and mouse, and provided expression tables processed using a pipeline for downloading (13). JingleBells provided BAM files of immune-related scRNA-seq datasets for visualization of reads (14). scRNASeqDB collected human single cell transcriptome datasets and help researchers to query and visualize gene expression in human single cells (15). However, all of them focused on collecting scRNA-seq datasets, a dedicated database devoted to deciphering the functional states of cancer single cells is still lacking.
Therefore, we developed CancerSEA, a dedicated database that aims to comprehensively decode distinct functional states of cancer cells at the single-cell level. As of July 2018, the database contains 41 900 cancer single cells in 25 human cancers with 14 manually curated cancer-related functional states (including stemness, invasion, metastasis, proliferation, EMT, angiogenesis, apoptosis, cell cycle, differentiation, DNA damage, DNA repair, hypoxia, inflammation and quiescence). By characterizing these functional state activities of each cancer cell, CancerSEA provides an atlas of cancer single-cell functional states and associates protein-coding genes (PCGs) and lncRNAs with these functional states at single-cell level for promoting mechanistic understanding of functional differences of cancer cells. We expect that this elaborate database can serve as an important and valuable resource for facilitating the exploration of the tumor heterogeneity.
MATERIALS AND METHODS
Data collection, curation and processing
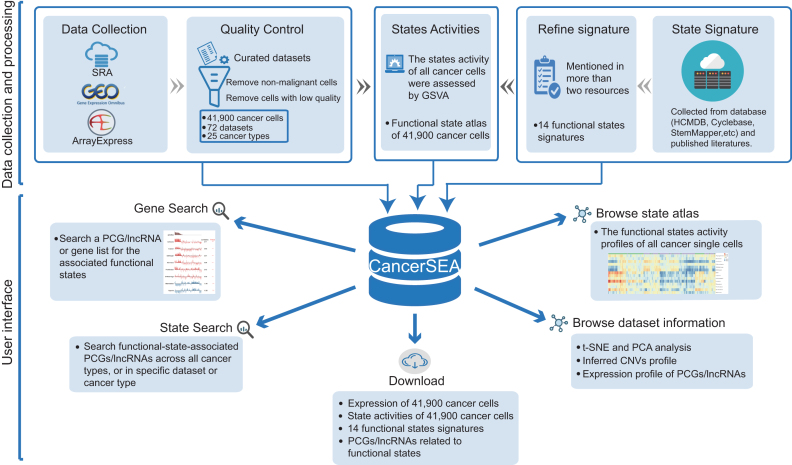
We systematically collected cancer-related scRNA-seq datasets in human from Sequence Read Archive (SRA), Gene Expression Omnibus (GEO) and ArrayExpress based on the following keywords: (‘single cell’ OR ‘single-cell’ OR ‘single cells’ OR ‘single-cells’) AND (‘transcriptomics’ OR ‘transcriptome’ OR ‘RNA-seq’ OR ‘RNA-sequencing’ OR ‘RNA sequencing’ OR ‘scRNA-seq’ OR ‘scRNA seq’) AND (‘tumor’ OR ‘tumour’ OR ‘cancer’ OR ‘carcinoma’ OR ‘neoplasm’ OR ‘neoplastic’). A total of 49 cancer-related scRNA-seq datasets involving 128 518 single cells were obtained initially (Supplementary Table S1). Among them, 28 offered raw FASTQ sequencing files, and the rest provided the expression matrix data. All datasets were collected before July 2018. All single cells in these datasets were analyzed through expression quantification, quality control, and characterization of functional states (Figure 1, Supplementary Figure S1).
Figure 1.
Overview of CancerSEA database. All scRNAseq datasets were collected from SRA, GEO and ArrayExpress, and were manually annotated and curated. For quality control, we removed non-malignant single cells and cells with low quality. In addition, we collected and refined 14 functional states signatures. State activities of cancer cells were assessed by GSVA. All data resource were deposited in CancerSEA, and displayed in web pages (gene search, state search, browse, download).
For each cancer-related scRNA-seq dataset, we carefully read the original paper, if available, and extracted the corresponding metadata, including the cancer types and sources (i.e., tissue, cell line, patient-derived xenograft (PDX) and circulating tumor cell (CTC)). Also, we obtained the cell groups (referring to patient ID, culture condition or cell phenotype) from the supplementary tables of the paper, and labeled each cell with the information of cell groups. These ‘cell group’ tags can help users to explore these datasets in specific patients and cell phenotypes. When a dataset contains different cancer types (or different types of sources), we manually divided it into multiple sub-datasets. Then 72 (sub)-datasets were generated (Supplementary Table S1).
For scRNA-seq datasets with raw sequencing files, an in-house bioinformatics pipeline was adopted for quality control and expression quantification (Supplementary Figure S1). Briefly, we used the NCBI SRA Toolkit (version 2.8.2) to obtain FASTQ files. Sequencing quality was assessed by FastQC (version 0.11.2). Then, for sequencing data, we trimmed adapter sequences and filtered low-quality reads (quality value lower than 20) using the Trim Galore (version 0.4.4). Expression quantification of transcripts was performed by salmon (version 0.9.1) with optional parameters -k (k = 31 for long reads and k = 15 for short reads), –gcBias, –seqBias and others default parameters by indexing the GENCODE (Release 28, GRCh38) reference transcriptome in quasi-mapping-based mode. The expression abundance values of genes (including PCGs and lncRNAs) were summarized by using R package tximport (version 1.6.0). The TPM (transcripts per million) values of PCGs and lncRNAs were used for subsequent analysis. For scRNA-seq datasets with only expression matrix, we directly converted the expression values to TPM/CPM values using a custom script. Expression values were log2 transformed with an offset of 1.
Quality control of single cells
Considering the low sensitivity and high technical noise of scRNA-seq assay, we carried out two steps for quality control of single cells. First, in order to ensure that all single cells were cancer cells without mixing normal cells from tumor microenvironment, we removed non-malignant single cells using the following criteria: (i) if the original papers have included the information about whether cells are malignant or not, only the malignant cells were remained; (ii) using an RNAseq-inferred copy number variation (CNV) approach (5) to distinguish malignant cells from non-malignant cells. After that, 68 708 malignant single cells remained. Second, we calculated two quality measures for each cell, including the number of genes with detectable expression (i.e. expression levels are greater than 0) and the average expression level of 87 housekeeping genes collected in (16). We then excluded cells with the number of expressed genes fewer than 1000 or with the average housekeeping expression <2. PCGs or lncRNAs with detectable expression in at least 10% or 5% cells (the minimum number of expressed cells should be greater than 10) were retained, respectively. At last, 41 900 cancer cells derived from 72 single-cell datasets from 25 cancer types were remained, involving 18 895 PCGs and 15 571 lncRNAs.
Characterizing functional states of cancer single cells
After reviewing almost all cancer single-cell sequencing studies, we concluded 14 crucial functional states of cancer cells, including stemness, invasion, metastasis, proliferation, EMT, angiogenesis, apoptosis, cell cycle, differentiation, DNA damage, DNA repair, hypoxia, inflammation and quiescence. To characterize these functional states for cancer single cells, we built the corresponding gene signatures through searching literatures and known databases (including some general databases, such as Gene Ontology (17) and MSigDB (18), and some specialized databases, such as Cyclebase (19), HCMDB (20) and StemMapper (21)) (Supplementary Table S2). For most of the signatures, the collected genes that were mentioned in more than two resources were kept. While for the invasion signature, genes mentioned in more than two invasion-associated terms collected from MSigDB were retained. Then, through functional annotations and literature searching, genes that negatively affect the corresponding functional states were removed.
Based on these signatures, the activities of 14 functional states across cancer single cells in each dataset were evaluated using Gene Set Variation Analysis (GSVA) with the GSVA package in R (22). In brief, for each gene, we first performed a non-parametric kernel estimation of its cumulative density function and then calculated an expression-level statistic to normalize expression profiles to a common scale. The expression-level statistic can reflect whether a gene is highly or lowly expressed in a specific cell in the context of the cell population distribution. Then, in each cell, the expression-level statistics of all genes were converted to normalized ranks. Next, we used the Kolmogorov–Smirnov like random walk statistic, similar to the GSEA method, to summarize the expression-level rank statistics of a given signature gene set into a final enrichment score (i.e. GSVA score), which is used to characterize the signature activity. At last, the enrichment scores of 14 signatures across cells in all scRNA-seq data were calculated. Then, for each single-cell dataset derived from tumor tissue, PDX and CTC, we identified significant correlations between gene expressions and functional state activities using Spearman's rank correlation test with Benjamini & Hochberg false discovery rate (FDR) correction for multiple comparisons (correlation > 0.3 and FDR < 0.05). Due to the low amount of mRNA within individual cells and sequencing technical noise, there is an excessive number of zeros in scRNA-seq data. During the calculation of gene-state associations, only cells with detectable expression of the genes of interest were used by setting the parameter ‘na.action’ to na.omit, and at least 30 cancer single cells were required.
Basic expression analysis of single cell datasets
For each dataset, we performed several basic analysis of high-quality cancer single-cell expression, including PCA and t-SNE analysis, hierarchical clustering of highly variable PCGs/lncRNAs and inferred CNVs. PCA and t-SNE analysis were conducted by ‘prcomp’ and ‘Rtsne’ function in R, respectively. Highly variable PCGs/lncRNAs were extracted by ‘FindVariableGenes’ function in Seurat R package with default parameters, which were visualized by ‘heatmap.2’ R function. Inferred CNVs of all cells were clustered by average-linkage hierarchical clustering and were visualized by ‘heatmap.2’ R function.
Database construction
CancerSEA is freely available to the research community at http://biocc.hrbmu.edu.cn/CancerSEA or http://202.97.205.69/CancerSEA and requires no registration or login. CancerSEA is deployed using tomcat (version 7.0.47) and tested in Mozilla Firefox, Google Chrome, and Apple Safari browsers. All the data are stored and queried by MySQL (version 5.6.40). Tables are visualized by Datatables (version 1.10.16). Graph charts are generated by HighCharts (version 6.1.0), d3 (version 5.4.0) and in-house R scripts.
RESULTS
Contents of CancerSEA
The current version of CancerSEA contains 14 functional states of 41 900 cancer single cells from 25 cancer types, involving 140 cancer patients, 76 cell lines with different conditions, 13 and 51 cancer patient-derived PDXs and CTCs, respectively. The average number of cancer single cells per cancer type is 1676, with alveolar rhabdomyosarcoma (ARMS) having the largest number of single cells (n = 6875) and neuroblastoma (NB) has the smallest number (n = 36). CancerSEA contains a total of 121 scRNA-seq expression profiles, including 72 PCG expression profiles and 49 lncRNA expression profiles. The average numbers of PCGs and lncRNAs per cell are, respectively, 4911 and 817, involving a total of 18 895 PCGs and 15 571 lncRNAs.
Database feature and utility
CancerSEA depicts a cancer single-cell state atlas and allows to query the relationships between genes (including PCGs and lncRNAs) and the 14 functional states. The easy-to-use interface provides an access for searching, browsing, visualizing and downloading data. The online user guide illustrates several use cases of CancerSEA.
Gene and gene list search
For a gene of interest, CancerSEA provides its relationships to the 14 functional states in different cancers. It allows to input a gene (PCG or lncRNA) in the ‘Search’ page (Search PCG/lncRNA for the associated functional states), with gene name or Ensemble ID (Figure 2A). After clicking the ‘Search’ button, the query results including four sections will be displayed. The first section ‘Basic information of the input gene (list)’ displays the basic annotations of the gene of interest, including gene symbol, alias, Ensembl ID, functional description and external links including Entrez Gene database and Ensemble database. When clicking the ‘Gene Symbol/ID’ of the gene, a box plot characterizing the expression patterns of the gene across all the datasets in which it expressed will be shown (Figure 2B). The section ‘Relevance across 14 functional states in distinct cancers’ contains two parts. The first parts ‘Correlation plot’ shows the average correlations between the expression of the gene of interest and the activity of each functional state across all single-cell datasets in different cancers with an interactive bubble chart. The upper bar plot provides a summarized association across all cancer types for each functional state by showing the number of single-cell datasets in which the gene is significantly related to the corresponding state. It can quickly and clearly indicate which functional states are the most relevant. When clicking the bar of a specific functional state, the bubbles having significant correlations in at least one dataset in the corresponding cancer type will be highlighted. When positioning the mouse on a highlighted bubble, the detailed information (including the correlation value and the corresponding p-value) in each dataset will be popped up, which informs users which datasets are the ‘significant datasets’(Figure 2C). The second part ‘Correlation data table ’ shows the detailed information of functional relevance in each dataset (Figure 2D). In the third section, users can learn more in-depth information about the functional relevance in specific cancer. Through selecting a cancer type in the drop-down box in the third section or clicking a cancer type in the left side of the bubble chart in the second section, a result table is used to display the basic information of all single-cell datasets in the selected cancer type and the corresponding correlations with the 14 functional states (Figure 2E). When clicking on the plus icon in a result row of a specific dataset, more details about the correlations are displayed. The ranked expression of the gene of interest in the selected dataset and the corresponding activities of functional states with statistical significance are shown with a series of bar charts. Users can choose different correlation cutoffs and p-values to filter functional states, and move the mouse over the bar chart of a state to obtain the functional relevance scatter plot. The last section ‘Functional relevance in distinct cell groups’ shows the detailed functional relevance in each specific cell group of a specific dataset through clicking different dataset tabs. It also contains a box plot showing the expression distribution of the input gene in this selected dataset and a t-SNE plot of all single cells with colors representing the expression levels of the input gene (Figure 2F).
Figure 2.
Functional relevance of a gene or gene list. (A) The ‘Search’ page of CancerSEA. (B) The basic annotations and expression pattern of SOX4. (C) Relevance of SOX4 across 14 functional states in distinct cancers. The size of the bubble represents the average correlation strength. The bar chart shows the number of datasets in which SOX4 is significantly related to the corresponding state. The red color indicates positive correlation, and the blue one indicates negative correlation. (D) The correlation data table shows the detailed information of all functional associations with SOX4 in each dataset. (E) Detailed functional relevance in HNSCC and in a specific cell group (F). In the scatter plot, the x-axis indicates the expression of SOX4, and the y-axis indicates the activity of the functional state.
CancerSEA also allows to input a gene list for querying the correlations between the average expression levels of the gene list and the activities of functional states (Figure 2A, Supplementary Methods). Users can input the gene list of their interest, or select pre-specified lists (including cancer hallmark-related functions/pathways from GO (8,23,24), MSigDB (18), KEGG and Reactome (25,26)). In addition, CancerSEA also allows uploading a comma-separated TXT file with all the genes in one line. The mapped and unmapped genes in the gene list will be presented in the first section ‘Basic information of the input gene (list)’. The basic annotations and expression pattern of each detected gene will be returned by clicking the gene symbol/ID".
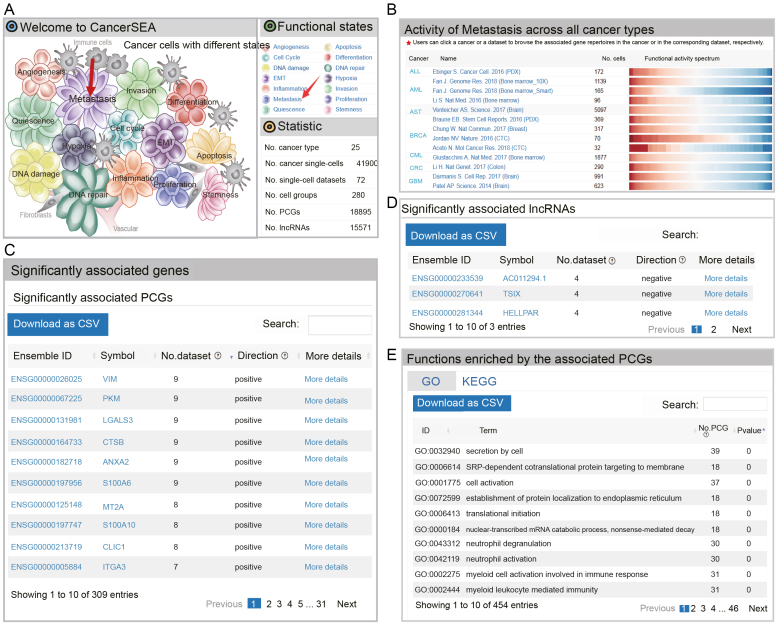
Functional state search
CancerSEA allows to query a functional state of interest to obtain the PCG/lncRNA catalogs that are highly related to the functional state at single-cell resolution in the ‘Home’ page and ‘Search’ page. In the ‘Home’ page, users can click the ‘state’ hyperlinks embedded in the cell-tree image ‘Cancer Cells with Different States’ and the ‘Functional states’ panel in the top right-hand window (Figure 3A).In the result page, the first panel shows the activity profiles of the selected functional state across different cancer types and the second panel allows users to browse and download the repertoires of PCGs/lncRNAs that are frequently associated with the state across different cancer types (significantly related to the query state at least in four scRNA-seq datasets) (Figure 3B–D). When clicking ‘More details’, users can be redirected to the corresponding ‘gene search’ result page. External links to Entrez Gene and Ensemble databases are also provided by clicking the ‘gene symbol’ and ‘Ensemble ID’, respectively. Functions and pathways enriched by these related PCGs (hypergeometric test, P < 0.05) are displayed in the third panel (Figure 3E). Furthermore, when clicking the cancer types in the first panel, users can acquire cancer-type specific PCG/lncRNA repertoires that are associated with the functional state of interest. The cancer-type specific PCG/lncRNA repertoires can also be obtained by selecting the cancer type and functional state in the ‘Search’ page (Search functional state for the related PCG/lncRNA repertoires) (Figure 2A). In addition, when clicking the names of single-cell datasets in the first panel, the PCG/lncRNA lists that are associated with the state in the selected dataset will be displayed.
Figure 3.
Activity spectrums of a functional state and its associated PCG/lncRNA repertoires. (A) The ‘Home’ page of CancerSEA. (B) Activity spectrums of metastasis across different cancer types. (C) PCGs and (D) lncRNAs frequently related to metastasis across different cancer types. (E) GO and KEGG terms enriched by the associated PCGs.
Functional state atlas and detailed single-cell dataset information
Functional state atlas of all cancer single cells is comprehensively presented in the ‘Browse’ page. Users can browse the functional state profiles of specific cancer by clicking the cancer type in the cancer-dataset hierarchical navigation menu (Figure 4). The hierarchical clustering heatmaps of state activity of all datasets in the selected cancer type are listed in the right panel, showing extensive functional heterogeneity across cancer cells. In addition, by clicking the dataset name in the navigation menu, users can select a dataset of interest to further browse the detailed information, including ‘Detailed description’, ‘Functional state profile’, ‘Cell distribution’, ‘Expression patterns of PCGs/lncRNAs’ and ‘Inferred CNV heatmap’. The ‘Detailed description’ section contains relevant publication information, cancer type, accession number, number of cells, cell group, source, and description. The ‘Functional state profile’ section shows the state activity heatmap. Visualization of the selected dataset using t-SNE and PCA analysis is displayed in the ‘Cell distribution’ section with point colors representing different cell groups. In the ‘Expression pattern of PCGs/lncRNAs’ section, expression heatmaps of highly variable PCGs/lncRNAs are displayed. In the ‘Inferred CNV heatmap’ section, users can easily view the CNVs pattern across the cancer cells.
Figure 4.
Functional state atlas and detailed dataset information in browse page. (A) Functional state atlas in specific cancer. Detailed information of selected dataset: basic information (B), functional state profile (C), t-SNE and PCA analysis (D), expression of PCG/lncRNA (E), inferred CNV profile (F).
Data download
All data in CancerSEA can be downloaded in the ‘Download’ page, containing the functional state profiles and PCG/lncRNA expression profiles for each single-cell dataset as well as functional state signatures. Users can select datasets of interest by searching for keywords, such as cancer type, dataset ID (i.e. ExpID) and source. By clicking ‘PCG/lncRNA’, users can download the corresponding expression profiles (before and after quality control) in a compressed file. Clicking on the icon of state score will download the functional state profile of the selected dataset. The detailed information of 14 functional state signatures is also provided in the ‘Download for signature profiles' section, including signature title, description, number of genes, sources, functional state signature genes and exact links to MsigDB gene lists, GO terms, PubMed literature or other databases.
Example application
To demonstrate the utility and potential application of CancerSEA, we used SOX4 and VIM as examples to query their associated functional states. SOX4 has been confirmed that its up-regulation confers cancer cell stemness properties (27). As expected, in the result page, the expression of SOX4 shows strong positive correlations with stemness state across almost all cancer types (Figure 2C), especially in head and neck squamous cell carcinoma (HNSCC), high-grade glioma (HGG) and non-small cell lung cancer (NSCLC). When focusing on HNSCC in the ‘Functional relevance in HNSCC’, we observed that SOX4 had a significantly high correlation of 0.62 (P ≤ 0.001) in the dataset ‘Puram SV. Cell. 2017 (Oral Cavity)’ (Figure 2E). The t-SNE plot of 2105 single cells in the dataset displays strong tumor heterogeneity with an extremely high expression of SOX4 in a subset of cells (Figure 2F). Like SOX4, we searched VIM, a type III intermediate filament protein, in CancerSEA and found that VIM is significantly related to EMT and metastasis states in most cancers (Supplementary Figure S2), consistent with previous that VIM is essential for cell attachment, migration and epithelial-mesenchymal transition (EMT) (28).
Also, we queried metastasis-related genes at single-cell resolution in the ‘Home’ page (Figure 3A). In the result page, we observed strong metastasis heterogeneity across all cancer types in the functional activity spectrums (Figure 3B). We obtained 165 genes (including 162 PCGs and 3 lncRNAs) that are correlated with metastasis in at least 4 scRNA-seq datasets (Supplementary Tables S3 and S4). Especially, in the top 10 genes with the most significant datasets, 9 (including VIM (29), LGALS3 (30), CTSB (31), ANXA2 (32), S100A6 (33), MT2A (34), S100A10 (35), CLIC1 (36), ITGA3 (37)) have been widely confirmed to be related with cancer metastasis (Figure 3C). When focusing on BRCA we obtained metastasis-related genes specific to BRCA (Supplementary Figure S3). The top one gene S100A10 (35) has been reported to be involved in the process of breast cancer cell adhesion to endothelial cells, indicating an involvement of it in breast cancer cell interactions during metastasis. These results suggest that CancerSEA is a reliable and useful database for users to query the relevant functional states of genes in cancer single cells.
SUMMARY AND FUTURE PERSPECTIVES
Tumours are complex ecosystems composed of cells with heterogeneous functional states, leading to the frequent recurrence of cancers. Single-cell sequencing technology provides an opportunity to decipher the functional heterogeneity of cancer cells at single-cell resolution. Thus, CancerSEA, a database for cancer single-cell functional state atlas, comes into being. To our best knowledge, CancerSEA is the first dedicated database to decode the cancer cell functional states at single-cell resolution. Through deciphering the 14 functional states in 41 900 cancer single cells of 25 human cancer types, CancerSEA helps to understand the molecular mechanisms of functional heterogeneity of cancer cells. We believe that CancerSEA will be a useful resource for cancer research.
CancerSEA devotes to deciphering the functional states of cancer cells at the single-cell level. It portrays the activity spectrums of 14 cancer-related functional states in 41 900 single cells of 25 cancer types, which show an unexpected functional heterogeneity among different cancer cells. CancerSEA allows users to query PCGs or lncRNAs for their relevant functional states at single cell resolution. It also allows searching for the associated functional states for a gene list. Furthermore, for each functional state, it provides highly associated PCG/lncRNA repertoires across all cancer types, in a specific cancer type, and in individual cancer single-cell datasets. In addition, CancerSEA stores 72 cancer single-cell datasets containing 121 expression profiles (including 72 and 49 expression profiles for PCGs and lncRNAs, respectively) and provides a quick access to download these expression profiles, their corresponding functional state spectrums, as well as the gene signatures of the 14 functional states.
CancerSEA is a useful resource that will facilitate to understand the functional heterogeneity of cancer cells. In the future, we will continue to work on this database in the following directions: (i) updating the database regularly to keep up with the upcoming scRNA-seq data through keyword search on a weekly basis; (ii) combining single-cell multi-omics data, such as genome, epigenome and proteome, to decode the functional states of cancer single cells; (iii) Refining the functional states into more specialized states (e.g. dividing the state of cancer metastasis into two sub-states including regional lymph nodes metastasis and distant metastasis); and (iv) supplementing new functional states (e.g. drug sensitivity or resistance). Through our efforts, we expect that CancerSEA will contribute to understanding the functional heterogeneity of cancer, and even to the diagnosis and treatment of cancer.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National High Technology Research and Development Program of China (863 Program) [2014AA021102, in part]; National Program on Key Basic Research Project (973 Program) [2014CB910504]; National Natural Science Foundation of China [61473106, 61573122]; China Postdoctoral Science Foundation [2016M600260]; Wu lien-teh Youth Science Fund Project of Harbin Medical University [WLD-QN1407]; Special Funds for the Construction of Higher Education in Heilongjiang Province [UNPYSCT-2016049]; Heilongjiang Postdoctoral Foundation [LBH-Z16098]. Funding for open access charge: National High Technology Research and Development Program of China (863 Program) [2014AA021102, in part]; National Program on Key Basic Research Project (973 Program) [2014CB910504]; National Natural Science Foundation of China [61473106, 61573122]; China Postdoctoral Science Foundation [2016M600260]; Wu lien-teh Youth Science Fund Project of Harbin Medical University [WLD-QN1407]; Special Funds for the Construction of Higher Education in Heilongjiang Province [UNPYSCT-2016049]; Heilongjiang Postdoctoral Foundation [LBH-Z16098].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kreso A., Dick J.E.. Evolution of the cancer stem cell model. Cell Stem Cell. 2014; 14:275–291. [DOI] [PubMed] [Google Scholar]
- 2. Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A. et al. . Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010; 467:1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J. et al. . Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011; 17:1086–1093. [DOI] [PubMed] [Google Scholar]
- 4. Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L. et al. . An integrated genomic analysis of human glioblastoma multiforme. Science. 2008; 321:1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014; 344:1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giustacchini A., Thongjuea S., Barkas N., Woll P.S., Povinelli B.J., Booth C.A.G., Sopp P., Norfo R., Rodriguez-Meira A., Ashley N. et al. . Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017; 23:692–702. [DOI] [PubMed] [Google Scholar]
- 7. Venteicher A.S., Tirosh I., Hebert C., Yizhak K., Neftel C., Filbin M.G., Hovestadt V., Escalante L.E., Shaw M.L., Rodman C. et al. . Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017; 355:1391–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 9. Meacham C.E., Morrison S.J.. Tumour heterogeneity and cancer cell plasticity. Nature. 2013; 501:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H., Luo S., Zhang X., Liao J., Quan F., Zhao E., Zhou C., Yu F., Yin W., Zhang Y. et al. . SEECancer: a resource for somatic events in evolution of cancer genome. Nucleic Acids Res. 2018; 46:D1018–D1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A. et al. . mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009; 6:377–382. [DOI] [PubMed] [Google Scholar]
- 12. Tirosh I., Venteicher A.S., Hebert C., Escalante L.E., Patel A.P., Yizhak K., Fisher J.M., Rodman C., Mount C., Filbin M.G. et al. . Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016; 539:309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abugessaisa I., Noguchi S., Bottcher M., Hasegawa A., Kouno T., Kato S., Tada Y., Ura H., Abe K., Shin J.W. et al. . SCPortalen: human and mouse single-cell centric database. Nucleic Acids Res. 2018; 46:D781–D787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ner-Gaon H., Melchior A., Golan N., Ben-Haim Y., Shay T.. JingleBells: a repository of immune-related single-cell RNA-sequencing datasets. J. Immunol. 2017; 198:3375–3379. [DOI] [PubMed] [Google Scholar]
- 15. Cao Y., Zhu J., Jia P., Zhao Z.. scRNASeqDB: a database for RNA-Seq based gene expression profiles in human single cells. Genes. 2017; 8:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H. 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. et al. . Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016; 352:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. The Gene Ontology, C. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017; 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P.. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015; 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Santos A., Wernersson R., Jensen L.J.. Cyclebase 3.0: a multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res. 2015; 43:D1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng G., Ma Y., Zou Y., Yin A., Li W., Dong D.. HCMDB: the human cancer metastasis database. Nucleic Acids Res. 2018; 46:D950–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pinto J.P., Machado R.S.R., Magno R., Oliveira D.V., Machado S., Andrade R.P., Braganca J., Duarte I., Futschik M.E.. StemMapper: a curated gene expression database for stem cell lineage analysis. Nucleic Acids Res. 2018; 46:D788–D793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanzelmann S., Castelo R., Guinney J.. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013; 14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanahan D., Weinberg R.A.. The hallmarks of cancer. Cell. 2000; 100:57–70. [DOI] [PubMed] [Google Scholar]
- 24. Plaisier C.L., Pan M., Baliga N.S.. A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers. Genome Res. 2012; 22:2302–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. et al. . The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018; 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen H., Blijlevens M., Yang N., Frangou C., Wilson K.E., Xu B., Zhang Y., Zhang L., Morrison C.D., Shepherd L. et al. . Sox4 expression confers bladder cancer stem cell properties and predicts for poor patient outcome. Int. J. Biol. Sci. 2015; 11:1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang W., Yi M., Zhang R., Li J., Chen S., Cai J., Zeng Z., Li X., Xiong W., Wang L. et al. . Vimentin is a crucial target for anti-metastasis therapy of nasopharyngeal carcinoma. Mol. Cell Biochem. 2018; 438:47–57. [DOI] [PubMed] [Google Scholar]
- 29. Richardson A.M., Havel L.S., Koyen A.E., Konen J.M., Shupe J., Wiles W.G.t., Martin W.D., Grossniklaus H.E., Sica G., Gilbert-Ross M. et al. . Vimentin is required for lung adenocarcinoma metastasis via heterotypic tumor cell-cancer-associated fibroblast interactions during collective invasion. Clin. Cancer Res. 2018; 24:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoshimura A., Gemma A., Hosoya Y., Komaki E., Hosomi Y., Okano T., Takenaka K., Matuda K., Seike M., Uematsu K. et al. . Increased expression of the LGALS3 (galectin 3) gene in human non-small-cell lung cancer. Genes Chromosomes Cancer. 2003; 37:159–164. [DOI] [PubMed] [Google Scholar]
- 31. Vasiljeva O., Papazoglou A., Kruger A., Brodoefel H., Korovin M., Deussing J., Augustin N., Nielsen B.S., Almholt K., Bogyo M. et al. . Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006; 66:5242–5250. [DOI] [PubMed] [Google Scholar]
- 32. Wang T., Yuan J., Zhang J., Tian R., Ji W., Zhou Y., Yang Y., Song W., Zhang F., Niu R.. Anxa2 binds to STAT3 and promotes epithelial to mesenchymal transition in breast cancer cells. Oncotarget. 2015; 6:30975–30992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lyu X., Li H., Ma X., Li X., Gao Y., Ni D., Shen D., Gu L., Wang B., Zhang Y. et al. . High-level S100A6 promotes metastasis and predicts the outcome of T1-T2 stage in clear cell renal cell carcinoma. Cell Biochem. Biophys. 2015; 71:279–290. [DOI] [PubMed] [Google Scholar]
- 34. Kim H.G., Kim J.Y., Han E.H., Hwang Y.P., Choi J.H., Park B.H., Jeong H.G.. Metallothionein-2A overexpression increases the expression of matrix metalloproteinase-9 and invasion of breast cancer cells. FEBS Lett. 2011; 585:421–428. [DOI] [PubMed] [Google Scholar]
- 35. Myrvang H.K., Guo X., Li C., Dekker L.V.. Protein interactions between surface annexin A2 and S100A10 mediate adhesion of breast cancer cells to microvascular endothelial cells. FEBS Lett. 2013; 587:3210–3215. [DOI] [PubMed] [Google Scholar]
- 36. Ye Y., Yin M., Huang B., Wang Y., Li X., Lou G.. CLIC1 a novel biomarker of intraperitoneal metastasis in serous epithelial ovarian cancer. Tumour Biol. 2015; 36:4175–4179. [DOI] [PubMed] [Google Scholar]
- 37. Nagata M., Noman A.A., Suzuki K., Kurita H., Ohnishi M., Ohyama T., Kitamura N., Kobayashi T., Uematsu K., Takahashi K. et al. . ITGA3 and ITGB4 expression biomarkers estimate the risks of locoregional and hematogenous dissemination of oral squamous cell carcinoma. BMC Cancer. 2013; 13:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.