Abstract
The Biological General Repository for Interaction Datasets (BioGRID: https://thebiogrid.org) is an open access database dedicated to the curation and archival storage of protein, genetic and chemical interactions for all major model organism species and humans. As of September 2018 (build 3.4.164), BioGRID contains records for 1 598 688 biological interactions manually annotated from 55 809 publications for 71 species, as classified by an updated set of controlled vocabularies for experimental detection methods. BioGRID also houses records for >700 000 post-translational modification sites. BioGRID now captures chemical interaction data, including chemical–protein interactions for human drug targets drawn from the DrugBank database and manually curated bioactive compounds reported in the literature. A new dedicated aspect of BioGRID annotates genome-wide CRISPR/Cas9-based screens that report gene–phenotype and gene–gene relationships. An extension of the BioGRID resource called the Open Repository for CRISPR Screens (ORCS) database (https://orcs.thebiogrid.org) currently contains over 500 genome-wide screens carried out in human or mouse cell lines. All data in BioGRID is made freely available without restriction, is directly downloadable in standard formats and can be readily incorporated into existing applications via our web service platforms. BioGRID data are also freely distributed through partner model organism databases and meta-databases.
INTRODUCTION
Biological interaction networks, as aggregated from a plethora of individual protein or genetic interactions, as well as interactions of RNA, DNA, membranes, carbohydrates and small molecule metabolites, serve as a framework for understanding gene–phenotype relationships and the mechanistic basis for all cellular functions (1,2). The characterization of molecular and functional interactions between genes, their products and biomolecules has been instrumental in interpreting genetic associations related to cancer and other diseases in a myriad of different contexts (3–6). These efforts have been tremendously accelerated by the development of unbiased high-throughput (HTP) methods for the detection of gene–phenotype relationships, protein interactions, genetic interactions and chemical interactions. Such methods have been progressively refined to increase coverage and resolution, and newer techniques are generating other types of biological data that had not been previously available at such a large scale (7). In particular, recent genome-wide genetic screens based on CRISPR/Cas9 genome editing technology have enabled the rapid characterization of gene–phenotype relationships both in cell lines derived from a variety of tissue types and in vivo mouse models (8,9). CRISPR/Cas9 approaches have also been devised to allow systematic exploration of gene–gene interactions in human cells (10,11). These comprehensive maps of gene function promise to further accelerate biomedical research and drug discovery (12,13).
The biological network paradigm has been used to facilitate drug target selection, interpret drug resistance or off-target effects, and forms the basis for targeted therapies and personalized medicine (14,15). An on-going challenge, however, is the unstructured nature of the biomedical literature, i.e., free form text, that cannot be easily parsed for computationally tractable data elements such as protein or genetic interactions. A primary goal of biomedical data curation is thus to convert text-, figure- and table-based experimental information from the biomedical literature into discrete, consistently structured records that can be easily parsed, combined and computed. To this end, the accurate annotation of protein, genetic and other forms of interaction data from the literature by a host of databases and meta-databases has expedited the formulation of both intuitive and more formal models of cellular functions (16), as well as the interpretation of complex genome-wide association studies for a wide variety of disease phenotypes (17).
The Biological General Repository for Interaction Datasets (BioGRID: https://thebiogrid.org) was first developed as an open-access centralized repository for protein and genetic interaction data reported in the biomedical literature (18). Since its inception in 2003, BioGRID has amassed almost 1.6 million biological interactions supported by published experimental data in humans and other major model organisms including the bacterium Escherichia coli, the budding yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe, the plant Arabidopsis thaliana, the nematode worm Caenorhabditis elegans, the fruit fly Drosophila melanogaster, the zebrafish Danio rerio, and the mouse Mus musculus, among many others. BioGRID has also grown in scope to include the curation of post-translational modifications (PTMs) and the annotation of chemical interactions between genes/proteins and bioactive small molecules. BioGRID curation is governed by controlled experimental vocabularies and guided by text mining methods. BioGRID data content is updated and freely distributed to the biomedical community as monthly releases, as well as through partnerships with model organism databases (MODs) such as Saccharomyces Genome Database (SGD) (19) or WormBase (20), various meta-databases for interaction data, and general data portals, such as NCBI (21) or UniProt (22). Since the previous update (23), a new resource within BioGRID called the Open Repository for CRISPR Screens (ORCS) has been developed to house and distribute large-scale CRISPR screen datasets across multiple model organism species (see https://orcs.thebiogrid.org). BioGRID thus provides the biological, biomedical and computational biology research communities with a rigorously annotated resource to help drive discovery in fundamental and clinical research.
DATABASE GROWTH AND STATISTICS
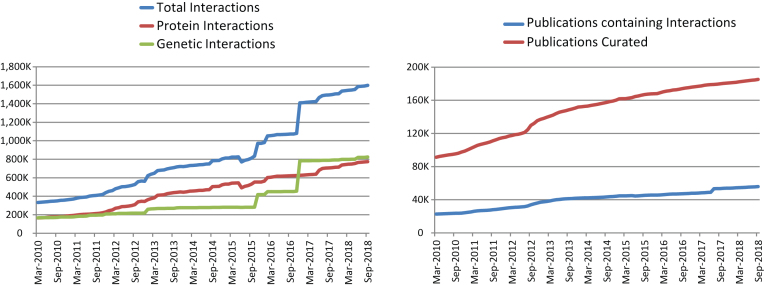
Since our 2017 update in the NAR Database Issue (23), the number of curated interactions housed in BioGRID has increased by 32%. As of September 2018 (version 3.4.164), BioGRID contained 1 295 777 interactions derived from HTP studies and 302 911 interactions derived from low-throughput (LTP) studies for a total of 1 598 688 (1 238 062 non-redundant) interactions. These correspond to 774 460 (578 582 non-redundant) protein interactions and 824 228 (675 685 non-redundant) genetic interactions (Table 1; Figure 1). These data were directly extracted from 55 809 manually annotated peer-reviewed publications (1437 HTP and 54 372 LTP studies) identified from the biomedical literature by keyword searches, text-mining approaches, and direct user submissions. All interactions reported in BioGRID are directly supported by experimental evidence that is categorized according to a structured set of interaction types that map to the experimental detection methods in the PSI-MI 2.5 standard (24). BioGRID also currently contains data on 726 378 protein PTMs (419 472 non-redundant) from 4742 publications, an increase of ∼600 000 PTMs since our previous update, as derived primarily from HTP studies.
Table 1.
Increase in BioGRID data content
| September 2016 (3.4.140) | September 2018 (3.4.164) | ||||||
|---|---|---|---|---|---|---|---|
| Organism | Type | Nodes | Edges | Publications | Nodes | Edges | Publications |
| Arabidopsis thaliana | PI | 9479 | 41 918 | 2168 | 9571 | 42 635 | 2283 |
| GI | 246 | 298 | 125 | 304 | 350 | 154 | |
| Caenorhabditis elegans | PI | 3277 | 6341 | 190 | 3281 | 6350 | 193 |
| GI | 1123 | 2330 | 31 | 1130 | 2336 | 34 | |
| Drosophila melanogaster | PI | 8236 | 38 638 | 454 | 8855 | 54 593 | 2792 |
| GI | 1042 | 9979 | 1 482 | 2958 | 13 440 | 4156 | |
| Escherichia coli | PI | 108 | 109 | 17 | 2161 | 12 917 | 26 |
| GI | 4000 | 166 137 | 15 | 4009 | 171 245 | 16 | |
| Homo sapiens | PI | 20 914 | 365 547 | 25 383 | 22 800 | 449 842 | 27 631 |
| GI | 1577 | 1663 | 283 | 2169 | 5229 | 322 | |
| Mus musculus | PI | 11 892 | 38 163 | 3529 | 12 958 | 44 575 | 3744 |
| GI | 275 | 309 | 176 | 336 | 377 | 192 | |
| Saccharomyces cerevisiae | PI | 6299 | 131 659 | 8074 | 6897 | 164 530 | 9112 |
| GI | 5719 | 212 092 | 7880 | 5956 | 572 320 | 8887 | |
| Schizosaccharomyces pombe | PI | 2946 | 12 817 | 1247 | 2984 | 13 134 | 1334 |
| GI | 3208 | 57 847 | 1459 | 3377 | 59 038 | 1551 | |
| Other organisms | ALL | 9688 | 14 814 | 2250 | 11 307 | 17 319 | 2609 |
| Total | ALL | 65 031 | 1 072 173 | 47 223 | 69 216 | 1 598 688 | 55 809 |
Data is drawn from monthly release 3.4.140 and 3.4.164 of BioGRID. Nodes refer to genes/proteins, edges refer to interactions. PI, protein (physical) interactions; GI, genetic interactions.
Figure 1.
Increase in data content of BioGRID from March 2010 (release 2.0.62) to September 2018 (release 3.4.164). Left panel shows the increase of annotated protein interactions (red), genetic interactions (green) and total interactions (blue). Right panel shows the number of curated publications that contained protein or genetic interaction data (blue) versus the total number of publications examined by curators (red).
In 2018, Google Analytics reported that BioGRID received on average 114 151 page views and 12 100 unique visitors per month. We estimate that these page views correspond to perusal of ∼24 million interactions by BioGRID users in 2018. These statistics do not include the widespread dissemination of BioGRID records by various partner databases, which include the MODs SGD (19), PomBase (25), Candida Genome Database (CGD) (26), WormBase (20), FlyBase (27), the Arabidopsis Information Resource (TAIR) (28), ZFIN (29) and Mouse Genome Database (MGD) (30) and the meta-database resources NCBI (21), UniProt (22), Pathway Commons (31), STRING (32) and others. In 2018, the BioGRID user base was located primarily in the USA (28%), followed by China (13%), India (7%), United Kingdom (6%), Germany (5%), Canada (4%), Japan (4%), France (3%) and all other countries (30%).
CURATION STRATEGY AND SPECIFIC PROJECTS
All curation activity in BioGRID continues to be controlled by an internal dedicated database called the Interaction Management System (IMS), which is used to administer triaged lists of publications for curation for different projects, to standardize all aspects of curation based on controlled vocabularies for experimental evidence and gene names, and to track individual curator contributions. BioGRID now contains interaction data for 71 different model species, an increase of five species from the previous update. As BioGRID now maintains annotation support for 350 species, an increase of over 100 species since the previous update, the database is well positioned to rapidly incorporate data for additional new species as opportunities arise.
BioGRID continues to maintain complete coverage of the primary literature for the main model yeasts S. cerevisiae (now at 736 850 total interactions and 535 436 non-redundant interactions) and S. pombe (now 72 172 total interactions and 58 711 non-redundant interactions). These datasets are also redistributed through SGD (19) and PomBase (25). Extensive curation of protein interactions is also carried out for the model plant A. thaliana (28), now undertaken in collaboration with the BAR database (see below). Other model organism curation is carried out in conjunction with the respective MODs but is not comprehensive due to limitations in curation capacity.
In order to maximize data content and facilitate access to large-scale interaction datasets across species, BioGRID endeavors to curate all publications that contain HTP protein and genetic interaction data. For example, BioGRID annotated almost 13 000 cell envelope protein interactions from an HTP study on a mass spectrometry-based protein interaction network for E. coli (33). In another example, 326 790 binary and 19 847 ternary genetic interactions detected in S. cerevisiae by synthetic genetic array (SGA) screens were curated from two recent publications (34,35). With respect to human data, 84 295 protein interactions have been curated since the previous update, including 32 761 new interactions reported in the BioPlex 2.0 dataset based on an affinity capture-mass spectrometry pipeline (36). Other large-scale human protein interaction data types added to BioGRID include 8744 interactions generated by BioID proximity labeling/capture followed by mass spectrometric identification, as reported in 25 publications. Genetic interactions detected in human cell lines by large-scale CRISPR/Cas9 screens have also been curated by BioGRID (see CRISPR/Cas9 screen section below). BioGRID curators frequently work with authors for deposition and/or release of large datasets prior to publication. Pre-publication data records are fully archived and searchable but are excluded from BioGRID downloads until conversion into full BioGRID records upon publication of the dataset.
The colossal and ever-increasing human biomedical literature, now at 18 million publications deposited in PubMed, presents an impasse for the limited throughput of manual curation approaches. This problem is exacerbated by the fact that only a fraction of candidate publications returned by PubMed queries contain experimentally validated interaction data, such that curators spend considerable effort on inspection of non-relevant publications (Figure 1). This problem can be partly alleviated by the use of text-mining approaches to rank publications for the likelihood of containing interaction data. Although automated information extraction systems are still inferior to expert manual curation based on precision/recall metrics (37,38), natural language processing (NLP) methods can boost manual annotation throughput (39). BioGRID is a longstanding participant in the BioCreative consortium that aims to develop and benchmark biomedical text-mining approaches (40). Since the previous update, BioGRID has contributed to the generation of high-quality reference sets for annotating PubMed abstracts and full text articles (41) and for extraction of protein interactions that are disrupted by natural or synthetic mutations (Doğan et al., in press).
Given that complete coverage of the literature is not feasible, the BioGRID curation strategy focuses in part on deep curation for specific themed projects on critical biological processes and/or specific diseases. A themed project begins with expert consultation and PubMed literature searches to define an extensive set of candidate publications. The publication set is prioritized with an algorithm that uses NLP to extract syntactic features and machine learning to rank abstracts based on higher-order features (42). The ranked publications are then curated, and the gene list recursively expanded based on interaction datasets. Such themed curation projects on biological processes include inflammation, chromatin modification, autophagy, the ubiquitin-proteasome system (UPS), the DNA damage response (DDR), phosphorylation-based signaling and stem cell regulators. Themed curation projects focused on particular diseases include cardiovascular disease and hypertension, glioblastoma (GBM), Fanconi Anemia (FA), diabetes and prevalent infectious diseases, such as tuberculosis and HIV.
We have continued to expand coverage in each current themed curation project. For example, in the UPS project we have compiled 596 293 sites (312 296 non-redundant) of ubiquitin modification on ∼10 000 human proteins and 44 074 sites on ∼3600 yeast proteins, an increase of over 3.5× for human sites and 1.2× for yeast sites compared to the previous BioGRID update. Most of these sites are drawn from HTP mass spectrometry studies that detect the presence of a GG ubiquitin remnant on substrate peptides (43). We have also curated an additional 76 304 interactions associated with proteins and enzymes of the UPS. Similarly, for the autophagy and DDR projects we have added a further 1845 and 2710 interactions respectively. Our disease-themed project on GBM, an aggressive and largely intractable form of brain cancer with limited treatment options (44), has progressed in collaboration with experts in the Stand Up to Cancer (SU2C) Stem Cell team (see www.standup2cancer.ca). A set of 56 GBM-associated genes known to be either mutated or of altered copy number in patient-derived tumor samples (45,46) has yielded a curated network of 12 200 interactions from 3173 publications so far. Biological interactions for all extant themed projects are updated through general BioGRID curation and in periodic dedicated curation drives.
Two new themed projects have recently been undertaken in collaboration with groups supported by the Biomedical Data Translator (see https://ncats.nih.gov/translator). In one project, BioGRID curators have captured interactions associated with the FA pathway, which helps to mediate the DDR and is implicated in a variety of human cancers (47). In consultation with FA experts, BioGRID curators assembled a core list of 53 DDR genes associated with the 20 known core FA genes, originally defined by genetic complementation groups in human patients. Using these gene lists as entry points, we have curated 12 960 interactions from over 2200 publications. A second new themed project associated with the Biomedical Data Translator has focused on Maturity Onset Diabetes of the Young (MODY), an autosomally inherited disease characterized by genetic defects in pancreatic β-cells that compromise insulin production (48). At present, 14 genes are genetically linked to various MODY subtypes and four of these genes (HNF1A, HNF4A, HNF1B and GCK) are known to account for >90% of MODY cases (49). From these 14 entry points, a MODY network of 483 protein interactions has been curated from 149 publications to date. The FA and MODY interaction datasets will be used as inputs and benchmarks for predictive computational methods being developed through the Biomedical Data Translator initiative.
MODEL ORGANISM DATABASE AND META-DATABASE PARTNERS
In addition to collaborating with experts in themed curation project efforts, BioGRID actively works together with MOD and meta-database resources in order to facilitate the widespread propagation of BioGRID records. The BioGRID curation and software teams will work with all interested collaborators on curation of interaction data in order to maximize curation efficiency and impact. This process also provides an opportunity for cross-validation of shared records. Any interaction record within BioGRID that originates from an external resource without modification is clearly attributed as such and hyperlinked to the original source database throughout the BioGRID website search portal and in all associated download files.
This type of partnership is illustrated by data sharing with FlyBase, the MOD for the fruit fly D. melanogaster (27). In the 3.4.150 build of BioGRID, we incorporated a comprehensive update from FlyBase that validated >48 000 previously curated D. melanogaster interactions and incorporated an additional 19 000 interactions that had not yet been curated by BioGRID. All of these interactions are clearly marked throughout BioGRID as having the source ‘FlyBase’. In a continuation of this collaboration, a subsequent update from FlyBase will add ∼8000 additional interactions in an upcoming release of BioGRID. In another example, an on-going collaboration with the S. pombe database, PomBase (25) aims to share manually curated protein and genetic interactions with BioGRID in order to minimize duplication of curation effort. Recent new collaborations have been forged with emerging databases, such as the Bio-Analytic Resource for Plant Biology (BAR), which will help to disseminate BioGRID plant interaction data and reciprocally provide BioGRID with ∼13 000 A. thaliana interactions (50). BioGRID also works closely with the Gene Ontology (GO) consortium (51) as opportunities arise, for example in the use of GO interaction evidence codes to direct BioGRID curation.
GENETIC INTERACTION CURATION
The unambiguous representation of genetic interactions is challenging due to both the complex phenotypes that may be monitored and the specific genetic context of an interaction, which may involve alleles of multiple interacting genes. To reconcile and unify the various genetic interaction terminologies used within different model organism research communities, BioGRID has collaborated with WormBase (20) to develop a new standardized Genetic Interactions Structured Terminology or GIST (Grove et al., in preparation). The GIST has been designed to precisely specify genetic interactions using a universal genetic interaction terminology and is supported by other MODs, including SGD (19), CGD (26), PomBase (25), ZFIN (29), FlyBase (27) and TAIR (28). Implementation of GIST across the different MODs will aid the interpretation of genetic interactions, as well as the integration of large volumes of genetic interaction data across multiple species. In order to accommodate all possible genetic interaction scenarios, the GIST has been organized in a modular format using a structured set of genetic terms that are completely independent from any phenotype(s) that might be linked to the interaction. To effectively describe complex phenotypes that arise in all species from yeast to metazoans, including humans, the GIST is designed to be used in conjunction with all relevant species- or tissue-specific phenotype ontologies such that the type of genetic interaction is curated as a separate entity with each specific phenotype that is scored. This approach allows BioGRID and the MODs to make use of deep species-specific phenotype ontologies across model organisms and humans, including the Ascomycete Phenotype Ontology (52), Uberon (53), the Human Phenotype Ontology (54), and the Monarch Initiative (55). As much as possible, the GIST has been designed to allow reconciliation of existing terms used by different resources. For example, of the various yeast genetic interactions currently annotated in BioGRID, 11 of the existing BioGRID terms map to 7 of the new GIST terms to allow for automated back-mapping of more than 572 300 LTP and HTP yeast genetic interactions associated with over 600 unique phenotypes (52). BioGRID will implement the GIST for forward curation of genetic interactions in human and model organisms, including yeast, worm, fly, mouse and zebrafish. The use of standardized GI terms within the GIST framework will also facilitate the cross-species integration of large genetic interaction datasets produced by HTP methods.
CHEMICAL INTERACTIONS
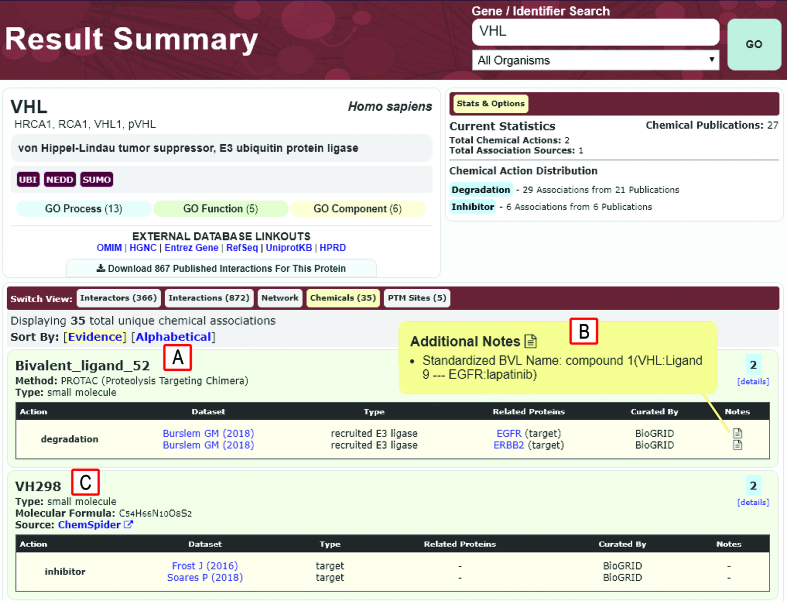
Comparatively few data resources combine chemical–protein interaction data with relevant protein interactions but include STITCH (56), ConsensusPathDB (57), SuperTarget (58) and IntAct (59). To extend our curation breadth to chemical interactions and facilitate network-based approaches to drug discovery, BioGRID has incorporated chemical–protein interaction records from DrugBank (60) and now manually curates small molecule–gene and –protein interactions. In order to incorporate chemical–protein interaction data into BioGRID, a minimal interoperable set of fields compatible with the various annotation systems used across different chemical databases was developed. We examined the content of major chemical interaction databases, including DrugBank (60), BindingDB (61), CTD (62), PharmGKB (63), ChEMBL (64) and others to determine the fields common to each resource. Based on this analysis, a minimal unified record structure was designed that contains: the target protein with both UniProt and GeneID identifiers; generic chemical name, synonyms and/or brand name; the class of agent, such as small molecule or biologic; the structural formula of the agent; CAS and/or ATC identifiers; the molecular action or effect of the agent; associated citations; and the original database source. This minimal record structure allows for efficient import of data into BioGRID and effective interoperability between multiple chemical databases. Relevant database sources for all of the associated records are clearly cited with linkouts to each database, thereby allowing users the option of directly accessing the original source of data for more detailed information. BioGRID has imported manually curated chemical–target data records from DrugBank (60), which contains >10 560 experimental and approved drugs and >4 490 proteins. The downloadable DrugBank files were parsed and drug-target interactions mapped to the minimal unified chemical record structure in BioGRID. The automated mapping of data was validated by extensive curator review to resolve any inconsistencies and ensure data integrity. Currently, BioGRID contains 27 785 chemical interactions manually curated by DrugBank involving 5035 small molecules and 2527 protein targets from 21 organisms, including human, HIV-1, Candida albicans and Escherichia coli. The vast majority of the curated chemical interactions involve human proteins, which represent 92% of the current collection. All chemical-gene/protein interactions can be found in the results summary page (Figure 2), rendered in the on-line BioGRID viewer, and downloaded in standard formats.
Figure 2.
Example of result summary page for chemical interactions of the E3 ubiquitin ligase VHL. (A) Details for Bivalent_ligand_52, a PROTAC composed of a ligand for VHL and a ligand for the degradation targets EFGR and ERBB2 (68). (B) Additional notes display the BVL name in a standardized format based on details provided in the original paper. (C) Chemical-protein interactions curated by BioGRID also include other small molecule inhibitors of UPS enzymes, in this case VH298 as an inhibitor of VHL. External links to ChemSpider provide additional chemical information.
Recently, BioGRID curators have manually curated interactions for over 140 chemical inhibitors/activators of human enzymes involved in the ubiquitin-proteasome system (UPS). Conjugation of the small protein modifier ubiquitin controls the stability, localization and/or activity of much of the proteome (65). These small molecules target a broad spectrum of UPS-related proteins including the core cascade of E1, E2 and E3 enzymes that mediate substrate ubiquitination, proteasome subunits and deubiquitinating enzymes (DUBs). Aside from conventional drug-like inhibitors/activators of UPS enzymes, BioGRID has curated novel bi-functional molecules designed to bridge heterologous substrates to E3 ubiquitin ligases to induce the degradation of specific target proteins. These bivalent ligands (BVLs) are known as PROTACs (protein-targeting chimeric molecules), SNIPERs (specific and non-genetic IAP-dependent protein erasers) and HaloPROTACs (66,67). In general, these compounds consist of two covalently linked ligands that recruit a specific E3 ubiquitin ligase to a target protein, thereby inducing target ubiquitination and proteolysis. Due to the unusual nature of these bivalent compounds, new record structures were devised to capture the key molecular attributes and mechanisms of action. These new standardized fields, as displayed in the Chemical View for the recruited E3 ligase, include the following: internally assigned BVL designation (e.g., Bivalent_ligand_#), Method (e.g., PROTAC/SNIPER/HaloPROTAC), Type (e.g., small molecule/polypeptidic), Action (e.g., degradation), Dataset (e.g., PubMed identifier), Interaction Type (e.g., recruited E3 ligase), Related Proteins (e.g., target protein name), and a Standardized BVL name that appears as an additional note in the details section. This nomenclature system can also be used to describe compounds that directly stabilize E3–substrate interactions, such as the pthalimide class of immunomodulatory (IMiD) drugs (67).
For visualization in a network graph, BVL-type molecules are assigned an internal display designation in the format ‘Bivalent_ligand_#’, with the number sequentially incremented for each additional curated BVL. This designation allows display of complex names and immediately identifies the compound in question as a bivalent ligand. The original published descriptions for BVLs are also displayed in the format ‘Compound name(Recruited E3:E3 Ligand - Target: Target Ligand)’ in the Chemical View details for each relevant E3 and target protein. For example, a particular PROTAC that targets the epidermal growth factor receptor (EGFR) for degradation (68) has been curated in BioGRID as Bivalent_ligand_52 with the standardized BVL name ‘compound 1 (VHL:Ligand 9 – EGFR:lapatinib)’ to indicate that it is called ‘compound 1’ in the original publication and is composed of two linked moieties, a ligand for the E3 enzyme subunit VHL named Ligand 9 and a ligand for the EGFR called lapatinib. The BioGRID Chemical View display has been modified to show entities pertinent to BVLs, i.e. the E3 ubiquitin ligase, the bivalent small molecule ligand, and the target protein (Figure 2). The viewer displays BVL information on the relevant protein result pages for the E3 enzyme and degradation target. PROTACS were only curated if experimentally confirmed to cause degradation of the intended target. If a single PROTAC (i.e. with the same E3- and target-binding moieties) was shown to degrade multiple targets, then each E3-target-BVL relationship was curated as a single BVL designation with all targets listed. To date, 62 different PROTAC-like molecules that target 116 proteins have been annotated from 46 different publications by BioGRID curators. This set of 167 curated chemical–protein interactions represents most if not all available published BVL-type compounds to date.
CRISPR/CAS9 SCREEN CURATION
The development of the budding yeast deletion collection almost 20 years ago enabled a new era of systematic high-throughput screens that revolutionized the mapping of gene–phenotype relationships (69). Subsequently, RNAi-mediated knockdown approaches in model organisms and mammalian cell lines enabled conceptually similar genome-wide screens, but these methods were hampered by incomplete knockdown and off-target effects. Recent development of complex genome-wide knockout libraries based on precise CRISPR/Cas9 sequence-specific endonuclease technology has enabled true loss-of-function genetic screens in mouse and human cell lines (8). Cas9 expressed in cell lines can be programmed with a complex library pool of single-guide RNAs (referred to as sgRNAs or gRNAs) to efficiently generate double strand breaks at targeted loci across the genome and thereby yield a pool of loss-of-function mutants due to error prone repair of the break; the cell line pool can then be used to carry out a systematic selection screen for viability or any other desired phenotype (8,70). The Cas9 nuclease has also been engineered to allow large-scale transcriptional activation and repression screens (71). The high fidelity, efficiency and relative simplicity of CRISPR/Cas9-based genome-wide screens has led to a deluge of publications on phenotypic screens in cell lines derived from humans and other species.
As CRISPR/Cas9 genome-wide experiments are still in their infancy, experimental methods and data analysis vary substantially from one publication to another. We thus developed a working minimal information about CRISPR/Cas9 screens (MIACS) record structure to represent common parameters shared among more than 100 distinct screens published to date. The BioGRID standard includes the variables sgRNA library name, Cas9 variant (CRISPRn, CRISPRi, CRISPRa), methodology, enzyme, cell line, cell type, organism, experimental set up, duration, selection conditions, screen type, phenotype, throughput, screen format, score type, analysis method and reported significance thresholds. To ensure curation consistency, we utilized terms from multiple established ontologies including EFO (72), BTO (73) and CLO (74), and developed CRISPR screen-specific controlled vocabularies for each MIACS category based on pilot curation of the original genome-wide screens (75–77). Our CRISPR curation strategy is gene-based rather than at the individual gRNA-level and therefore includes original gene-level quantitative data for each published screen. Curators thus capture details on original scoring schemes and analytical methods, which currently include BAGEL (78), CasTLE (79), CERES (80), MAGeCK (81), RANKS (82) and others. Score types and confidence indicators (p-, q- and/or FDR values) are reported as in the original source publication, with hits assigned according to the reported significance thresholds. When no clear cut-offs are provided in the publication, significance thresholds are inferred based on the number of hits reported or by assigning a conventional p/q/FDR value of <0.05. Datasets are then organized to provide a ranked display list of genes for the screen. A description of standard vocabularies and CRISPR screen curation can be found on the BioGRID help page.
AN OPEN REPOSITORY FOR CRISPR SCREENS (ORCS) at BIOGRID
To house and distribute comprehensive collections of CRISPR screen datasets across multiple model organism species we have developed the Open Repository for CRISPR Screens (ORCS) within BioGRID (https://orcs.thebiogrid.org). BioGRID ORCS provides a unified warehouse for all published CRISPR screen data and a straightforward user-friendly interface for searching, filtering and downloading of CRISPR screen datasets. Recently established repositories such as GenomeCRISPR (83) and PICKLES (84) provide raw screen data, author-processed data and/or re-scored data. To maintain consistency with authors' published conclusions, ORCS reports only published scores for screen data. ORCS displays results at the publication-, screen- and gene-level with original scores and significance thresholds, along with information about associated analytical methods and other metadata when available. Current screen formats in ORCS include negative and positive selection screens based on viability and other phenotypic readouts in conjunction with nuclease-mediated knockout (CRISPRn), transcriptional activation (CRISPRa) and transcriptional inactivation (CRISPRi) library designs. To date, BioGRID ORCS has annotated 505 screens from human and mouse cell lines drawn from 36 publications that in total applied 14 different statistical methods.
BioGRID ORCS searches can be performed by identifier (gene name, sequence identifier, third-party database identifier), by publication (PubMed ID, author name, keyword), and by controlled vocabulary terms in more than a dozen MIACS categories. All results are presented in an easy to navigate tabular format and are internally hyperlinked to associated BioGRID records to allow recursive searches (Figure 3). Upon clicking on any identifier, publication or screen result, users are taken to a details page that shows curated scores, gene annotations and manually assigned controlled vocabulary terms. Screens can also be visualized by a line graph that depicts an overall score distribution for the entire screen. In addition, results can be filtered to provide more focused datasets for inspection. Genes that scored significantly within a screen are highlighted within all search results throughout the site. All screen data available on the BioGRID ORCS website are freely available for download (see https://downloads.thebiogrid.org) in multiple tab-delimited formats and also as the original supplementary files associated with the publication. Custom datasets can also be generated on-the-fly to include only those identifiers, publications or screens of interest to the user.
Figure 3.
Example of screen summary result page in BioGRID ORCS. (A) Annotated screen details. (B) Score distribution graph. (C) Screen search and filter functions. (D) Sort function for screen scores and annotation. Genes scored as significant in the original publication are designated by ‘Yes’ in the hit column.
For developers, we have built a comprehensive BioGRID ORCS web service with the necessary mechanisms for automated retrieval of BioGRID ORCS screen datasets via standard software tools and platforms. Detailed documentation on how to utilize these interfaces can be found in the BioGRID Wiki (https://wiki.thebiogrid.org/doku.php/orcs:webservice). We have also generated a series of simple open source example programs in Python to illustrate different approaches (https://github.com/BioGRID/ORCS-REST-EXAMPLES). Downloads, web service datasets and example programs are freely accessible to all parties under the MIT license (https://en.wikipedia.org/wiki/MIT_License).
BioGRID ORCS curation and data content will be tightly integrated with interaction data elsewhere in BioGRID. For example, to date, 13 papers curated in BioGRID ORCS also contain protein and/or genetic interactions curated in BioGRID. Reciprocal internal hyper-links between ORCS and BioGRID for all genes and shared PMIDs are provided when applicable. High-throughput CRISPR-based genetic interaction datasets for human and other species will become prevalent as multiplex CRISPR screening technologies are refined and expanded. Only a handful of such CRISPR-based genetic interaction screens have been published so far. For example, a recently curated publication identified ∼3000 human genetic interactions in two different cancer cell lines based on a CRISPRi approach in which 458 query genes were crossed to each other and control genes, resulting in the systematic perturbation of 222 784 gene pairs (11). High confidence negative and positive genetic interactions were identified using a stringent cutoff score and 5% FDR, and all genetic interactors with their corresponding scores were uploaded in BioGRID. The integration of CRISPR-based genetic interaction network data with phenotypic screens will undoubtedly provide many new insights into gene function and genetic network structure.
DATABASE AND INFRASTRUCTURE IMPROVEMENTS
We have continued to enhance usability throughout the entirety of the BioGRID web interface. Recent improvements to the underlying software and hardware have allowed an increase in page load speeds of 30%, thereby ensuring that users obtain results in a timely manner even under peak load conditions. Moreover, database improvements and upgrades to latest software versions have decreased search result load times, even for large wildcard style searches, which encourages users to test different search terms with minimal time commitment. We have continued to improve our graphical user interfaces (GUI) to ensure all result views are straightforward and easy to comprehend, particularly for new users of the website. With respect to underlying database architecture, we continue our migration toward a microservice-based architecture that will underpin BioGRID 4.0 (see Future Developments) and all other BioGRID projects. This structure will improve scalability and facilitate the development of on-the-fly filtering, custom download generation, automated curation pipelines, text-mining enhancements, multi-platform accessibility for mobile devices and the web-based network viewer.
To reduce ambiguity caused by inconsistent gene nomenclature that pervades the biomedical literature, all BioGRID tools rely on a comprehensive annotation system that is designed to collapse redundant results and correct for common non-standard nomenclature pitfalls. This strategy allows BioGRID to present a comprehensive set of synonyms for all genes and a unified search result for users. The inclusion of synonyms can also help the user disambiguate different gene functions. The BioGRID annotation system combines many online resources that include Entrez Gene, UniProt, Ensembl, RefSeq, HGNC, SGD, CGD, MGI, FlyBase, WormBase, TAIR, PubMed and GenBank to aid in this process. Our latest annotation updates now support more than 77 million systematic names, aliases, official symbols and external identifiers from Ensembl, UniProt, NCBI, Entrez-Gene, GenBank, SGD, PomBase, WormBase, FlyBase, MGD, HGNC, MGD, TAIR, VectorBase, BeeBase, ZFIN and HPRD, among other sources. When applicable, relevant results within the site are hyperlinked to these associated resources providing an accessible means of retrieving additional details for any individual gene or associated publication. This underlying annotation resource underpins all BioGRID tools and technologies including newly released projects such as BioGRID ORCS. For instance, although screens currently reported in ORCS represent only two different organisms, H. sapiens and M. musculus, the flexible annotation platform will allow expansion to additional model organism species as screen datasets are reported in the literature.
Since moving all BioGRID project websites, databases, and scripts to the cloud in previous years (see 2013, 2015 and 2017 NAR updates), BioGRID has supported a consistent increase in usage while continuing to maintain >99.99% uptime accessibility on all systems. As usage has increased, the resources needed to meet demand have been increased in parallel. Since the previous update, we have doubled the CPU, storage, and memory available on all BioGRID servers and continued to add additional servers when required. Recently, we migrated all BioGRID pre-generated download files, such as monthly interaction updates, to a cloud-based content delivery network (CDN) that provides rapid and decentralized access to all files anywhere in the world via a 40 Gigabit network infrastructure. This enhancement ensures that BioGRID download files are readily and rapidly accessible in all contexts, such as for manual downloads or script-based retrieval for more complex computational pipelines.
While BioGRID does not record any personal information about users, we have recently improved security and privacy of all communications with BioGRID websites, tools, web services and resources, by completing a top-to-bottom transition to Secure Sockets Layer (SSL) support for all user-facing projects. This transition enforces top-of-the-line encryption across the entirety of the BioGRID project space, ensuring communication between users and our websites is secure and private. For users still accessing BioGRID resources via older http-based communication, we recommend that links and connections be updated to the https versions (e.g., https://thebiogrid.org).
The most recent major point release of BioGRID (version 3.5, release date October 2018) includes a newly-designed BioGRID project page format that serves as a unified entry point to project-specific data. Project pages enable access to all data associated with a particular curation theme through the use of tags that identify every gene associated with a given project. Advanced search filters allow interrogation of interaction data within the project theme, and search result annotation has been redesigned to include detailed popups that provide publication information and experimental evidence. Statistics for projects have been improved to support a graphical pie-chart display that can be customized by the user. Project pages are formatted in a new responsive layout model that automatically adapts to support users with both large display and small screen dimensions, such as mobile devices. An initial themed project page has been released for the curation of protein and genetic interactions of all kinases, phosphatases and associated subunits in the budding yeast S. cerevisiae, termed the kinome (Figure 4, see http://yeastkinome.thebiogrid.org). This project began as a systematic HTP mass spectrometry-based study that reported 1844 interactions for all proteins in the kinome (85). The kinome project dataset now extends to 97 397 genetic and protein interactions, as well as 3853 post-translational modifications curated from over 4700 publications (86). This new project page replaces a previous static site for the original dataset (www.yeastkinome.org) and will be updated monthly with version control through on-going S. cerevisiae curation. Project pages for other curation themes will be progressively implemented through addition of gene-tag classifiers for all genes associated with each theme-based curation project.
Figure 4.
S. cerevisiae kinome project page at BioGRID. (A) Description of project with hyperlinks to resources and downloads. (B) Project-level statistics. (C) Searchable project gene list and annotation.
DATA DISSEMINATION
All BioGRID data records can be searched via standard web search page interfaces or downloaded in a number of standardized tabular (tab, tab2 and mitab) and structured (PSI-MI 1.0 XML, PSI-MI 2.5 XML, JSON) formats (https://downloads.thebiogrid.org). The BioGRID (https://wiki.thebiogrid.org/doku.php/biogridrest) and BioGRID ORCS (https://wiki.thebiogrid.org/doku.php/orcs:webservice) REST web services support over 1000 active projects worldwide that perform over 100 000 queries per month with an average return of ∼3.5 million interactions per month. For example, the REST service enables the direct comparison of all data in BioGRID to real time experimental data in the ProHits mass spectrometry LIMS (87). The IMEx consortium PSICQUIC API interface (88) also currently sends >170 000 queries per month to BioGRID from third party plugins. In addition to our work with the MODs on various curation projects, BioGRID datasets are also made available via third party meta-databases, resources, and query tools. For example, BioGRID interaction data is available as a hyperlink for all gene and protein entries in the widely-used NCBI and UniProt databases, respectively (21,22). Other major meta-database resources that disseminate BioGRID data include STRING (32), Pathway Commons (31), Gene Mania (89), InnateDB (90) and FlyAtlas (91) (see https://wiki.thebiogrid.org/doku.php/partners for full list). BioGRID data is also now disseminated through the Network Data Exchange (92), which allows users to visualize and explore networks drawn from BioGRID records (see https://goo.gl/Zu6bTe).
We have continued to update BioGRID Wiki documentation on all tools and resources (see http://wiki.thebiogrid.org). In early 2016, we released two protocol papers that outline key functions in step-by-step processes to aid new users in using the platform (93,94). BioGRID also continues to maintain an active e-mail help desk to assist users and facilitate the direct deposition of large datasets (biogridadmin@gmail.com). Finally, all new source code has been deposited at our GitHub organizational page (https://github.com/BioGRID) and we continue to update both our Twitter feed (https://twitter.com/biogrid) and YouTube channel (https://www.youtube.com/user/TheBioGRID) with the latest BioGRID news and feature updates.
FUTURE DEVELOPMENTS
BioGRID will continue to annotate protein, genetic and chemical interaction data from the primary biomedical literature with a particular focus on HTP protein and genetic interaction data, large-scale CRISPR screen data, and themed human curation projects. The BioGRID curation pipeline will be further enhanced with improved text-mining tools in conjunction with text-mining groups and the BioCreative consortium. Collaborations with diverse database partners, including MODs, phenotype databases, and chemical databases will serve to disseminate BioGRID curation data and foster cooperative curation efforts. We will continue to provide resources and support for the propagation of BioGRID data through partner databases. Major new improvements are anticipated for BioGRID and the linked BioGRID ORCS resource as we work towards BioGRID 4.0 as a comprehensive renewal of the database infrastructure and user interface. A revision of the Interaction Management System is nearing completion and will specifically facilitate curation of complex experimental techniques and higher-order interaction data types across all BioGRID projects. Through these efforts, BioGRID will continue to strive to provide a wide range of curated biological interaction data for the biomedical research community.
DATA AVAILABILITY
All data, software and resources referred to in this publication are available at the following URLs:
http://yeastkinome.thebiogrid.org/
https://downloads.thebiogrid.org/
https://webservice.thebiogrid.org/
ACKNOWLEDGEMENTS
The authors thank John Aitchison, Brenda Andrews, Gary Bader, Anastasia Baryshnikova, Andre Bernards, Judy Blake, Charlie Boone, Stephen Burley, Fiona Coutinho, Mike Cook, Peter Dirks, Jennifer Dougherty, Andrew Emili, Russ Finley, Michael Gilson, Anne-Claude Gingras, Gustavo Gluzman, Chris Grove, Steve Gygi, Melissa Haendel, Wade Harper, Peter Hornbeck, Eva Huala, Sui Huang, Trey Ideker, Igor Jurisica, Thom Kaufman, James Knight, Theo Knijnenburg, Jianzhu Ma, Chris Mungal, Chad Myers, Nick Provart, Ivan Sadowski, Paul Sternberg, Xiaojing Tang, Olga Troyanskaya, Monte Westerfield, John Wilbur, David Wishart, Val Wood, Floris Schoeters, Helen Yu, Mike Yu and Cathy Wu for collaborations, support, discussions and/or access to pre-publication datasets.
FUNDING
National Institutes of Health Office of Research Infrastructure Programs [R01OD010929 to M.T., K.D.]; National Center For Advancing Translational Sciences of the National Institutes of Health [OT3TR002026 to M.T.; S. Huang, P.I.]; Genome Canada/Genome Quebec/Ontario Genomics Institute Largescale Applied Proteomics [OGI-069 to M.T.; A.-C. Gingras, co-P.I.]; Stand Up To Cancer Canada [to M.T.; P. Dirks, P.I.]; Canada Research Chair in Systems and Synthetic Biology [to M.T.]. Funding for open access charge: National Institute of Health [R01OD010929].
Conflict of interest statement. None declared.
REFERENCES
- 1. Yeger-Lotem E., Sharan R.. Human protein interaction networks across tissues and diseases. Front. Genet. 2015; 6:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma J., Yu M.K., Fong S., Ono K., Sage E., Demchak B., Sharan R., Ideker T.. Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods. 2018; 15:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hofree M., Shen J.P., Carter H., Gross A., Ideker T.. Network-based stratification of tumor mutations. Nat. Methods. 2013; 10:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sahni N., Yi S., Taipale M., Fuxman Bass J.I., Coulombe-Huntington J., Yang F., Peng J., Weile J., Karras G.I., Wang Y. et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015; 161:647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Srivas R., Shen J.P., Yang C.C., Sun S.M., Li J., Gross A.M., Jensen J., Licon K., Bojorquez-Gomez A., Klepper K. et al. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol. Cell. 2016; 63:514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W., Ma J., Ideker T.. Classifying tumors by supervised network propagation. Bioinformatics. 2018; 34:i484–i493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Snider J., Kotlyar M., Saraon P., Yao Z., Jurisica I., Stagljar I.. Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 2015; 11:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shalem O., Sanjana N.E., Zhang F.. High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 2015; 16:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chow R.D., Chen S.. Cancer CRISPR screens in vivo. Trends Cancer. 2018; 4:349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen J.P., Zhao D., Sasik R., Luebeck J., Birmingham A., Bojorquez-Gomez A., Licon K., Klepper K., Pekin D., Beckett A.N. et al. Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat. Methods. 2017; 14:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horlbeck M.A., Xu A., Wang M., Bennett N.K., Park C.Y., Bogdanoff D., Adamson B., Chow E.D., Kampmann M., Peterson T.R. et al. Mapping the genetic landscape of human cells. Cell. 2018; 174:953–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keenan A.B., Jenkins S.L., Jagodnik K.M., Koplev S., He E., Torre D., Wang Z., Dohlman A.B., Silverstein M.C., Lachmann A. et al. The library of integrated network-based cellular signatures NIH program: system-Level cataloging of human cells response to perturbations. Cell Syst. 2018; 6:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurata M., Yamamoto K., Moriarity B.S., Kitagawa M., Largaespada D.A.. CRISPR/Cas9 library screening for drug target discovery. J. Hum. Genet. 2018; 63:179–186. [DOI] [PubMed] [Google Scholar]
- 14. Berg E.L. Systems biology in drug discovery and development. Drug Discov. Today. 2014; 19:113–125. [DOI] [PubMed] [Google Scholar]
- 15. Hood L., Friend S.H.. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat. Rev. Clin. Oncol. 2011; 8:184–187. [DOI] [PubMed] [Google Scholar]
- 16. Mitra K., Carvunis A.R., Ramesh S.K., Ideker T.. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 2013; 14:719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Califano A., Butte A.J., Friend S., Ideker T., Schadt E.. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat. Genet. 2012; 44:841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Breitkreutz B.J., Stark C., Tyers M.. The GRID: The general repository for interaction datasets. Genome Biol. 2003; 4:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Skrzypek M.S., Nash R.S., Wong E.D., MacPherson K.A., Hellerstedt S.T., Engel S.R., Karra K., Weng S., Sheppard T.K., Binkley G. et al. Saccharomyces genome database informs human biology. Nucleic Acids Res. 2018; 46:D736–D742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee R.Y.N., Howe K.L., Harris T.W., Arnaboldi V., Cain S., Chan J., Chen W.J., Davis P., Gao S., Grove C. et al. WormBase 2017: molting into a new stage. Nucleic Acids Res. 2018; 46:D869–D874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016; 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A. et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017; 45:D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerrien S., Orchard S., Montecchi-Palazzi L., Aranda B., Quinn A.F., Vinod N., Bader G.D., Xenarios I., Wojcik J., Sherman D. et al. Broadening the horizon-level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007; 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDowall M.D., Harris M.A., Lock A., Rutherford K., Staines D.M., Bahler J., Kersey P.J., Oliver S.G., Wood V.. PomBase 2015: updates to the fission yeast database. Nucleic Acids Res. 2015; 43:D656–D661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skrzypek M.S., Binkley J., Binkley G., Miyasato S.R., Simison M., Sherlock G.. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017; 45:D592–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gramates L.S., Marygold S.J., Santos G.D., Urbano J.M., Antonazzo G., Matthews B.B., Rey A.J., Tabone C.J., Crosby M.A., Emmert D.B. et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 2017; 45:D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012; 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Howe D.G., Bradford Y.M., Eagle A., Fashena D., Frazer K., Kalita P., Mani P., Martin R., Moxon S.T., Paddock H. et al. The Zebrafish Model Organism Database: new support for human disease models, mutation details, gene expression phenotypes and searching. Nucleic Acids Res. 2017; 45:D758–D768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith C.L., Blake J.A., Kadin J.A., Richardson J.E., Bult C.J. Mouse Genome Database, Group . Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. 2018; 46:D836–D842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur O., Anwar N., Schultz N., Bader G.D., Sander C.. Pathway commons, a web resource for biological pathway data. Nucleic Acids Res. 2011; 39:D685–D690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Babu M., Bundalovic-Torma C., Calmettes C., Phanse S., Zhang Q., Jiang Y., Minic Z., Kim S., Mehla J., Gagarinova A. et al. Global landscape of cell envelope protein complexes in Escherichia coli. Nat. Biotechnol. 2018; 36:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D. et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016; 353:aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuzmin E., VanderSluis B., Wang W., Tan G., Deshpande R., Chen Y., Usaj M., Balint A., Mattiazzi Usaj M., van Leeuwen J. et al. Systematic analysis of complex genetic interactions. Science. 2018; 360:eaao1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H. et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017; 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murugesan G., Abdulkadhar S., Natarajan J.. Distributed smoothed tree kernel for protein-protein interaction extraction from the biomedical literature. PLoS One. 2017; 12:e0187379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salwinski L., Licata L., Winter A., Thorneycroft D., Khadake J., Ceol A., Aryamontri A.C., Oughtred R., Livstone M., Boucher L. et al. Recurated protein interaction datasets. Nat. Methods. 2009; 6:860–861. [DOI] [PubMed] [Google Scholar]
- 39. Mottin L., Gobeill J., Pasche E., Michel P.A., Cusin I., Gaudet P., Ruch P.. neXtA5: accelerating annotation of articles via automated approaches in neXtProt. Database (Oxford). 2016; 2016:baw098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirschman L., Yeh A., Blaschke C., Valencia A.. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005; 6:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Islamaj Dogan R., Kim S., Chatr-Aryamontri A., Chang C.S., Oughtred R., Rust J., Wilbur W.J., Comeau D.C., Dolinski K., Tyers M.. The BioC-BioGRID corpus: full text articles annotated for curation of protein-protein and genetic interactions. Database (Oxford). 2017; 2017:baw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim S., Wilbur W.J.. Classifying protein-protein interaction articles using word and syntactic features. BMC Bioinformatics. 2011; 12:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heap R.E., Gant M.S., Lamoliatte F., Peltier J., Trost M.. Mass spectrometry techniques for studying the ubiquitin system. Biochem. Soc. Trans. 2017; 45:1137–1148. [DOI] [PubMed] [Google Scholar]
- 44. Dirks P.B. Brain tumor stem cells: the cancer stem cell hypothesis writ large. Mol. Oncol. 2010; 4:420–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H. et al. The somatic genomic landscape of glioblastoma. Cell. 2013; 155:462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mackay A., Burford A., Carvalho D., Izquierdo E., Fazal-Salom J., Taylor K.R., Bjerke L., Clarke M., Vinci M., Nandhabalan M. et al. Integrated molecular Meta-Analysis of 1,000 pediatric High-Grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017; 32:520–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nalepa G., Clapp D.W.. Fanconi Anaemia and cancer: An intricate relationship. Nat. Rev. Cancer. 2018; 18:168–185. [DOI] [PubMed] [Google Scholar]
- 48. Firdous P., Nissar K., Ali S., Ganai B.A., Shabir U., Hassan T., Masoodi S.R.. Genetic testing of maturity-onset diabetes of the young current status and future perspectives. Front Endocrinol. (Lausanne). 2018; 9:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shields B.M., Hicks S., Shepherd M.H., Colclough K., Hattersley A.T., Ellard S.. Maturity-onset diabetes of the young (MODY): How many cases are we missing?. Diabetologia. 2010; 53:2504–2508. [DOI] [PubMed] [Google Scholar]
- 50. Waese J., Provart N.J.. The Bio-Analytic resource for plant biology. Methods Mol. Biol. 2017; 1533:119–148. [DOI] [PubMed] [Google Scholar]
- 51. The Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017; 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Engel S.R., Balakrishnan R., Binkley G., Christie K.R., Costanzo M.C., Dwight S.S., Fisk D.G., Hirschman J.E., Hitz B.C., Hong E.L. et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010; 38:D433–D436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mungall C.J., Torniai C., Gkoutos G.V., Lewis S.E., Haendel M.A.. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012; 13:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Groza T., Kohler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T. et al. The human phenotype ontology: Semantic unification of common and rare disease. Am. J. Hum. Genet. 2015; 97:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McMurry J.A., Kohler S., Washington N.L., Balhoff J.P., Borromeo C., Brush M., Carbon S., Conlin T., Dunn N., Engelstad M. et al. Navigating the phenotype frontier: the Monarch Initiative. Genetics. 2016; 203:1491–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M.. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016; 44:D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Herwig R., Hardt C., Lienhard M., Kamburov A.. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016; 11:1889–1907. [DOI] [PubMed] [Google Scholar]
- 58. Hecker N., Ahmed J., von Eichborn J., Dunkel M., Macha K., Eckert A., Gilson M.K., Bourne P.E., Preissner R.. SuperTarget goes quantitative: Update on drug-target interactions. Nucleic Acids Res. 2012; 40:D1113–D1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N. et al. The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014; 42:D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J.. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016; 44:D1045–D1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J.. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 2017; 45:D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barbarino J.M., Whirl-Carrillo M., Altman R.B., Klein T.E.. PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018; 10:e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nowotka M.M., Gaulton A., Mendez D., Bento A.P., Hersey A., Leach A.. Using ChEMBL web services for building applications and data processing workflows relevant to drug discovery. Expert Opin. Drug Discov. 2017; 12:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Oh E., Akopian D., Rape M.. Principles of ubiquitin-dependent signaling. Annu. Rev. Cell Dev. Biol. 2018; 34:137–162. [DOI] [PubMed] [Google Scholar]
- 66. Huang X., Dixit V.M.. Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Res. 2016; 26:484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cromm P.M., Crews C.M.. Targeted protein degradation: From chemical biology to drug discovery. Cell Chem. Biol. 2017; 24:1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Burslem G.M., Smith B.E., Lai A.C., Jaime-Figueroa S., McQuaid D.C., Bondeson D.P., Toure M., Dong H., Qian Y., Wang J. et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem. Biol. 2018; 25:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999; 285:901–906. [DOI] [PubMed] [Google Scholar]
- 70. Wang T., Lander E.S., Sabatini D.M.. Large-scale single guide RNA library construction and use for CRISPR–Cas9-Based genetic screens. Cold Spring Harb. Protoc. 2016; 2016:doi:10.1101/pdb.top086892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sanjana N.E. Genome-scale CRISPR pooled screens. Anal. Biochem. 2017; 532:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Malone J., Holloway E., Adamusiak T., Kapushesky M., Zheng J., Kolesnikov N., Zhukova A., Brazma A., Parkinson H.. Modeling sample variables with an experimental factor ontology. Bioinformatics. 2010; 26:1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gremse M., Chang A., Schomburg I., Grote A., Scheer M., Ebeling C., Schomburg D.. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011; 39:D507–D513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sarntivijai S., Lin Y., Xiang Z., Meehan T.F., Diehl A.D., Vempati U.D., Schurer S.C., Pang C., Malone J., Parkinson H. et al. CLO: the cell line ontology. J. Biomed. Semantics. 2014; 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR–Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science. 2014; 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W.. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014; 509:487–491. [DOI] [PubMed] [Google Scholar]
- 78. Hart T., Moffat J.. BAGEL: A computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics. 2016; 17:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Morgens D.W., Deans R.M., Li A., Bassik M.C.. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016; 34:634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S. et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017; 49:1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S.. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014; 15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bertomeu T., Coulombe-Huntington J., Chatr-Aryamontri A., Bourdages K.G., Coyaud E., Raught B., Xia Y., Tyers M.. A High-Resolution Genome-Wide CRISPR/Cas9 viability screen reveals structural features and contextual diversity of the human cell-essential proteome. Mol. Cell Biol. 2018; 38:e00302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rauscher B., Heigwer F., Breinig M., Winter J., Boutros M.. GenomeCRISPR - a database for high-throughput CRISPR/Cas9 screens. Nucleic Acids Res. 2017; 45:D679–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lenoir W.F., Lim T.L., Hart T.. PICKLES: the database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res. 2018; 46:D776–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Breitkreutz A., Choi H., Sharom J.R., Boucher L., Neduva V., Larsen B., Lin Z.Y., Breitkreutz B.J., Stark C., Liu G. et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010; 328:1043–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sadowski I., Breitkreutz B.J., Stark C., Su T.C., Dahabieh M., Raithatha S., Bernhard W., Oughtred R., Dolinski K., Barreto K. et al. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database (Oxford). 2013; 2013:bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu G., Zhang J., Choi H., Lambert J.P., Srikumar T., Larsen B., Nesvizhskii A.I., Raught B., Tyers M., Gingras A.C.. Using ProHits to store, annotate, and analyze affinity purification-mass spectrometry (AP-MS) data. Curr. Protoc. Bioinformatics. 2012; 8:doi:10.1002/0471250953.bi0816s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. del-Toro N., Dumousseau M., Orchard S., Jimenez R.C., Galeota E., Launay G., Goll J., Breuer K., Ono K., Salwinski L. et al. A new reference implementation of the PSICQUIC web service. Nucleic Acids Res. 2013; 41:W601–W606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T. et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010; 38:W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., Winsor G.L., Hancock R.E., Brinkman F.S., Lynn D.J.. InnateDB: systems biology of innate immunity and beyond-recent updates and continuing curation. Nucleic Acids Res. 2013; 41:D1228–D1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leader D.P., Krause S.A., Pandit A., Davies S.A., Dow J.A.T.. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 2018; 46:D809–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pratt D., Chen J., Welker D., Rivas R., Pillich R., Rynkov V., Ono K., Miello C., Hicks L., Szalma S. et al. NDEx, the network data exchange. Cell Syst. 2015; 1:302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Oughtred R., Chatr-aryamontri A., Breitkreutz B.J., Chang C.S., Rust J.M., Theesfeld C.L., Heinicke S., Breitkreutz A., Chen D., Hirschman J. et al. Use of the BioGRID database for analysis of yeast protein and genetic interactions. Cold Spring Harb. Protoc. 2016; 2016:doi:10.1101/pdb.prot088880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Oughtred R., Chatr-aryamontri A., Breitkreutz B.J., Chang C.S., Rust J.M., Theesfeld C.L., Heinicke S., Breitkreutz A., Chen D., Hirschman J. et al. BioGRID: a resource for studying biological interactions in yeast. Cold Spring Harb. Protoc. 2016; 2016:doi:10.1101/pdb.top080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data, software and resources referred to in this publication are available at the following URLs:
http://yeastkinome.thebiogrid.org/
https://downloads.thebiogrid.org/
https://webservice.thebiogrid.org/