Abstract
Ancient mitochondrial DNA is used for tracing human past demographic events due to its population-level variability. The number of published ancient mitochondrial genomes has increased in recent years, alongside with the development of high-throughput sequencing and capture enrichment methods. Here, we present AmtDB, the first database of ancient human mitochondrial genomes. Release version contains 1107 hand-curated ancient samples, freely accessible for download, together with the individual descriptors, including geographic location, radiocarbon dating, and archaeological culture affiliation. The database also features an interactive map for sample location visualization. AmtDB is a key platform for ancient population genetic studies and is available at https://amtdb.org.
INTRODUCTION
Ancient DNA (aDNA) is a genetic material obtained from ancient specimens, and unlike modern DNA, undergoes fragmentation and post-mortem damages caused mainly by environmental factors (1). Ancient DNA studies, conducted in the last 30 years, have confirmed that while maintaining appropriate procedures, we are able to recover genetic material from ancient specimens. Until recently, the majority of human aDNA studies were focused mainly on mitochondrial DNA (mtDNA) thanks to the fact that mtDNA is present in cells in a higher copy number than the nuclear genome, and therefore it is often the only genetic marker that can be recovered from poorly preserved samples. Due to its maternal inheritance, high mutation rate, absence of recombination and population-level variability, it is a useful tool for reconstructing the past demographic events (2). Despite the long-standing interest in ancient mtDNA, it was only in the past few years, when a high number of complete mt genomes were made available, alongside with the development of the high-throughput sequencing, often combined with the capture enrichment methods.
Mitochondrial DNA, often as a part of nuclear genome studies, was used to reconstruct demographic events that took place in pre-LGM (Last Glacial Maximum) and post-LGM era in Europe (3,4), to trace demographic changes that shaped past and modern populations mtDNA variation (5–13), including the influence of Neolithization process (14–23), and Steppe migrations (24–26). Moreover, mtDNA was used in several kinship studies as a molecular marker which excludes direct maternal kinship between ancient individuals (27–31).
Although there are currently available modern mtDNA databases, e.g. EMPOP (32), MITOMAP (33), HmtDB (34), and mtDB (35), there is no database that would be dedicated specifically to ancient mt genomes. A database concentrated primarily on ancient DNA is the Online Ancient Genome Repository (https://www.oagr.org.au). OAGR is the database primarily for samples generated (or collaborated on) by the Australian Centre for Ancient DNA, University of Adelaide, and includes both human SNP markers data and microbiome data. Our AmtDB is filling this gap by consistent way of mapping the published aDNA samples from different sources, and providing the associated metadata in standard, uniform, easily-downloadable-and-usable way, together with the mt genomes sequences and links to other resources. While our primary focus lies on ancient mtDNA, the metadata itself can be easily used in ancient genomic, archaeological or anthropological studies.
Database overview and functionality
The AmtDB database, as of initial version v1.000, contains 1107 samples. For 887 of these samples we provide the full mt sequences in FASTA format. For all samples, we offer metadata in form of additional descriptors. Although we utilize custom scripts for semi-automated data retrieval, all provided data are hand-curated and checked.
Authors of the aDNA studies usually provide the mtDNA sequences in three different ways, or in any combination of there of:
As complete mtDNA sequences deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) database (labeled as fasta).
As results from high-throughput sequencing, in the form of SAM/BAM files deposited in an appropriate database, i.e. European Nucleotide Archive (https://www.ebi.ac.uk/ena) or Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) (labeled as bam).
In haplotype format, i.e. list of changed position in comparison to rCRS (36) or RSRS (37) (labeled as reconstructed).
In case of available FASTA from GenBank, we provide this sequence in the database. Otherwise, we either reconstruct the mt sequence from the haplotype using the Haplosearch (https://haplosearch.com) tool (38), or preferably reconstruct the mt sequence from provided SAM/BAM files, merging multiple files (per individual) with the use of SAMtools (39). The bioinformatics pipeline in this procedure includes mapping the merged reads as single-end reads against the rCRS with BWA software (40) and collapsing duplicate sequence reads with identical start and end coordinates using FilterUniqueSAMCons.py script (41). Consensus sequences are built using ANGSD toolkit (42). In these samples we also display the average sequence depth (coverage). User can filter all samples according to the mt sequence source discussed above (fasta, bam, reconstructed).
More details about mtDNA reconstruction pipeline can be found in our previous publication (15) and in AmtDB documentation (https://amtdb.org/help).
Besides the mt sequences, the AmtDB contains additional information about the samples, the metadata. The samples can be selected and browsed based on primary ID and alternative ID(s), several geographic location descriptors, latitude and longitude in decimal degrees, archaeological site, or group of archaeological cultural background descriptors. We also supply a comment column, which may contain additional info for the sample, usually information about relationship, uncertainties, or important notes that do not fit into other category and might be valuable for researchers. Biological variables include sex, mt haplogroup, Y chromosomal haplogroup, and Y chromosomal haplotype. For sample age related information, we use calibrated BCE or CE ((Before) Common Era) dates wherever possible. For the radiocarbon dated samples, we provide the precise min. and max. values of the 95.4% probability interval for calibrated (B)CE date, uncalibrated BP (Before Present) age, and radiocarbon laboratory and sample code. For samples that are not directly 14C dated, but other samples from the same layer are, we provide calibrated (B)CE age of the layer. For samples, that were dated only according to the material culture associated with the sample, we use uncalibrated (B)CE age. Our database search engine allows to filter 14C dated samples only. For each sample we also provide publication reference, DOI based reference link and link to sample (mt) sequence depository.
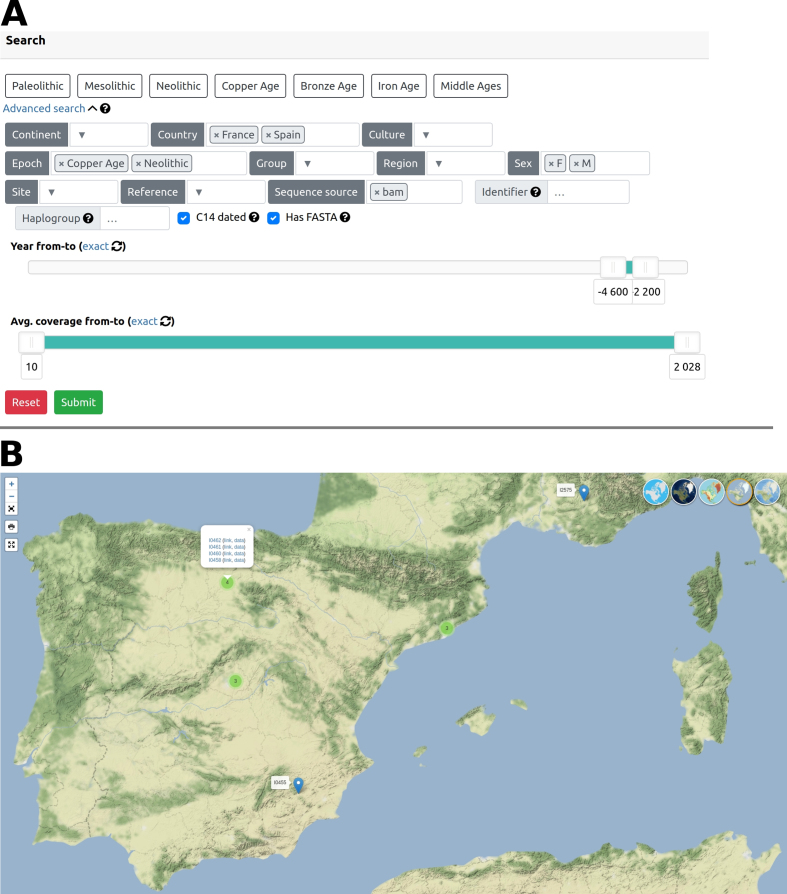
Focal point of our simple, clear and user-friendly interface with advanced search options (Figure 1A) is the visualization of the filtered samples on an interactive world map (Figure 1B). Samples on the map can be clustered together by their distance and smaller clusters are created when the map is zoomed in. Tooltip with sample links appears when cluster is right clicked. Maps are available in several graphical overlays (political, physical, satellite or blind map), and are ready for download together with all provided sequences and metadata, without registration.
Figure 1.
AmtDB (https://amtdb.org) advanced search overview. (A) The database was filtered for Neolithic and Copper Age samples from France and Spain. These samples also have following attributes: known sex (‘F’ or ‘M’), age between 4600 and 2200 BCE, complete mtDNA available, mt reconstructed (sequence source) from BAM files with average coverage at least 10, and are radiocarbon dated. (B) Visualization of the search results on a map, showing unclustered and clustered samples (clustering can be toggled on and off). Tooltip with sample links can be displayed for each sample or cluster (here shown for four samples cluster from Arroyal I site in Burgos, Spain).
CONCLUSION
The database is currently in initial operational capability phase, v1.000, and will get 2–3 major updates per year, concentrating on adding more published samples into the database. We believe the community of ancient human populations researchers will find AmtDB useful, as to our best knowledge, there is no comparable database in terms of usability and data content.
DATA AVAILABILITY
The Ancient human mitochondrial genomes database can be found at https://amtdb.org.
FUNDING
Ministry of Education, Youth and Sports of the Czech Republic [ELIXIR-CZ project LM2015047, part of the international ELIXIR infrastructure, under the Projects CESNET, LM2015042]; Polish National Science Center [2014/12/W/NZ2/00466]. Funding for open access charge: Ministry of Education, Youth and Sports of the Czech Republic.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pääbo S. Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplification. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramakrishnan U., Hadly E.A.. Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Mol. Ecol. 2009; 18:1310–1330. [DOI] [PubMed] [Google Scholar]
- 3. Posth C., Renaud G., Mittnik A., Drucker D.G., Rougier H., Cupillard C., Valentin F., Thevenet C., Furtwängler A., Wißing C. et al. Pleistocene mitochondrial genomes suggest a single major dispersal of non-africans and a late glacial population turnover in Europe. Curr. Biol. 2016; 26:827–833. [DOI] [PubMed] [Google Scholar]
- 4. Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A. et al. The genetic history of Ice Age Europe. Nature. 2016; 534:200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brandt G., Haak W., Adler C.J., Roth C., Szecsenyi-Nagy A., Karimnia S., Moller-Rieker S., Meller H., Ganslmeier R., Friederich S. et al. Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science. 2013; 342:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brotherton P., Haak W., Templeton J., Brandt G., Soubrier J., Jane Adler C., Richards S.M., Der Sarkissian C., Ganslmeier R., Friederich S. et al. Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nat. Commun. 2013; 4:1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gallego-Llorente M., Connell S., Jones E.R., Merrett D.C., Jeon Y., Eriksson A., Siska V., Gamba C., Meiklejohn C., Beyer R. et al. The genetics of an early Neolithic pastoralist from the Zagros, Iran. Sci. Rep. 2016; 6:31326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kılınç G.M., Omrak A., Özer F., Günther T., Büyükkarakaya A.M., Bıçakçı E., Baird D., Dönertaş H.M., Ghalichi A., Yaka R. et al. The demographic development of the first farmers in Anatolia. Curr. Biol. 2016; 26:2659–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lazaridis I., Nadel D., Rollefson G., Merrett D.C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K. et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016; 536:419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Omrak A., Günther T., Valdiosera C., Svensson E.M., Malmström H., Kiesewetter H., Aylward W., Storå J., Jakobsson M., Götherström A.. Genomic evidence establishes anatolia as the source of the European neolithic gene pool. Curr. Biol. 2016; 26:270–275. [DOI] [PubMed] [Google Scholar]
- 11. Haber M., Doumet-Serhal C., Scheib C., Xue Y., Danecek P., Mezzavilla M., Youhanna S., Martiniano R., Prado-Martinez J., Szpak M. et al. Continuity and admixture in the last five millennia of levantine history from ancient canaanite and Present-Day lebanese genome sequences. Am. J. Hum. Genet. 2017; 101:274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathieson I., Alpaslan-Roodenberg S., Posth C., Szécsényi-Nagy A., Rohland N., Mallick S., Olalde I., Broomandkhoshbacht N., Candilio F., Cheronet O. et al. The genomic history of southeastern Europe. Nature. 2018; 555:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olalde I., Brace S., Allentoft M.E., Armit I., Kristiansen K., Booth T., Rohland N., Mallick S., Szécsényi-Nagy A., Mittnik A. et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018; 555:190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haak W., Balanovsky O., Sanchez J.J., Koshel S., Zaporozhchenko V., Adler C.J., Der Sarkissian C.S.I., Brandt G., Schwarz C., Nicklisch N. et al. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 2010; 8:e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chyleński M., Juras A., Ehler E., Malmström H., Piontek J., Jakobsson M., Marciniak A., Dabert M.. Late Danubian mitochondrial genomes shed light into the Neolithisation of Central Europe in the 5th millennium BC. BMC Evol. Biol. 2017; 17:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skoglund P., Malmstrom H., Raghavan M., Stora J., Hall P., Willerslev E., Gilbert M.T.P., Gotherstrom A., Jakobsson M.. Origins and genetic legacy of neolithic farmers and Hunter-Gatherers in Europe. Science. 2012; 336:466–469. [DOI] [PubMed] [Google Scholar]
- 17. Skoglund P., Malmstrom H., Omrak A., Raghavan M., Valdiosera C., Gunther T., Hall P., Tambets K., Parik J., Sjogren K.-G. et al. Genomic diversity and admixture differs for Stone-Age scandinavian foragers and farmers. Science. 2014; 344:747–750. [DOI] [PubMed] [Google Scholar]
- 18. Bollongino R., Nehlich O., Richards M.P., Orschiedt J., Thomas M.G., Sell C., Fajkošová Z., Powell A., Burger J.. 2000 years of parallel societies in stone age central europe. Science. 2013; 342:479–481. [DOI] [PubMed] [Google Scholar]
- 19. Gamba C., Jones E.R., Teasdale M.D., McLaughlin R.L., Gonzalez-Fortes G., Mattiangeli V., Domboróczki L., Kővári I., Pap I., Anders A. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 2014; 5:5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G., Castellano S., Lipson M. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014; 513:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lipson M., Szécsényi-Nagy A., Mallick S., Pósa A., Stégmár B., Keerl V., Rohland N., Stewardson K., Ferry M., Michel M. et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 2017; 551:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saag L., Varul L., Scheib C.L., Stenderup J., Allentoft M.E., Saag L., Pagani L., Reidla M., Tambets K., Metspalu E. et al. Extensive farming in estonia started through a Sex-Biased migration from the steppe. Curr. Biol. 2017; 27:2185–2193. [DOI] [PubMed] [Google Scholar]
- 23. Mittnik A., Wang C.-C., Pfrengle S., Daubaras M., Zariņa G., Hallgren F., Allmäe R., Khartanovich V., Moiseyev V., Tõrv M. et al. The genetic prehistory of the Baltic Sea region. Nat. Commun. 2018; 9:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allentoft M.E., Sikora M., Sjögren K.-G., Rasmussen S., Rasmussen M., Stenderup J., Damgaard P.B., Schroeder H., Ahlström T., Vinner L. et al. Population genomics of Bronze Age Eurasia. Nature. 2015; 522:167–172. [DOI] [PubMed] [Google Scholar]
- 25. Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015; 522:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Barros Damgaard P., Marchi N., Rasmussen S., Peyrot M., Renaud G., Korneliussen T., Moreno-Mayar J.V., Pedersen M.W., Goldberg A., Usmanova E. et al. 137 ancient human genomes from across the Eurasian steppes. Nature. 2018; 557:369–374. [DOI] [PubMed] [Google Scholar]
- 27. Lee E.J., Renneberg R., Harder M., Krause-Kyora B., Rinne C., Mueller J., Nebel A., von Wurmb-Schwark N.. Collective burials among agro-pastoral societies in later Neolithic Germany: perspectives from ancient DNA. J. Archaeol. Sci. 2014; 51:174–180. [Google Scholar]
- 28. Juras A., Chyleński M., Krenz-Niedbała M., Malmström H., Ehler E., Pospieszny Ł., Łukasik S., Bednarczyk J., Piontek J., Jakobsson M. et al. Investigating kinship of Neolithic post-LBK human remains from Krusza Zamkowa, Poland using ancient DNA. Forensic Sci. Int. Genet. 2017; 26:30–39. [DOI] [PubMed] [Google Scholar]
- 29. Haak W., Brandt G., de Jong H.N., Meyer C., Ganslmeier R., Heyd V., Hawkesworth C., Pike A.W.G., Meller H., Alt K.W.. Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18226–18231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Naumann E., Krzewińska M., Götherström A., Eriksson G.. Slaves as burial gifts in Viking Age Norway? Evidence from stable isotope and ancient DNA analyses. J. Archaeol. Sci. 2014; 41:533–540. [Google Scholar]
- 31. Malmström H., Vretemark M., Tillmar A., Durling M.B., Skoglund P., Gilbert M.T.P., Willerslev E., Holmlund G., Götherström A.. Finding the founder of Stockholm – A kinship study based on Y-chromosomal, autosomal and mitochondrial DNA. Ann. Anat. - Anat. Anzeiger. 2012; 194:138–145. [DOI] [PubMed] [Google Scholar]
- 32. Parson W., Dür A.. EMPOP–a forensic mtDNA database. Forensic Sci. Int. Genet. 2007; 1:88–92. [DOI] [PubMed] [Google Scholar]
- 33. Lott M.T., Leipzig J.N., Derbeneva O., Michael Xie H., Chalkia D., Sarmady M., Procaccio V., Wallace D.C.. MtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 2013; 44:1.23.1–1.23.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clima R., Preste R., Calabrese C., Diroma M.A., Santorsola M., Scioscia G., Simone D., Shen L., Gasparre G., Attimonelli M.. HmtDB 2016: data update, a better performing query system and human mitochondrial DNA haplogroup predictor. Nucleic Acids Res. 2017; 45:D698–D706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ingman M., Gyllensten U.. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006; 34:D749–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N.. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999; 23:147–147. [DOI] [PubMed] [Google Scholar]
- 37. Behar D.M, van Oven M., Rosset S., Metspalu M., Loogväli E.-L., Silva N.M., Kivisild T., Torroni A., Villems R.. A ‘Copernican’ reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012; 90:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fregel R., Delgado S.. HaploSearch: A tool for haplotype-sequence two-way transformation. Mitochondrion. 2011; 11:366–367. [DOI] [PubMed] [Google Scholar]
- 39. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meyer M., Kircher M.. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010; 2010:pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 42. Korneliussen T.S., Albrechtsen A., Nielsen R.. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014; 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Ancient human mitochondrial genomes database can be found at https://amtdb.org.