Abstract
Epigenome-Wide Association Study (EWAS) has become increasingly significant in identifying the associations between epigenetic variations and different biological traits. In this study, we develop EWAS Atlas (http://bigd.big.ac.cn/ewas), a curated knowledgebase of EWAS that provides a comprehensive collection of EWAS knowledge. Unlike extant data-oriented epigenetic resources, EWAS Atlas features manual curation of EWAS knowledge from extensive publications. In the current implementation, EWAS Atlas focuses on DNA methylation—one of the key epigenetic marks; it integrates a large number of 329 172 high-quality EWAS associations, involving 112 tissues/cell lines and covering 305 traits, 1830 cohorts and 390 ontology entities, which are completely based on manual curation from 649 studies reported in 401 publications. In addition, it is equipped with a powerful trait enrichment analysis tool, which is capable of profiling trait-trait and trait-epigenome relationships. Future developments include regular curation of recent EWAS publications, incorporation of more epigenetic marks and possible integration of EWAS with GWAS. Collectively, EWAS Atlas is dedicated to the curation, integration and standardization of EWAS knowledge and has the great potential to help researchers dissect molecular mechanisms of epigenetic modifications associated with biological traits.
INTRODUCTION
Epigenome-Wide Association Study (EWAS) has become a powerful approach to identify epigenetic variations associated with biological traits (1,2). With the rapid advancement of high-throughput sequencing technologies, epigenetic variations—especially variations in DNA methylation, have been extensively reported to be associated with a wide range of traits in human, including not only phenotypes and diseases, but also behaviors and environmental exposures. Specifically, phenotypic traits, such as aging and Body Mass Index (BMI), have been found to be strongly associated with DNA methylation (3,4). Environmental exposures and behaviors, such as air pollution and smoking, can affect global as well as gene-specific DNA methylation (5,6). Furthermore, some disease-related epigenetic signatures have been used as early diagnostic and/or prognostic biomarkers (7–9). As a growing body of EWAS evidence supports the use of epigenetic modifications as biomarkers for human healthcare and disease treatment, it is thus highly needed to have a comprehensive collection of EWAS knowledge by integrating associations of epigenetic variations with diverse biological traits.
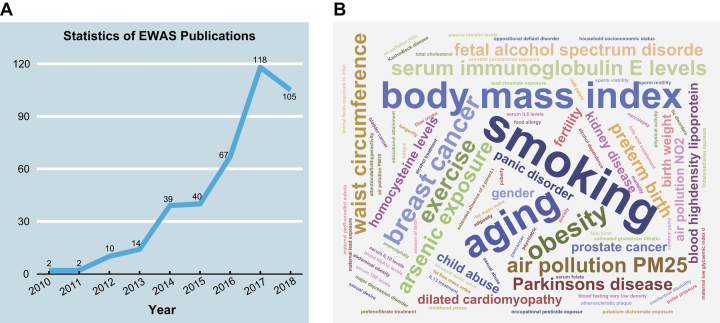
Toward this end, valuable efforts have been made in developing epigenetic databases that integrate different types of epigenetic data. However, none of them is specialized for integrating EWAS knowledge as well as associated information. For example, DiseaseMeth and MethHC collect annotations of aberrant DNA methylation only in human diseases, ignoring associations with other important phenotypes, behaviors and environmental exposures (10–12). MethBank and International Human Epigenome Consortium (IHEC) only incorporate epigenome data, without collecting associations of epigenetic data with traits (13–15). GWAS Catalog, a popular resource of genetic research, focuses only on published Genome-Wide Association Study (GWAS) data (16,17). Moreover, to our knowledge (as of 20 September 2018), there are two unpublished database resources, namely, EWAS Catalog (http://www.ewascatalog.org/) and EWASdb (http://www.bioapp.org/ewasdb/), which are devoted to the integration of EWAS associations. However, the former seems still under construction (as accessed on 20 September 2018) and the latter provides summary-level EWAS information for different traits or genes. Although more and more studies have shown the significant potential of epigenetic modifications in precision medicine, there still lacks a specialized resource for systematically integrating EWAS associations, especially considering the ever-increasing number of EWAS publications (Figure 1).
Figure 1.
(A) Statistics of EWAS publications. (B) Word cloud of traits.
Here we present EWAS Atlas (http://bigd.big.ac.cn/ewas/), a curated knowledgebase of EWAS. As one of core resources in the BIG Data Center (18–20), EWAS Atlas is devoted to providing a comprehensive collection of high-quality EWAS associations in support of systematic investigations of complex molecular mechanisms associated with different biological traits. Unlike extant data-oriented epigenetic resources, EWAS Atlas features manual curation of EWAS knowledge from extensive publications and accordingly incorporates a large number of high-quality EWAS data and a diversity of traits and ontology entities. EWAS Atlas provides open access to all curated data and thus would serve as a valuable resource for the global research community.
IMPLEMENTATION
EWAS Atlas is built with Spring boot (http://spring.io/), a mature and convention-over-configuration Model-View-Controller (MVC) framework, deployed in a Centos Linux 6.4 environment. In the back-end part, EWAS data is stored in MySQL (https://www.mysql.com/), a free and popular relational database management system. The web pages are constructed using HTML5 and rendered using Thymeleaf (https://www.thymeleaf.org/). Front-end interfaces are built using Bootstrap (https://getbootstrap.com/) with JQuery (https://jquery.com/) to provide responsive and user-friendly web pages. Furthermore, ECharts (http://echarts.baidu.com/) is used to provide interactive charting and data visualization. The trait enrichment tool is implemented by Python 3.5 (https://www.python.org/).
DATA CURATION AND DATABASE CONTENTS
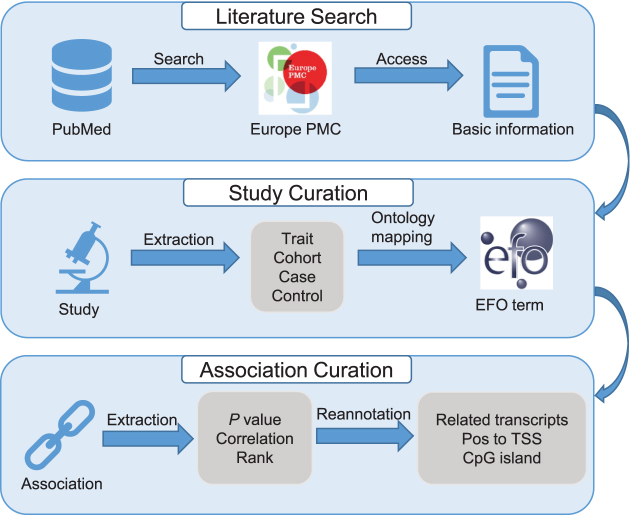
To provide high-quality information curated from EWAS publications, we set up a standardized curation process involving three major steps, viz., literature search, study curation and association curation (Figure 2). First, we perform literature search in PubMed using pre-defined keywords. Publications are eligible for inclusion in EWAS Atlas only if they contain necessary description on involved traits and significant EWAS associations. Consequently, a total of 401 publications are qualified and their other basic information (e.g. abstract, citation) are obtained automatically through Europe PMC API (https://europepmc.org/developers/) (21). The second is study curation, viz., manual curation of detailed study information from publications, including trait name(s), brief description of case and control groups, clinical and pathological characteristics of study populations. To unify the representation of biological traits, entities are mapped to the standardized terms in Experimental Factor Ontology (EFO) (22), which combines parts of several biological ontologies, such as Disease Ontology (DO) and Gene Ontology (GO). Finally, we conduct association curation to manually collect information of eligible associations (that should have P-value < 1.0E–4 or adjusted P-value < 0.05), including correlations between DNA methylation levels and experimental variables as well as their ranks in specific studies. Furthermore, considering various annotation systems adopted by different array platforms, all probes in 27K, 450K and 850K are re-annotated based on GENCODE release 28 (GRCh37) to maintain their consistency (23). The data curation in EWAS Atlas can be achieved by multiple curators through user-friendly web interfaces, enabling collaborative curation and enhancing the efficiency of the curation process.
Figure 2.
The curation model adopted by EWAS Atlas. It is noted that eligible associations should have P-value < 1.0E–4 or adjusted P-value < 0.05.
Based on the standardized curation process, EWAS Atlas integrates a large collection of 329 172 high-quality associations manually curated from 649 studies reported in 401 publications, involving 112 tissues/cell lines and covering 305 traits, 1830 cohorts and 390 ontology terms. Consequently, it is interestingly found according to the current collection in EWAS Atlas that the most extensively studied trait is smoking, involving 49 studies and 26 996 associations (http://bigd.big.ac.cn/ewas/browse?traitList=smoking) and that PTPRN2, which encodes receptor-type tyrosine-protein phosphatase, is associated with >90 different traits (http://bigd.big.ac.cn/ewas/browse?gene=PTPRN2). To facilitate users in browsing these data, EWAS Atlas provides five panels, where data are organized and presented in terms of trait, probe, gene, study and publication, respectively.
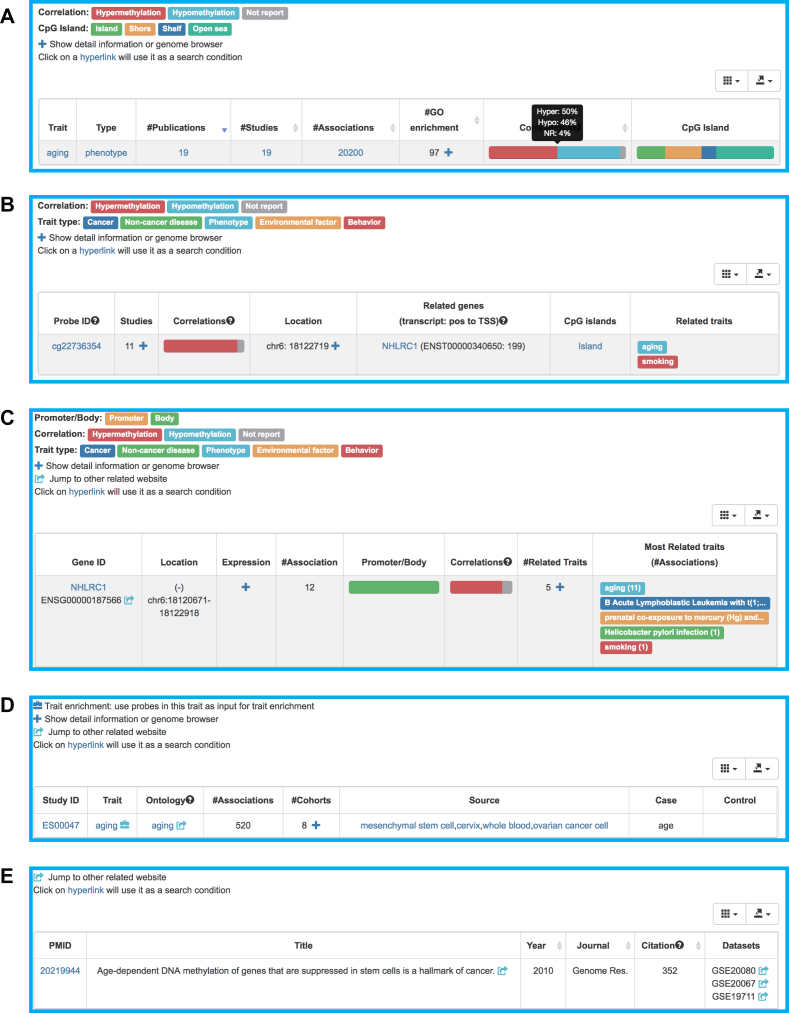
The trait panel provides an overview of all collected traits documented in EWAS Atlas. For each trait, both general details (name and mapped EFO terms) and summary data related to this trait are listed in a table form (Figure 3A). Specially, the percentages of biological different DNA methylation signatures (hyper/hypo) and CpG island relations (island, shore, shelf, open sea) are displayed in a visualized way. One trait may have multiple related probes that associate with different genes. To reveal biological processes for a specific trait, all relevant genes are collected for GO enrichment analysis. Thus, EWAS Atlas also provides a list of significantly enriched GO terms. The probe panel provides not only basic probe annotations (e.g. genomic coordinate, related transcripts, CpG island relation), but also summary-level descriptions of association data (e.g. occurrence frequency of a specific probe, percentages of hypo-/hyper-methylation). For each probe, details of individual associations, including study IDs, correlations, effect sizes and associated traits, are presented in a tab-separated table (Figure 3B). The gene panel contains not only general information (e.g. gene ID, genomic coordinate, tissue expression) but also summary-level EWAS association information (e.g., percentages of associations on promoter or gene body, related traits, number of associations). The study panel displays an overview of all EWAS studies, involving reported traits as well as brief descriptions of case and control groups. More importantly, an abundant collection of clinical and pathological characteristics of study populations, consisting of sample size, age, sex ratio and ancestry, are also curated from literature and integrated into EWAS Atlas (Figure 3C). Additionally, publication with its bibliographic details (title, year, journal, PubMed ID and citation) are collectively summarized in the publication panel (Figure 3D). In all four panels, hyperlinks to external databases, such as Gene Expression Omnibus (GEO) (24), European Nucleotide Archive (ENA) (25), PubMed, are provided to offer convenient access to additional information.
Figure 3.
Screenshot of web pages for (A) Trait, (B) Probe, (C) Gene, (D) Study and (E) Publication.
Powered by a large number of curated EWAS knowledge, EWAS Atlas provides an online tool for trait enrichment analysis (TEA). It allows users to submit customized probe(s)/trait as input and then explore trait-trait and trait-epigenome relationships. In the current version, the weighted Fisher's exact test is used to calculate the co-occurrence probability between input DNA methylation probes and trait-related DNA methylation probes, where the weight of each probe is the number of studies that reported this probe-trait association and equals 1 if absent. Based on this TEA tool, users can obtain TEA results, including significantly enriched traits and ontology terms. For example, when inputting reported DNA methylation sites related with cardiovascular risk (data originally from (26)), the TEA tool clearly shows that the most related trait is smoking; this result is well consistent with previous findings that smoking is one of the major cardiovascular risk factors, demonstrating its great potential utility in profiling the relationships among diverse traits (27,28).
To browse and query EWAS data in an efficient and friendly manner, EWAS Atlas is equipped with multiple filters to enable users to easily find traits, probes, associations, studies and publications of interest. Specifically, these filters cover a wide range of data items, including trait, gene symbol, probe ID, ontology, genomic coordinate, study ID, PMID, etc. As a consequence, it helps users to efficiently narrow down the query results. In addition, EWAS Atlas provides auto-suggestion functionality, which is able to provide a list of candidate terms according to users’ inputs. To fully benefit the global scientific community, all relevant data in EWAS Atlas are open access and publicly available at http://bigd.big.ac.cn/ewas/downloads. To ease data downloading, all query results that are displayed in web pages can be exported as a tab-delimited file. Furthermore, in order to enable programmatic access to EWAS data, a series of RESTful APIs (http://bigd.big.ac.cn/ewas/api) are implemented for automatic data retrieval, simply by specifying different types of identifiers (e.g. study ID, gene symbol, PMID, probe ID).
DISCUSSION AND FUTURE DEVELOPMENTS
EWAS Atlas, to our knowledge, is the first knowledgebase integrating a comprehensive collection of epigenome variations associated with phenotypes, diseases, behaviors and environmental exposures. It features manual curation of EWAS knowledge from extensive publications and accordingly incorporates a large number of high-quality EWAS data and a diversity of traits and ontology entities. Considering the great potential of epigenetic modifications in precision medicine, EWAS Atlas would be of great utility in dissecting complex molecular mechanisms associated with various diseases and promoting the development of novel diagnostics and therapeutics. With the rapid increase of EWAS studies, future efforts include regular update to incorporate EWAS associations from latest published literatures. Meanwhile, different types of epigenetic marks (e.g. RNA modification (29), histone modification) will be expanded and included in EWAS Atlas. Moreover, efforts will be also devoted to linking EWAS associations with GWAS associations. We also call for worldwide collaborations to work together to build EWAS Atlas into a valuable resource covering more comprehensive associations and traits and further providing potential guidance for human healthcare and disease treatment.
ACKNOWLEDGEMENTS
We thank a number of users for reporting bugs and providing suggestions as well as the anonymous reviewers for their valuable comments on this work.
FUNDING
The Strategic Priority Research Program of the Chinese Academy of Sciences [XDA19050302, XDB13040500]; National Key Research Program of China [2017YFC0907502]; National Programs for High Technology Research and Development (863 Program) [2015AA020108, 2012AA020409]; National Natural Science Foundation of China [31200978]; 13th Five-year Informatization Plan of Chinese Academy of Sciences [XXH13505-05]; International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008]; ‘100-Talent Program’ of Chinese Academy of Sciences. Funding for open access charge: The Strategic Priority Research Program of the Chinese Academy of Sciences [XDA19050302, XDB13040500].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rakyan V.K., Down T.A., Balding D.J., Beck S.. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011; 12:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lappalainen T., Greally J.M.. Associating cellular epigenetic models with human phenotypes. Nat. Rev. Genet. 2017; 18:441–451. [DOI] [PubMed] [Google Scholar]
- 3. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dick K.J., Nelson C.P., Tsaprouni L., Sandling J.K., Aissi D., Wahl S., Meduri E., Morange P.E., Gagnon F., Grallert H. et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014; 383:1990–1998. [DOI] [PubMed] [Google Scholar]
- 5. Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M.. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet. 2013; 22:843–851. [DOI] [PubMed] [Google Scholar]
- 6. Panni T., Mehta A.J., Schwartz J.D., Baccarelli A.A., Just A.C., Wolf K., Wahl S., Cyrys J., Kunze S., Strauch K. et al. Genome-Wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ. Health Perspect. 2016; 124:983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Story Jovanova O., Nedeljkovic I., Derek S., Walker R.M., Liu C., Luciano M., Bressler J., Brody J., Drake A.J., Evans K.L. et al. DNA methylation signatures of depressive symptoms in Middle-aged and elderly persons: meta-analysis of multiethnic Epigenome-wide studies. JAMA Psychiatry. 2018; 75:949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hannon E., Schendel D., Ladd-Acosta C., Grove J., i P.-B.A.S.D.G., Hansen C.S., Andrews S.V., Hougaard D.M., Bresnahan M., Mors O. et al. Elevated polygenic burden for autism is associated with differential DNA methylation at birth. Genome Med. 2018; 10:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lv J., Chen K.. Broad H3K4me3 as a novel epigenetic signature for normal development and disease. Genomics Proteomics Bioinformatics. 2016; 14:262–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lv J., Liu H., Su J., Wu X., Liu H., Li B., Xiao X., Wang F., Wu Q., Zhang Y.. DiseaseMeth: a human disease methylation database. Nucleic Acids Res. 2012; 40:D1030–D1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiong Y., Wei Y., Gu Y., Zhang S., Lyu J., Zhang B., Chen C., Zhu J., Wang Y., Liu H. et al. DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017; 45:D888–D895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang W.Y., Hsu S.D., Huang H.Y., Sun Y.M., Chou C.H., Weng S.L., Huang H.D.. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015; 43:D856–D861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zou D., Sun S., Li R., Liu J., Zhang J., Zhang Z.. MethBank: a database integrating next-generation sequencing single-base-resolution DNA methylation programming data. Nucleic Acids Res. 2015; 43:D54–D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li R., Liang F., Li M., Zou D., Sun S., Zhao Y., Zhao W., Bao Y., Xiao J., Zhang Z.. MethBank 3.0: a database of DNA methylomes across a variety of species. Nucleic Acids Res. 2018; 46:D288–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stunnenberg H.G., Epigenome International Human, Hirst M.. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell. 2016; 167:1897. [DOI] [PubMed] [Google Scholar]
- 16. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014; 42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. BIG Data Center Members Database Resources of the BIG Data Center in 2018. Nucleic Acids Res. 2018; 46:D14–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Data Center Members BIG. The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017; 45:D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo J. GSA and BIGD: Filling the gap of bioinformatics resource and service in china. Genomics Proteomics Bioinformatics. 2017; 15:11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Europe, P.M.C.C. Europe PMC: a full-text literature database for the life sciences and platform for innovation. Nucleic Acids Res. 2015; 43:D1042–D1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malone J., Holloway E., Adamusiak T., Kapushesky M., Zheng J., Kolesnikov N., Zhukova A., Brazma A., Parkinson H.. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010; 26:1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silvester N., Alako B., Amid C., Cerdeno-Tarraga A., Cleland I., Gibson R., Goodgame N., Ten Hoopen P., Kay S., Leinonen R. et al. Content discovery and retrieval services at the European Nucleotide Archive. Nucleic Acids Res. 2015; 43:D23–D29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernandez-Sanles A., Sayols-Baixeras S., Curcio S., Subirana I., Marrugat J., Elosua R.. DNA methylation and Age-Independent cardiovascular risk, an Epigenome-Wide approach: the REGICOR study (REgistre GIroni del COR). Arterioscler. Thromb. Vasc. Biol. 2018; 38:645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ambrose J.A., Barua R.S.. The pathophysiology of cigarette smoking and cardiovascular disease: an update. J. Am. Coll. Cardiol. 2004; 43:1731–1737. [DOI] [PubMed] [Google Scholar]
- 28. Bazzano L.A., He J., Muntner P., Vupputuri S., Whelton P.K.. Relationship between cigarette smoking and novel risk factors for cardiovascular disease in the United States. Ann. Intern. Med. 2003; 138:891–897. [DOI] [PubMed] [Google Scholar]
- 29. Xiong X., Yi C., Peng J.. Epitranscriptomics: toward a better understanding of RNA modifications. Genomics Proteomics Bioinformatics. 2017; 15:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]