Abstract
The Genome Database for Rosaceae (GDR, https://www.rosaceae.org) is an integrated web-based community database resource providing access to publicly available genomics, genetics and breeding data and data-mining tools to facilitate basic, translational and applied research in Rosaceae. The volume of data in GDR has increased greatly over the last 5 years. The GDR now houses multiple versions of whole genome assembly and annotation data from 14 species, made available by recent advances in sequencing technology. Annotated and searchable reference transcriptomes, RefTrans, combining peer-reviewed published RNA-Seq as well as EST datasets, are newly available for major crop species. Significantly more quantitative trait loci, genetic maps and markers are available in MapViewer, a new visualization tool that better integrates with other pages in GDR. Pathways can be accessed through the new GDR Cyc Pathways databases, and synteny among the newest genome assemblies from eight species can be viewed through the new synteny browser, SynView. Collated single-nucleotide polymorphism diversity data and phenotypic data from publicly available breeding datasets are integrated with other relevant data. Also, the new Breeding Information Management System allows breeders to upload, manage and analyze their private breeding data within the secure GDR server with an option to release data publicly.
INTRODUCTION
The Genome Database for Rosaceae (GDR, https://www.rosaceae.org) is the central repository and data-mining resource for genomics, genetics and breeding data of Rosaceae, an economically and nutritionally important crop family that includes almond, apple, apricot, blackberry, cherry, peach, pear, plum, raspberry, rose and strawberry. Composed of species with a wide variety of form, habit, function and ploidy, the Rosaceae family also provides a useful system for studying plant biology (1).
First established in 2003, GDR initially provided web interfaces and analysis tools for emerging genomic and genetic data such as genus-specific EST unigene sets, linkage maps and genetic markers (2,3). With the availability of whole genome sequence data and large-scale phenotypic and genotypic data, GDR, as reported in our year 10 update, markedly expanded with data and functionality (4). In the past 5 years we have witnessed another big change in the volume and type of data generated by the Rosaceae research community (Table 1). These data include multiple versions of whole genome assemblies and annotations for each major crop species, increased RNA-Seq data, multiple single-nucleotide polymorphism (SNP) arrays for major crops, increased publications on quantitative trait loci (QTLs) and genetic maps, especially using SNPs and more breeding programs and projects that utilize SNP genotyping. We have performed new types of analyses, developed the Rosaceae Trait Ontology that is tightly linked to the Plant Trait Ontology (TO) (5) and focused on integration of data across databases, organisms and data types. The value-added efforts undertaken in data analyses, curation and integration were combined with development and enhancement of new and existing search interfaces and tools to enable more efficient sharing and reuse of the pivotal data generated by the community. This report describes the GDR with a focus on new data and functionalities accumulated and developed in the past 5 years.
Table 1.
Comparison of number of GDR entries between 2008, 2013 and 2018 by data type
| Number of entries by year | ||||
|---|---|---|---|---|
| Data type | 2008 | 2013 | 2018 | Details |
| Genome | 0 | 5 | 21 | Multiple versions of whole genome assemblies and annotations from seven major crops and 14 species |
| Gene and mRNA | 0 | 236191 genes | 528890 genes and 585574 mRNAs | Genes and mRNAs from whole genome assemblies and parsed from NCBI nucleotide sequences |
| Transcript | 90337 | 200467 | 1065226 RefTrans and 1412519 Unigenes | RefTrans for six genus and unigene V5 for six genus and the rosaceae family |
| Marker | 1700 | 2229311 | 3285775 | Including 127877 for Fragaria, 2613842 for Malus, 470747 for Prunus, 1593 for Pyrus, 71716 for Rosa and 8743 for Rubus |
| Genetic Map | 37 | 84 | 313 | Including 27 for Fragaria, 108 for Malus, 168 for Prunus, 9 for Pyrus, 15 for Rosa and 13 for Rubus |
| QTL | 0 | 1195 | 3799 | QTL and MTL associated with 392 agronomic traits |
| MTL | 27 | 52 | 103 | |
| Species | <100 | 516 | 1967 | Data available for 1967 species, specific species pages with hyperlinks to various data and tools for 13 major species |
| Germplasm | 0 | 8613 | 14411 | Including 1799 for Fragaria, 4635 for Malus, 7367 for Prunus, 465 for Pyrus, 145 for Rosa, and 42 for Rubus |
| Phenotype data | 0 | 578 568 | 878426 | 878426 measurements include 389191 publicly available data and the rest available for the RosBreed project team members |
| Genotype data | 0 | 28 296 | 10806569 | SNP genotype data from 10787946 measurements using 12229 markers for 6 species, SSR genotype data from 18623 measurements using 5044 markers for 10 species |
| Publication | 2447 | 5182 | 7449 | 7499 publications on genomic, genetic and breeding research in Rosaceae |
Data and web interface
The GDR interface has been re-designed to provide easier access points to data and tools such as the Major Genera Quick Start and Tools Quick Start featured on the homepage. The Major Genera Quick Start allows users to view which types of data are available for a genus of interest and provides links to access these data. Similarly, Species pages under the species navigation menu provide the same information for major species. The Tools Quick Start is organized into genomics, genetics and breeding sections; each section provides links to appropriate pages to access available data and tools. New features that can quickly familiarize users to GDR data and functionality include the dynamic data overview page where users can browse the current data types and numbers in GDR and short video tutorials. Tutorials are available for site overview, species pages, Breeding Information Management System (BIMS) and all of the search pages. GDR is implemented in Tripal (https://tripal.info), an open-source, resource efficient database platform (6,7). Below, we describe the currently available data and interfaces, with a focus on new features.
Genomics data
Whole genome sequence and associated data
Currently available in GDR are multiple versions of whole genome assemblies and annotations from seven major crops and 14 species: Fragaria vesca, Fragaria × ananassa, Fragaria iinumae, Fragaria nipponica, Fragaria nubicola, Fragaria orientalis, Malus × domestica, Prunus avium, Prunus persica, Potentilla micrantha, Pyrus communis, Rosa multiflora, Rosa chinensis and Rubus occidentalis. To standardize the names of genome assemblies and annotations, GDR uses and recommends a naming protocol: [Genus] [species] genome v[assembly version].a[annotation-version].
Twenty one genome assemblies are now available in the GDR. Four strawberry genome assemblies, including the newest v4.0 (8), are available for the diploid woodland strawberry, F. vesca, which serves as the reference genome for the Fragaria genus, in addition to two genome assemblies for the octoploid cultivated strawberry Fragaria × ananassa and four wild diploid Fragaria species (9). For both the F. vesca genome v1.1 (10) and v2.0 (11), additional annotations are available: v1.1.a2 (12) and v2.0.a2 (13), respectively. The genes from v1.1.a2 have also been aligned to the v2.0 genome, so all three gene annotations are available. The draft genome of P. micrantha (14), a species that does not develop fleshy fruit but is closely related to Fragaria, is also available. For Malus × domestica, a de novo assembly of a ‘Golden Delicious’ doubled-haploid apple tree (GDDH13) with 17 pseudomolecules is now available (15) along with three versions of assemblies for the heterozygous apple genome (16). For Prunus, in addition to two assemblies of peach, P. persica genome v1.0 (17) and v2.0 (18), the whole genome assembly of sweet cherry P. avium genome v1.0 (19) is available. For Pyrus, the draft genome of the European pear P. communis genome v1.0 (20) is available. For Rosa, two new assemblies from a double haploid R. chinensis plant (21,22) are available as well as a R. multiflora draft genome v1.0 (23). For Rubus, the chromosome-scale whole genome assembly of black raspberry R. occidentalis genome v1.1 (24) as well as the draft genome v1.0 (25) are available.
Additional data provided by the GDR team on these assemblies include computational annotation of predicted genes with homology to genes of closely related or model plant species and assignment of InterPro protein domains (26) and GO terms (27,28). The GDR team also performs synteny analysis to find conserved syntenic regions among the newest versions of all publicly available Rosaceae genomes using MCScanX (29). Some of the Rosaceae whole genome assembly data are available in NCBI with NCBI’s own gene annotation and naming convention. To help researchers compare two different gene annotation sets, GDR performs BLAT analysis between NCBI annotated genes and genes from the original genome assemblies.
Assemblies annotated as above and GDR functional and synteny annotations can be accessed in their respective genome pages, gene search pages, BLAST server and using graphical viewers such as JBrowse (30) and the Tripal Synteny Viewer (https://github.com/tripal/tripal_synview). Each species page provides a summary for the species along with a resources sidebar with hyperlinks to various data and tools for the species, and has a genome subsection that lists all genome assemblies for the species. Individual genome pages provide downloadable files, including Generic File Format and FASTA formats, for an assembly that include annotated gene predictions, homology and positions of repeats and genetic markers including SNPs. Additionally, lists of annotated functional terms and Microsoft Excel files of protein homologs mapped via BLAST+ (31) are available for downloading. These files contain hyperlinks to external databases as well as to GDR pages including JBrowse and gene or marker detail pages when applicable. The Search Genes and Transcripts page allows users to search for specific genes and sequences in the above assemblies and transcriptome datasets. The new search interface allows users to conduct various searches, by genus, species, dataset, gene/transcript name, genomic location and association with computationally inferred functionality such as protein names, GO terms, InterPro domains and KEGG pathway terms, all in one page. This search interface allows users to perform a query such as ‘Return all genes annotated with the word ‘resistance’ between 1.0 and 3.5 Mb on chromosome Pp02 of the peach genome v2.0’. Using the search site, users can download the results or proceed to the gene details page within the GDR. Recently added functionality in the Search Genes and Transcripts page includes ‘customized output’. This option allows users to customize the result table and the downloadable Excel file to include various functional annotation results. The gene details page has several links in the resources sidebar to display the sequence and its motif annotations, genome alignments and homologies to sequences of other species in the GDR and other databases. The alignment details provide links to view a gene in JBrowse. The new synteny section lists all the orthologs and paralogs in other genomes discovered by synteny analyses and provides hyperlinks to the gene pages and Synteny Viewer. Via the Synteny Viewer page (Figure 1A), accessible from the tools navigation menu, users can choose a scaffold/chromosome of one reference genome and choose multiple other genomes for comparison. The viewer then displays a clickable circular image, as well as a table, showing all the syntenic blocks between them (Figure 1B). Choosing one block either from the image or the table leads to a page where all the syntenic genes within the block are shown with the Expect value (E-value) of their homology (Figure 1C). Gene names are linked to a specific gene page (Figure 1D) where orthologs/paralogs in other genomes are available among other information on the genes such as associated function and genomic location with a link to JBrowse (Figure 1E). Conserved synteny data made available in the GDR thus allows users starting with one rosaceous genome to explore genes, anchored trait loci and genetic markers within orthologous regions of another rosaceous genome. Using JBrowse, users can view all the genomic features aligned to the genome, such as gene models, transcripts, repeats, SNPs, other genetic markers and genes from other model plant species. In addition, the multivariant viewer JBrowse plugin has been implemented to allow users to view SNPs present among sequence data from multiple germplasm individuals. For example, RosBREED resequencing data (23 accessions) aligned to the peach v1.0 genome is available in JBrowse through the plugin. The GDR has upgraded to the Tripal BLAST module (http://tripal.info/extensions/modules/tripal-blast-analysis), replacing the old BLAST and batch BLAST tools. The new BLAST enables results to link to the genome scaffolds in JBrowse and to the gene/transcript details in the GDR and on the NCBI. Predicted genes from whole genome sequences were used in the construction of PlantCyc (metabolic pathway) databases (32) using PathwayTools (33). Currently, four PlantCyc databases are available in the GDR: PeachCyc, AppleCyc, FragariaCyc and RubusCyc.
Figure 1.
Synteny Viewer in GDR. (A) Home page of Synteny Viewer allows users to choose a reference genome and a chromosome and multiple genomes for comparison. Users can also choose a synteny block ID. (B) A circular diagram and a table shows the synteny blocks between a chromosome of a reference genome and all chromosomes of another genome being compared. (C) A bar diagram and a table that shows all the genes in a syntenic block. The table displays E-value between the matching genes and the gene names have hyperlinks to the gene detail page. (D) A gene detail page with resource side bar and the hyperlink to JBrowse. (E) JBrowse around the mRNA of interest with tracks such as gene, mRNA, SNP and genes parsed from NCBI nucleotide database.
Transcriptome data
The GDR team now regularly performs an analysis that combines published RNA-Seq and dbEST datasets to create a reference transcriptome (RefTrans) for major species or genera and provides putative gene function identified by homology to known proteins. The RNA-Seq sequences from peer-reviewed publications were downloaded from the NCBI Short Read Archive (SRA) and subject to quality control using the Trimmomatic (v0.32, default parameters, 34) and custom Perl scripts. The remaining RNA-Seq reads were assembled de novo with Trinity (v2.6.6, 35) using default assembly parameters and a minimum coding length of 200 bases. Quality control of the ESTs included vector sequence screening (UniVec_Core,ftp://ftp.ncbi.nih.gov/pub/UniVec/) using cross_match (36), removal of tRNA/rRNA/snRNA sequences identified using tblastx (37) and Poly-A tail trimming. The filtered ESTs were assembled using the CAP3 program (P −90, 38). Bowtie (v 2.3.3) (39) was applied to multi-map the RNA-Seq reads and ESTs back to the assembled contigs and singlets. The contigs and singlets were clustered into genes using CH-HIT (v4.6.4, 40) and Corset (v1.0.7) (41) with default parameters. The longest isoform greater than 500 nt was selected to represent each Corset cluster and create the RefTrans sequences. The RefTran sequences are functionally characterized by pairwise comparison using the BLASTX algorithm against the Swiss-Prot and TrEMBL protein databases. Information on the top ten matches with an E-value of ≤ 1E-06 are recorded and stored in GDR together with the RefTrans sequences. InterPro domains and Gene Ontology assignments are made using InterProScan at the EBI through Blast2GO. Transcriptomes and their associated annotation are available to download in the transcriptome page that can be accessed from each species page, to search and download in the Search Genes and Transcripts page, to view on the genome in JBrowse and to perform similarity searches in the BLAST server. Currently available datasets include F. vesca GDR RefTrans v1.0, P. avium GDR RefTrans v1.0, P. persica GDR RefTrans v1.0, Malus × domestica GDR RefTrans v1.0 and Rubus GDR RefTrans v2.0. The GDR continues to provide unigene v5.0 for each genus (Prunus, Malus, Fragaria, Rosa and Pyrus), composed of singlets and contigs assembled from the publicly available Rosaceae ESTs downloaded from dbEST at NCBI. Unigene v5.0 for the entire Rosaceae is composed of the assembled contigs and singlets for the five genera.
GDR also hosts user-provided assembled transcripts such as a rose transcriptome, which are orthologous open reading frame sequences of roses used to construct the WagRhSNP 68K Axiom SNP array (42), black cherry Prunus serotina unigene v1.0 from Pennsylvania State University, and assembled transcript sequences of red raspberry, Rubus idaeus (43). In addition, the GDR provides links to RNA-Seq and DNA datasets available in the NCBI SRA for Rosaceae genus or major species.
Genetic maps, genetic diversity data, markers and trait loci
Genetic maps
With continuous effort on curating peer-reviewed, published data, GDR now contains 313 genetic maps for Rosaceae species, including 168 for Prunus, 108 for Malus, 27 for Fragaria, 15 for Rosa, 13 for Rubus and 9 for Pyrus. The data associated with genetic maps include mapped positions of molecular markers, QTLs and heritable phenotypic markers, as well as mapping population(s) and publication(s). GDR has a new graphic interface, MapViewer (http://tripal.info/extensions/modules/tripalmap) to view genetic maps. MapViewer allows users to view and compare maps from different populations and species, facilitating information transfer from well-studied species to less-studied ones. These comparisons are especially useful in Rosaceae due to well conserved synteny among the genomes of Rosaceae genera (1,44). While the functionality of MapViewer is similar to CMap (45), a commonly used tool in biological databases including the GDR, MapViewer is much more integrated with other pages, such as the map, marker and QTL pages. In addition, MapViewer allows users to zoom into specific regions of a linkage group, choose types of markers to be displayed and change the colors of the markers that are displayed.
Genetic diversity data
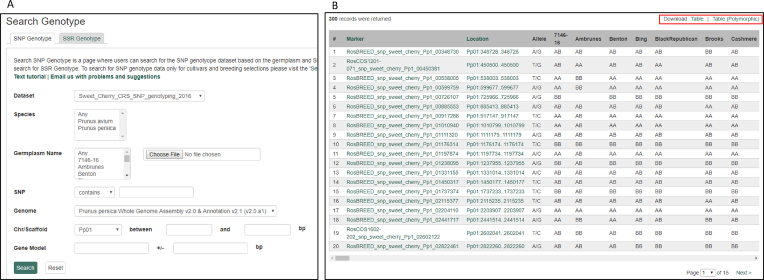
The GDR contains DNA polymorphism data from various published genetic diversity studies and public breeding projects such as RosBREED (46). Currently, data from 24 diversity projects are available: thirteen from Prunus, ten from Malus/Pyrus and one from Malacomeles (false serviceberry). The GDR provides separate search pages for the SNP and the SSR genotypic data to provide appropriate query and downloadable result tables depending on the data type. There are 19 and 5 datasets available for SSR and SNP genotype search pages, respectively. In the SNP genotype search page, users can filter results by dataset name, species, germplasm name, SNP name, genomic location and/or gene name (Figure 2A). Users can also upload a file with germplasm names. This filtering allows users to perform tailored querying, such as finding SNP polymorphisms around a gene of interest in a chosen set of germplasm. The results table provides SNP name, genomic location, allele and genotypic data of all of the germplasm chosen in the order of SNP location in the genome, so that users can view the genotype of each germplasm along the chromosome (Figure 2B). Users can download the genotypes for all markers displayed in the results page or the genotypes for only the markers that are polymorphic within the germplasm set chosen (Figure 2B).
Figure 2.
SNP Genotype Search Page in GDR. (A) Users can search SNP genotype data by dataset name, species, germplasm name, SNP name, genome location and/or gene name. Users can also upload a file with a list of germplasm names. (B) Search result table that shows SNP name, genome location, allele and the genotype data of all the germplasm chosen in the order of SNP location in the genome. The red square highlights the options to download the genotype for all the marker displayed in the result page or the genotype data that are polymorphic in the germplasm set chosen.
Genetic marker and SNP array data
The GDR provides details on more than 3 million genetic markers used in genetic map development, genetic diversity studies, genome wide association studies and SNP array development. Marker annotations include marker aliases, source germplasm, source description, primer sequences, polymerase chain reaction conditions, literature references and map position, where available. For SNPs, the marker details also include SNP array name, SNP array ID, dbSNP ID, alleles, flanking sequences and probes. SNP marker data available from GDR include those from array development projects such as the 9K (47), 20K (48) and 480K (49) arrays for apple, IRSC cherry 6K array (50), IPSC peach 9K array (51), 90K array for cultivated strawberry (52) and 68K array for rose (42). The SNP array data are available to download in Microsoft Excel format from the genome pages, to view in JBrowse as well as to query in the Marker Search page. The GDR provides a new ‘SNP Marker Search’ page in addition to the existing ‘Marker Search’ and ‘Search Nearby Markers’ pages. The search filter feature in the ‘Marker Search’ page includes marker name, marker type, the species from which the marker is developed, the species to which the marker is mapped, trait name and map position in the genetic map and genome. Filtering by trait name is a new feature that allows users to search for markers that are near and/or within QTLs using the associated trait name. The table in the results page shows marker name, alias, marker type, species, genetic map location and genome location. The downloaded file contains the same information, as well as the citation. The ‘SNP Marker Search’ is designed so that users can filter using array information as well as SNP name and genomic location. The results table is also specific for SNP, with alleles, SNP array information, genome location and flanking sequences. In both search pages, users can upload a file of marker names for querying. Another search interface, ‘Search Nearby Markers’, allows users to find markers near a targeted locus.
Trait locus data and Rosaceae Trait Ontology
The GDR contains detailed data on published trait loci: QTLs and MTLs (Mendelian Trait Loci). These data include 3799 QTLs and 103 MTLs that are associated with 392 horticultural traits such as powdery mildew disease resistance, fruit skin color, ripening time and volatile organic compound content. These trait terms constitute the Rosaceae Trait Ontology. The Rosaceae Trait Ontology has been built to standardize trait names and abbreviations for all trait data entered into the GDR and to connect trait terms to TO (5), with the goal of data integration across databases, organisms and data types. Each of these trait terms is either an existing TO term or a child term of one. One trait term can belong to multiple Root TO terms. New Rosaceae Trait Ontology terms, which do not exist in TO, have been submitted to the Trait Ontology consortium for inclusion. In addition to the Rosaceae Trait Ontology, QTLs are annotated in the GDR with aliases, curator-assigned QTL labels, published symbols, trait names, taxa, trait descriptions, screening methods, map positions, associated markers, statistical values, datasets, contact information and references. The search page for trait loci allows searching by trait locus type (QTL or MTL), species, trait category (root TO term), trait name, published symbol and GDR-assigned label.
Breeding data
Breeding data stored in the GDR include phenotypic, genotypic, germplasm and pedigree data from the RosBREED project (46), the Washington State University Apple Breeding Program (53) and private breeding programs. Publicly available breeding data can be queried using the ‘Search Trait Evaluation’ and ‘Search Genotype’ pages. The ‘Search Trait Evaluation’ page provides two tabs, for querying qualitative or quantitative traits. In each tab, users can filter the data by crop dataset name and trait cut-off values of up to three trait descriptors. The result table and downloadable file has germplasm name, species, the trait values chosen and the dataset name. The germplasm and the dataset name in the results table are linked to the detail page where other associated data can be accessed. The germplasm page has a resource sidebar for genotypic and phenotypic data, as well as an overview and associated images, where the data can be viewed and downloaded. In addition, the public breeding data can be queried and downloaded using the BIMS. BIMS is a new Tripal module (http://tripal.info/extensions/modules/tripal-bims) we developed to provide breeders and breeding project teams with tools to store, manage, archive and analyze their private or public breeding data. BIMS is compatible with Field Book (54), an open-source App for android phones and tablets, that allows researchers to replace hard-copy field books or spreadsheet files. BIMS allows breeders to upload data collected with the Field Book app directly to their private, secure database in the GDR into a consistent format. Having BIMS in the GDR gives breeders the opportunity to integrate their data with publicly available data and easily release some or all of their data publicly as needed. Currently, the sweet cherry phenotypic data from the RosBREED project is available for public access through BIMS. In BIMS, an accordion menu on the left side provides quick access to various functionalities. The ‘Data Import’ section provides data templates for users to enter their data and upload the data file themselves (Figure 3A). The ‘Search’ section allows users to search and save the list of germplasm individuals (‘accessions’) using any combination of properties and trait cut-off values: name, trial, location, cross, parent and trait values (Figure 3B). When a filter is applied to choose accessions, the rightmost section shows the number of accessions belonging to the filtered dataset. When a trait descriptor is chosen as a filter, the middle section shows a histogram along with the statistical values, such as maximum, minimum, mean and standard deviation, to show users the distribution of data points within the dataset chosen (Figure 3B). The list of the accession names chosen can be viewed and downloaded in a table with an option of adding more data on the accessions such as parents, cross number and trait values (Figure 3C). The list of accessions can be saved in user accounts and can be used to retrieve any data associated with the accessions. The ‘Data Analysis’ section allows users to choose two datasets, using the categories or saved accession lists, and compare the trait statistics between the two datasets (Figure 3D). This analysis function allows users to compare various traits of two sets of accessions such as progenies from two different crosses.
Figure 3.
BIMS in GDR. (A) ‘Template List’ subsection in ‘Data Import’ section provide downloadable templates for users to enter various breeding data. (B) Search’ section allows users to search and save the list of accessions using any combination of properties and trait cut of values: name, trial, location, cross, parent and trait values. The middle section shows the statistical information on the filtered dataset for the trait chosen and the right section shows the number of accessions filtered so far. (C) A page with the search result table. Users can add more columns in the table using ‘Column options’ and save/download the result table. (D) ‘Data Analysis’ section that allows users to choose two datasets, using the categories or saved accession lists and compare the trait statistics between the two datasets.
Community resources and data submission
The GDR continues to provide community-based resources under the community navigation menu, including pages for the US RosEXEC, RosIGI and conference, employment notices and mailing lists. The US RosEXEC (Rosaceae Genomics, Genetics and Breeding Executive Committee) and RosIGI (Rosaceae International Genomics Initiative) serve as communication and coordination focal points for the research community, with the former also operating as the Steering Committee for the GDR. The US RosEXEC and RosIGI pages provide official documents, minutes of quarterly meetings, membership lists and subcommittee information. Several mailing lists, in addition to the GDR mailing list, are available to serve the community with information for specific interests or purposes and the archives can be viewed through the message board sites.
While the GDR team actively curates data from publications, we encourage authors to officially archive their datasets by submitting their data at the time of manuscript submission. The Data Submission page under the Data navigation menu provides various data templates for genes, genetic maps, QTL maps, as well as genotypic and phenotypic data. In addition, the page has a link to a page where the recommended list of files for whole genome data submission are available.
CONCLUSION AND FUTURE DIRECTION
Recent availability of multiple genome assemblies and annotation data from seven major crops and 14 species in the Rosaceae family opened up the opportunity to investigate the evolution and biological basis of a wide range of plant forms and functions, as well as to share knowledge among major rosaceous crops to improve their performance. With the goal of facilitating these efforts, the GDR focused on integrating the wealth of the new genomic data with transcriptomic, genetic map, genetic marker, trait locus, phenotypic and genotypic data. To accommodate scientists’ data-mining needs that came with these new types and large volume of data, we made various new web interfaces available. The new web interfaces are either new Tripal modules that we developed, such as MapViewer, BIMS, Chado Loader, Chado Data Display and Chado Search modules (55) or Tripal modules that other database teams developed such as the Synteny Viewer and Tripal BLAST. The open-source database platform Tripal, allows us to meet emerging demands for storage, querying and display of new data types more efficiently and quickly.
During the last 5 years, we also made advances in curating data with more ontologies and incorporating ontologies in data query pages to facilitate data-sharing across different data types, species and databases. Future effort will include further developing MapViewer to integrate genome data and genetic maps, providing an enhanced querying interface, expanding analysis capabilities in BIMS through access to additional functionality, integrating Galaxy analysis platform through Tripal Galaxy module (https://www.drupal.org/project/tripal_galaxy), providing access to High Performance Computing and enabling cross-Tripal databases querying capability using Tripal Elasticsearch (56) and Tripal Exchange (http://tripal.info/tutorials/v3.x/web-services).
Use of the GDR, the community resource for Rosaceae genomics, genetics and breeding research, has continued to grow over the last 5 years. Between September 1, 2013 and August 31, 2018, the GDR had 256075 visits by 95259 unique visitors from 190 countries, who accessed 1547877 pages. The GDR is part of AgBioData (57) (https://www.agbiodata.org), a consortium working to improve standards and sustainability of genomics, genetics and breeding databases and further enable agricultural science.
ACKNOWLEDGEMENTS
The authors acknowledge with thanks their funding sources, the Rosaceae research community for providing data, support and feedback, the Tripal and GMOD community of developers for developing and sharing Tripal modules and code, the AgBioData Consortium and US Land Grant Universities for support.
FUNDING
USDA National Institute of Food and Agriculture Specialty Crop Research Initiative projects [2014-51181-2237, 2014-51181-22378]; USDA National Institute of Food and Agriculture National Research Support Project 10; NSF PGRP award #444573, NSF CIF21 DIBBs award #1443040; Washington Tree Fruit Research Commission, Washington State University; Clemson University. Funding for open access charge: Federal Grant; USDA NIFA SCRI Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shulaev V., Korban S.S., Sosinski B., Abbott A.G., Aldwinckle H.S., Folta K.M., Iezzoni A., Main D., Arus P., Dandekar A.M. et al. Multiple models for rosaceae genomics. Plant Physiol. 2008; 147:985–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jung S., Jesudurai C., Staton M., Du Z., Ficklin S., Cho I., Abbott A., Tomkins J., Main D.. GDR (Genome Database for Rosaceae): integrated web resources for Rosaceae genomics and genetics research. BMC Bioinformatics. 2004; 5:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung S., Staton M., Lee T., Blenda A., Svancara R., Abbott A., Main D.. GDR (Genome Database for Rosaceae): integrated web-database for Rosaceae genomics and genetics data. Nucleic Acids Res. 2008; 36:D1034–D1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jung S., Ficklin S.P., Lee T., Cheng C.-H., Blenda A., Zheng P., Yu J., Bombarely A., Cho I., Ru S. et al. The Genome Database for Rosaceae (GDR): year 10 update. Nucleic Acids Res. 2014; 42:D1237–D1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cooper L., Meier A., Laporte M.-A., Elser J.L., Mungall C., Sinn B.T., Cavaliere D., Carbon S., Dunn N.A., Smith B. et al. The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics. Nucleic Acids Res. 2018; 46:D1168–D1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanderson L.-A., Ficklin S.P., Cheng C.-H., Jung S., Feltus F.A., Bett K.E., Main D.. Tripal v1.1: a standards-based toolkit for construction of online genetic and genomic databases. Database (Oxford). 2013; 2013:bat075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ficklin S.P., Sanderson L.-A., Cheng C.-H., Staton M.E., Lee T., Cho I.-H., Jung S., Bett K.E., Main D.. Tripal: a construction toolkit for online genome databases. Database (Oxford). 2011; 2011:bar044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edger P.P., VanBuren R., Colle M., Poorten T.J., Wai C.M., Niederhuth C.E., Alger E.I., Ou S., Acharya C.B., Wang J. et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience. 2018; 7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirakawa H., Shirasawa K., Kosugi S., Tashiro K., Nakayama S., Yamada M., Kohara M., Watanabe A., Kishida Y., Fujishiro T. et al. Dissection of the octoploid strawberry genome by deep sequencing of the genomes of fragaria species. DNA Res. 2014; 21:169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shulaev V., Sargent D.J., Crowhurst R.N., Mockler T.C., Folkerts O., Delcher A.L., Jaiswal P., Mockaitis K., Liston A., Mane S.P. et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2010; 43:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tennessen J.A., Govindarajulu R., Ashman T.-L., Liston A.. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 2014; 6:3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darwish O., Shahan R., Liu Z., Slovin J.P., Alkharouf N.W.. Re-annotation of the woodland strawberry (Fragaria vesca) genome. BMC Genomics. 2015; 16:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., Wei W., Feng J., Luo H., Pi M., Liu Z., Kang C.. Genome re-annotation of the wild strawberry Fragaria vesca using extensive Illumina- and SMRT-based RNA-seq datasets. DNA Res. 2018; 25:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buti M., Moretto M., Barghini E., Mascagni F., Natali L., Brilli M., Lomsadze A., Sonego P., Giongo L., Alonge M. et al. The genome sequence and transcriptome of Potentilla micrantha and their comparison to Fragaria vesca (the woodland strawberry). Gigascience. 2018; 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daccord N., Celton J.-M., Linsmith G., Becker C., Choisne N., Schijlen E., van de Geest H., Bianco L., Micheletti D., Velasco R. et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017; 49:1099–1106. [DOI] [PubMed] [Google Scholar]
- 16. Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., Fontana P., Bhatnagar S.K., Troggio M., Pruss D. et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010; 42:833–839. [DOI] [PubMed] [Google Scholar]
- 17. The International Peach Genome Initiative Verde I., Abbott A.G., Scalabrin S., Jung S., Shu S., Marroni F., Zhebentyayeva T., Dettori M.T., Grimwood J. et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013; 45:487–494. [DOI] [PubMed] [Google Scholar]
- 18. Verde I., Jenkins J., Dondini L., Micali S., Pagliarani G., Vendramin E., Paris R., Aramini V., Gazza L., Rossini L. et al. The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics. 2017; 18:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shirasawa K., Isuzugawa K., Ikenaga M., Saito Y., Yamamoto T., Hirakawa H., Isobe S.. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017; 24:499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chagné D., Crowhurst R.N., Pindo M., Thrimawithana A., Deng C., Ireland H., Fiers M., Dzierzon H., Cestaro A., Fontana P. et al. The draft genome sequence of European pear (Pyrus communis L. ’Bartlett’). PLoS One. 2014; 9:e92644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hibrand L., Ruttink T., Hamama L., Kirov I., Lakhwani D., Zhou N.-N., Bourke P., Daccord N., Leus L., Schulz D. et al. A high-quality sequence of Rosa chinensis to elucidate genome structure and ornamental traits. Nat. Plants. 2018; 4:473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raymond O., Gouzy J., Just J., Badouin H., Verdenaud M., Lemainque A., Vergne P., Moja S., Choisne N., Pont C. et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018; 50:772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura N., Hirakawa H., Sato S., Otagaki S., Matsumoto S., Tabata S., Tanaka Y.. Genome structure of Rosa multiflora, a wild ancestor of cultivated roses. DNA Res. 2018; 25:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jibran R., Dzierzon H., Bassil N., Bushakra J.M., Edger P.P., Sullivan S., Finn C.E., Dossett M., Vining K.J., VanBuren R. et al. Chromosome-scale scaffolding of the black raspberry (Rubus occidentalis L.) genome based on chromatin interaction data. Hortic. Res. 2018; 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. VanBuren R., Bryant D., Bushakra J.M., Vining K.J., Edger P.P., Rowley E.R., Priest H.D., Michael T.P., Lyons E., Filichkin S.A. et al. The genome of black raspberry (Rubus occidentalis). Plant J. 2016; 87:535–547. [DOI] [PubMed] [Google Scholar]
- 26. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M. et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. The Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017; 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T. -h., Jin H., Marler B., Guo H. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L. et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schläpfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T. et al. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017; 173:2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caspi R., Dreher K., Karp P.D.. The challenge of constructing, classifying, and representing metabolic pathways. FEMS Microbiol. Lett. 2013; 345:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011; 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gordon D., Abajian C., Green P.. Consed: a graphical tool for sequence finishing. Genome Res. 1998; 8:195–202. [DOI] [PubMed] [Google Scholar]
- 37. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 38. Huang X., Madan A.. CAP3: A DNA sequence assembly program. Genome Res. 1999; 9:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu L., Niu B., Zhu Z., Wu S., Li W.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012; 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davidson N.M., Oshlack A.. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014; 15:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koning-Boucoiran C.F.S., Esselink G.D., Vukosavljev M., van ’t Westende W.P.C., Gitonga V.W., Krens F.A., Voorrips R.E., van de Weg W.E., Schulz D., Debener T. et al. Using RNA-Seq to assemble a rose transcriptome with more than 13,000 full-length expressed genes and to develop the WagRhSNP 68k Axiom SNP array for rose (Rosa L.). Front. Plant Sci. 2015; 6:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fuentes L., Monsalve L., Morales-Quintana L., Valdenegro M., Martínez J.-P., Defilippi B.G., González-Agüero M.. Differential expression of ethylene biosynthesis genes in drupelets and receptacle of raspberry (Rubus idaeus). J. Plant Physiol. 2015; 179:100–105. [DOI] [PubMed] [Google Scholar]
- 44. Jung S., Cestaro A., Troggio M., Main D., Zheng P., Cho I., Folta K.M., Sosinski B., Abbott A., Celton J.-M. et al. Whole genome comparisons of Fragaria, Prunus and Malus reveal different modes of evolution between Rosaceous subfamilies. BMC Genomics. 2012; 13:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Youens-Clark K., Faga B., Yap I.V., Stein L., Ware D.. CMap 1.01: a comparative mapping application for the Internet. Bioinformatics. 2009; 25:3040–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iezzoni A., Peace C., Main D., Bassil N., Coe M., Finn C., Gasic K., Luby J., Hokanson S., McFerson J. et al. RosBREED2: progress and future plans to enable DNA-informed breeding in the Rosaceae. Acta Hortic. 2017; doi:10.17660/ActaHortic.2017.1172.2. [Google Scholar]
- 47. Chagné D., Crowhurst R.N., Troggio M., Davey M.W., Gilmore B., Lawley C., Vanderzande S., Hellens R.P., Kumar S., Cestaro A. et al. Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS One. 2012; 7:e31745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bianco L., Cestaro A., Sargent D.J., Banchi E., Derdak S., Di Guardo M., Salvi S., Jansen J., Viola R., Gut I. et al. Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS One. 2014; 9:e110377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bianco L., Cestaro A., Linsmith G., Muranty H., Denancé C., Théron A., Poncet C., Micheletti D., Kerschbamer E., Di Pierro E.A. et al. Development and validation of the Axiom(®) Apple480K SNP genotyping array. Plant J. 2016; 86:62–74. [DOI] [PubMed] [Google Scholar]
- 50. Peace C., Bassil N., Main D., Ficklin S., Rosyara U.R., Stegmeir T., Sebolt A., Gilmore B., Lawley C., Mockler T.C. et al. Development and evaluation of a genome-wide 6K SNP array for diploid sweet cherry and tetraploid sour cherry. PLoS One. 2012; 7:e48305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Verde I., Bassil N., Scalabrin S., Gilmore B., Lawley C.T., Gasic K., Micheletti D., Rosyara U.R., Cattonaro F., Vendramin E. et al. Development and evaluation of a 9K SNP array for peach by internationally coordinated SNP detection and validation in breeding germplasm. PLoS One. 2012; 7:e35668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bassil N.V., Davis T.M., Zhang H., Ficklin S., Mittmann M., Webster T., Mahoney L., Wood D., Alperin E.S., Rosyara U.R. et al. Development and preliminary evaluation of a 90 K Axiom® SNP array for the allo-octoploid cultivated strawberry Fragaria × ananassa. BMC Genomics. 2015; 16:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Evans K. Apple Breeding in Pacific Northwest. Acta Hortic. 2013; 976:75–78. [Google Scholar]
- 54. Rife T.W., Poland J.A.. Field Book: an open-source application for field data collection on android. Crop. Sci. 2014; 54:1624–1627. [Google Scholar]
- 55. Jung S., Lee T., Cheng C.-H., Ficklin S., Yu J., Humann J., Main D.. Extension modules for storage, visualization and querying of genomic, genetic and breeding data in Tripal databases. Database (Oxford). 2017; 2017:bax092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen M., Henry N., Almsaeed A., Zhou X., Wegrzyn J., Ficklin S., Staton M.. New extension software modules to enhance searching and display of transcriptome data in Tripal databases. Database (Oxford). 2017; 2017:bax052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harper L., Campbell J., Cannon E.K.S., Jung S., Poelchau M., Walls R., Andorf C., Arnaud E., Berardini T.Z., Birkett C. et al. AgBioData consortium recommendations for sustainable genomics and genetics databases for agriculture. Database (Oxford). 2018; 2018:bay088. [DOI] [PMC free article] [PubMed] [Google Scholar]