Abstract
Mouse monoclonal antibodies with varying specificities against the Gag capsid of simian and human immunodeficiency virus (SIV/HIV) were generated by immunizing mice with whole inactivated SIVagmTYO-1. Monoclonal antibody AG3.0 showed the broadest reactivity recognizing the Gag capsid protein (p24–27) and Gag precursors p38, p55, and p150 of HIV-1, HIV-2, SIVmac, and SIVagm. Using overlapping peptides, the AG3.0 epitope was mapped in capsid to a sequence (SPRTLNA) conserved among HIV-1, HIV-2, SIVrcm, SIVsm/mac, and SIVagm related viruses. Because of its broad cross-reactivity, AG3.0 was used to develop an antigen capture assay with a lower detection limit of 100 pg/ml HIV-1 Gag p24. Interestingly, AG3.0 was found to have a faster binding on/off rate for SIVagmVer and SIVmac Gag than for SIVagmSab Gag, possibly due to differences outside the SPRTLNA motif. In addition, the ribonucleic acid (RNA) coding for AG3.0 was sequenced to facilitate the development of humanized monoclonal antibodies.
Keywords: AG3.0, epitope, Gag, capsid, p24, p27, human immunodeficiency virus, simian immunodeficiency virus
INTRODUCTION
Since the discovery of HIV-1 and HIV-2 (Barre-Sinoussi et al., 1983; Clavel et al., 1986; Gallo et al., 1983) an increasing number of related SIVs have been isolated from nonhuman primates. Serological evidence of SIV infection has been shown for at least 40 of the 69 different primate species in Africa and has been confirmed by sequence analysis in 32 (Aghokeng et al., 2010; Locatelli et al., 2008a). Complete SIV genome sequences are available for 20 species (Liegeois et al., 2009; Van Heuverswyn and Peeters, 2007; VandeWoude and Apetrei, 2006). The primate lentiviruses can presently be classified into 10 distinct lineages based upon phylogenetic relationships of full-length sequences: 1) SIVcpz from chimpanzees (Pan troglodytes) including HIV-1 and SIVgor from western gorillas (Gorilla gorilla) (Santiago et al., 2003; Van Heuverswyn et al., 2007; Van Heuverswyn et al., 2006); 2) SIVsm from sooty mangabeys (Cercocebus atys) including HIV-2 and SIVmac from macaques (Macaca spp.) (Clavel et al., 1986; Franchini et al., 1987; Hirsch et al., 1989); 3) SIVagm from African green monkeys (members of the Chlorocebus aethiops superspecies) (Allan et al., 1991; Hirsch et al., 1993b; Johnson et al., 1990); 4) SIVsyk from Sykes' monkeys (Cercopithecus albogularis) (Hirsch et al., 1993a); 5) SIVlho from L'Hoest monkeys (C. lhoesti) including SIVsun from sun-tailed monkeys (C. solatus) and SIVmnd-1 from mandrills (Mandrillus sphinx) (Beer et al., 1999; Hirsch et al., 1999; Tsujimoto et al., 1989); 6) SIVrcm from red-capped mangabeys (Cercocebus torquatus) including SIVdrl from drills (Mandrillus leucophaeus), and SIVmnd-2 from mandrills (Beer et al., 2001; Hu et al., 2003); 7) SIVdeb from DeBrazza monkeys (C. neglectus) (Bibollet-Ruche et al., 2004) including SIVden from Dent's mona monkeys (C. mona denti) (Dazza et al., 2005); 8) SIVgsn from greater spot-nosed monkeys (C. nictitans) including SIVmon from mona monkeys (C. mona), and SIVmus-1 and mus-2 from mustached monkeys (C. cephus) (Aghokeng et al., 2007; Courgnaud et al., 2003a); 9) SIVtal from talapoin monkeys (Miopithecus ogouensis) (Liegeois et al., 2006); and 10) SIVcol from Colobus monkeys (Colobus guereza) (Courgnaud et al., 2001). Some lineages, such as SIVcpz, are presumed to be the result of recombination between other lineages (SIVrcm and SIVgsn) (Bailes et al., 2003). Smaller fragments from other SIV-infected African primate species have also been sequenced and may indicate the existence of either separate lineages [SIVbkm from black mangabeys (Lophocebus aterrimus) (Takemura et al., 2005), SIV-ASC-Qu from Schmidt's guenon (C. ascanius schmidti) (Verschoor et al., 2004)], or reveal genetic relationships to the lineages mentioned above (SIVwrc, SIVolc to the SIVlho lineage) (Courgnaud et al., 2003b; Locatelli et al., 2008b).
For diagnostic purposes, monoclonal antibodies (mAbs) are useful to characterize immunodominant antigenic sites or functional domains on viral components. In addition, they are needed to develop a variety of virological assays, most importantly antigen capture assays for HIV and SIV. Moreover, measurements of SIV antigen in cells and tissues, such as kinetic studies, virus titrations, immunohistochemistry, and neutralization tests, require a reliable and fast method for measuring virus production. HIV/SIV capsid protein (p24–27) is the most abundant protein produced during virus replication (Veronese et al., 1988) and is found both inside infected cells and in virus particles released by infected cells forming a protective shell for viral RNA. Antigen-capture enzyme-linked immunosorbent assays (ELISAs) that determine the p24–27 amount in plasma or cell culture samples are frequently used in HIV/SIV research as endpoint for neutralization as well as antiviral assays. Besides antigen capture ELISA, there are other assays available for quantitating HIV/SIV in body or tissue culture fluids, such as viral RNA quantification or the measurements of reverse transcriptase (RT) activity. Compared to the antigen capture ELISA, those other methods have both advantages and disadvantages. The measurement of viral RNA is highly sensitive, but requires exact knowledge of the underlying sequence and necessitates storage of the samples at very low temperatures to prevent degradation of the viral RNA. On the other hand, quantification of RT enzyme activity is less sensitive than that of p24–27 protein determination because Gag is the major structural protein and is present at about 5,000 copies per virion (Briggs et al., 2004). In addition, inappropriate sample storage can also interfere with sensitivity of RT quantitation since enzymes are more sensitive to temperature changes than are proteins. Measurement of RT activity is also not specific for HIV/SIV since all retroviruses code for this enzyme. Compared to viral RNA and RT, p24–27 is a fairly stable protein, is specific for and also antigenically cross-reactive with a wide variety of HIVs and SIVs, and large numbers of samples can be processed on a single ELISA plate. Antigen capture assays for HIV and SIV are commercially available, but are expensive and can have limited utility for detecting a wide range of SIV and HIV types. Neutralization tests or virus titrations usually require processing of high quantities of samples, and thus it is desirable to develop cost-effective in-house assays that incorporate monoclonal antibodies broadly reactive to SIV and HIV. Furthermore, determinations can be made by detecting p24–27 expression in infected cells by means of immunohistochemical staining methodologies or western blot/radioimmunoprecipitation.
In this study, the generation and characterization of several mouse monoclonal antibodies against SIVagm is presented; one antibody (AG3.0) was broadly reactive to HIV-1, HIV-2/SIVsm/SIVmac and SIVagm by western blotting and radioimmunoprecipitation. The epitope recognized by AG3.0 mapped to a 7-mer peptide within the N-terminal portion of the Gag capsid, a highly conserved region among primate lentiviruses (HIV-1, HIV-2/SIVmac, SIVagm, and SIVrcm). The antibody was donated to the NIH AIDS Research and Reference Reagent Program (ARRRP; www.aidsreagent.org, catalog number 4121) and has been used in experimental studies by a number of investigators (Leblanc, Perez, and Hope, 2008; Potash et al., 1998; Sakuma et al., 2007; Schiavoni et al., 2004). As a next step, a p24–27 antigen capture assay comparable in sensitivity to immunostaining of infected cells was developed. Testing of representative virus strains from six primate lentiviral lineages by antigen capture assay revealed the broad spectrum binding of mAb AG3.0 in recognizing p24–27 species from HIV-1, HIV-2, SIVmac, SIVagm, and SIVrcm, but not SIVlho, SIVsun, and SIVsyk. The lower limit of quantitation of the assay was determined to be 100 pg/ml p24 Gag. Subsequently, both the heavy and light chains of AG3.0 were sequenced and annotated to facilitate humanized anti-p24 antibody development.
RESULTS
Generation of three monoclonal antibodies that react with SIV Gag
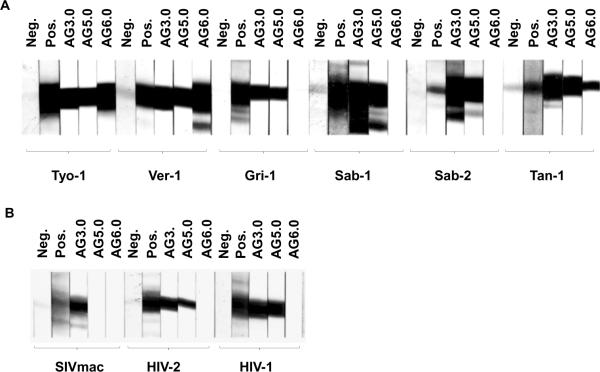
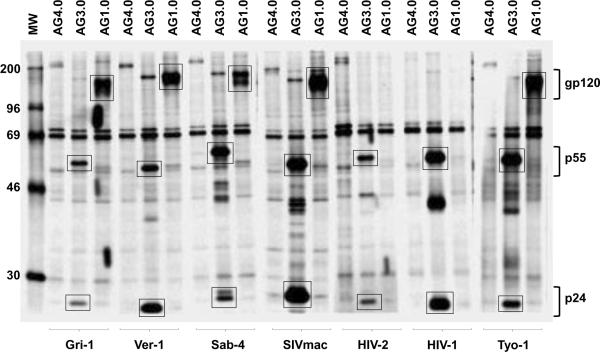
Mouse monoclonal antibodies against whole disrupted SIVagmTYO-1 were produced using a standard mouse-cell hybridoma technique. Hybridomas were screened for reactivity to SIVagmTYO-1 by whole virus ELISA. The specificity of positive hybridomas was further evaluated by immunoblotting and three hybridomas were selected, whose supernatants detected SIV Gag p27. The hybridomas were cloned by single cell endpoint dilution and were designated AG3.0, AG5.0, and AG6.0. All three monoclonal antibodies were determined to be of the immunoglobulin G type 1 (IgG1) isotype. As shown in Fig. 1, AG3.0 showed the broadest reactivity by immunoblotting and detected Gag p24–27 from all four SIVagm isolates (ver, gri, sab, tan) as well as from SIVmac, HIV-2 and HIV-1, AG5.0 reacted with all SIVagm subtypes, and HIV-1 and HIV-2 Gag proteins, but not with SIVmac Gag while AG6.0 only reacted with SIVagm Gag from vervet and tantalus monkeys. Additionally, radioimmunoprecipitation analysis (RIPA) was performed with both cell and virus lysates (Fig. 2). RIPA differs from immunoblot analysis in that radiolabeled native proteins in solution react with antibody whereas in immunoblotting viral proteins are mostly denatured due to the presence of sodium dodecyl sulfate (SDS). The monoclonal antibody AG3.0 reacted with p24–27 and the Gag precursor proteins p38, p55, and p160 from SIV- and HIV-infected cell lysates. Monoclonal antibody AG1.0, recognizing gp120 from SIVagm and SIVmac, and antibody AG4.0, an anti-cellular antibody, served as controls. Overall, AG3.0 was the most broadly cross-reacting monoclonal antibody recognizing the Gag capsid and precursor products of at least three different primate lentivirus lineages.
Figure 1. Western blot analysis.
Reactivity of different Gag-specific monoclonal antibodies against various purified isolates of SIV and HIV in western blot performed as described in the materials and methods. A: Isolates from the four subspecies of African green monkeys. B: Isolates from rhesus macaques and humans. Neg: Negative control sera from uninfected individuals of the relevant species. Pos: Positive polyclonal control sera from individuals of the relevant species infected with the corresponding virus.
Figure 2. Radioimmunoprecipitation analysis.
Binding of the AG3.0 monoclonal antibody to radiolabeled Gag protein from cells infected with various SIV and HIV isolates. The monoclonal antibodies AG1.0, recognizing gp120 from SIVagm and SIVmac, and AG4.0, recognizing a cell protein, were included as controls. Bands showing reactivity with p24–27, p55, and gp120 are marked with a box.
AG3.0 reacts with a 7-mer Gag peptide that is conserved between several lentivirus lineages
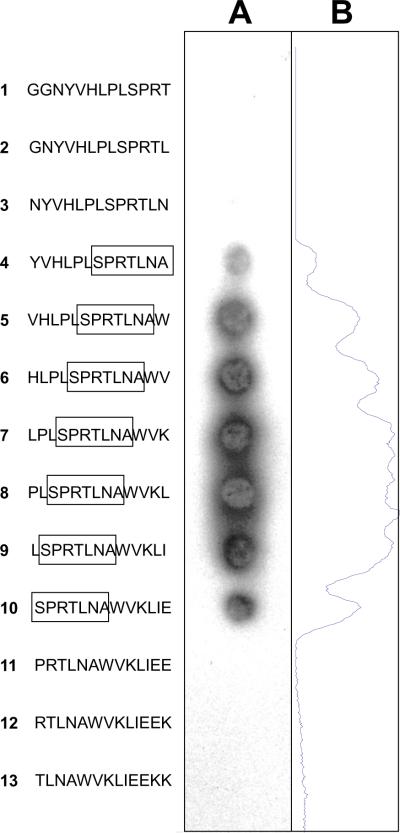
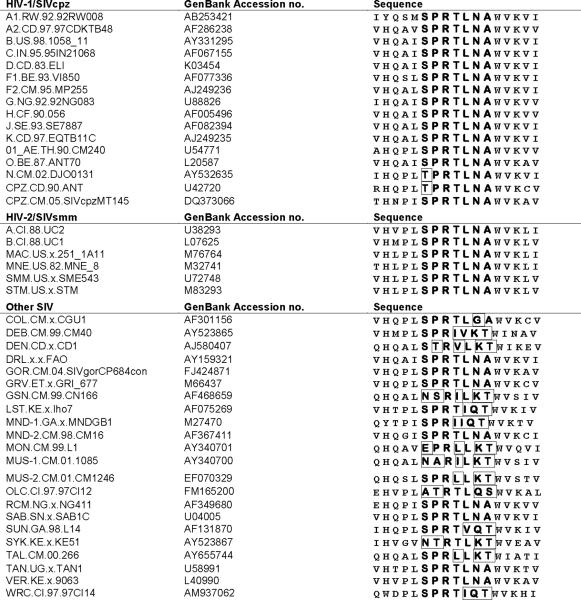
In order to determine the epitope recognized by AG3.0, we initially used a series of overlapping Gag p26 peptides provided by the EU AIDS Vaccine Integrated Project's Programme EVA for epitope mapping. Twenty-two 20-mer peptides with a 10 amino acid overlap were tested for reactivity with AG3.0 (Table 1). AG3.0 reacted with a single peptide spanning amino acid 11 to 30 from SIVmac251Gag (HLPLSPRTLNAWVKLIEEKK), but not with any of the surrounding peptides, indicating that the minimal AG3.0 epitope is located in the center of the peptide at the N-terminus of Gag capsid. Subsequent fine mapping using 13-mers with 12-mer overlaps covering the sequence GGNYVHLPLSPRTLNAWVKLIEEKK (SIVmac251Gag aa 141–165) revealed a minimal epitope with the sequence SPRTLNA. This was concluded from the fact that two of the peptides missing one amino acid at the beginning or the end of the epitope (NYVHLPLSPRTLN and PRTLNAVWKLIEE, Fig. 3) were not recognized by AG3.0. A sequence alignment between representatives of ten primate lentiviral lineages is shown in Table 2A and demonstrates that the epitope SPRTLNA is conserved between most SIVcpz, SIVsm, SIVagm, and SIVrcm related viruses. Only the HIV-1 N group and SIVcpz-ANT differ by one amino acid at the N-terminus (threonine instead of serine). This peptide also differs in the following viruses (conserved amino acids underlined): SIVlho (SPRTIQT), SIVsun (SPRTVQT), SIVmndGB1 (SPRIIQT), SIVcol (SPRTLGA), SIVsyk (PVRTLKT), SIVmon (EPRLLKT), SIVmus-1 (NARILKT), SIVmus-2 (SPRILKT), SIVgsn (NSRILKT), SIVdeb (SPRIVKT), SIVden (STRVLKT), and SIVtal (SPRLLKT). Generally, the N-terminus of the epitope was more conserved than the C-terminus among the ten primate lentivirus lineages. The only conserved amino acid among all simian and human immunodeficiency viruses described so far was arginine in position 3.
Table 1.
Rough epitope mapping of AG3.0
| SIVmac251 Gag Capsid Sequence | |||||
| 001 | PVQQIGGNYV | HLPLSPRTLN | AWVKLIEEKK | FGAEVVPGFQ | ALSEGCTPYD |
| 051 | INQMLNCVGD | HQAAMQIIRD | IINEEAADWD | LQHPQPAPQQ | GQLREPSGSD |
| 101 | IAGTTSSVDE | QIQWMYRQQN | PIPVGNIYRR | WIQLGLQKCV | RMYNPTNILD |
| 151 | VKQGPKEPFQ | SYVDRFYKSL | RAEQTDAAVK | NWMTQTLLIQ | NANPDCKLVL |
| 201 | KLGLVNPTLE | EMLTACQGVG | GPGQKARL | ||
| Peptidea | Sequence | Mean OD | StDev |
|---|---|---|---|
| 1 | 01–20 | 0.01 | 0.00 |
| 2 | 11–30 | 0.37 | 0.05 |
| 3 | 21–40 | 0.01 | 0.00 |
| 4 | 31–50 | 0.01 | 0.00 |
| 5 | 41–60 | 0.01 | 0.00 |
| 6 | 51–70 | 0.01 | 0.00 |
| 7 | 61–80 | 0.01 | 0.00 |
| 8 | 71–90 | 0.01 | 0.00 |
| 9 | 81–100 | 0.01 | 0.00 |
| 10 | 91–110 | 0.01 | 0.00 |
| 11 | 101–120 | 0.01 | 0.00 |
| 12 | 111–130 | 0.01 | 0.00 |
| 13 | 121–140 | 0.01 | 0.00 |
| 14 | 131–150 | 0.01 | 0.00 |
| 15 | 141–160 | 0.01 | 0.00 |
| 16 | 151–170 | 0.01 | 0.00 |
| 17 | 161–180 | 0.01 | 0.00 |
| 18 | 171–190 | 0.01 | 0.00 |
| 19 | 181–200 | 0.01 | 0.00 |
| 20 | 191–210 | 0.01 | 0.00 |
| 21 | 201–220 | 0.01 | 0.00 |
| 22 | 211–228 | 0.01 | 0.00 |
Peptides were ordered from the European AIDS Reagent Programme, reference ARP714.1–22; peptide 15, 16, and 18 have an additional N-terminal cysteine)
Figure 3. Fine epitope mapping of SIVmac Gag.
Binding of the AG3.0 monoclonal antibody to synthetic peptides spanning the putative epitope in Gag within the sequence GGNYVHLPLSPRTLNAWVKLIEEKK (SIVmac251 Gag aa 141–165). The peptides, 13-mers with a 12 amino acid overlap were synthesized directly onto a cellulose membrane and binding of AG3.0 visualized by enhanced chemiluminescence (A). A density plot along the membrane was carried out to aid analysis or reactivity (B).
Table 2A.
Alignment of Gag capsid N-terminus (B.FR.83.HXB2_LAI_IIIB aa 160–176)
AG3.0 antigen capture assay for detecting virus from three major lentiviral lineages
Antigen capture assays are useful tools for identifying and quantifying plasma virus levels in infected monkeys and for measuring virus growth in tissue culture. Since AG3.0 recognized three major lineages commonly used in AIDS research, a broadly reactive antigen capture assay was developed, which could detect a wide variety of HIV/SIV capsid antigens. In order to generate pure antibody material, IgG was purified from hybridoma supernatants using Protein G sepharose columns, and the optimal AG3.0 IgG concentration was determined as follows: An ELISA plate coated with twofold dilutions of AG3.0 was reacted with 0.2% Tween20 disrupted SIVagmVER.DE.x.AGM3 supernatant. Captured protein was detected using a polyclonal antibody from an SIV-infected monkey followed by goat anti-human immunoglobulin G-peroxidase conjugate and substrate. The optimal IgG concentration for coating ELISA plates was determined as 0.675 μg/well.
Next, virus supernatant from three different primate lentivirus lineages (HIV-1, HIV-2/SIVsm, and SIVagm) as well the chimeric virus SHIV 89.6 (HIV-1 89.6 env, tat, rev, vpu in an SIVmac239 backbone) were serially titrated in C8166 or Molt4clone8 cells (Table 3) and the amount of p24–27 content determined in an AG3.0 antigen capture assay. Wells were considered positive if the optical density exceeded the mean of the negative controls plus three times the standard deviation. In parallel, the virus-exposed cells were subjected to HIV/SIV-specific immunostaining. The mean tissue culture infectious dose (TCID50) was calculated using both endpoints, i.e., antigen capture assay optical densities of supernatants and HIV/SIV-specific immunostaining of cells. As shown in Table 3, the results of the antigen capture assay matched the results of the cellular immunostaining within the expected range of assay variability. These results suggested that the antigen capture assay is equivalent to immunostaining as endpoint analysis for virus titrations and virus neutralization assays.
Table 3.
Comparison of TCID50/ml calculations using AG3.0 antigen capture assay or HIV/SIV-specific immunostaining
| Log10 Titer (TCID50/ml) |
|||
|---|---|---|---|
| Endpoint analysis |
|||
| Virus | Titrated in | AG3.0 antigen capture assay of supernatants | HIV/SIV-specific immunostaining of cells |
| SHIV 89.6 | C8166 cells | 5.52 | 5.64 |
| HIV-2 UC3 | C8166 cells | 4.99 | 4.79 |
| SIVsmmPBj 1.9 | C8166 cells | 4.87 | 5.04 |
| SIVagm3 | C8166 cells | 6.54 | 6.48 |
| SIVagm3 | Molt4clone8 cells | 4.81 | 4.63 |
| SIVmac 32H | C8166 cells | 6.71 | 6.71 |
| HIV-1 LAI | C8166 cells | 4.69 | 4.39 |
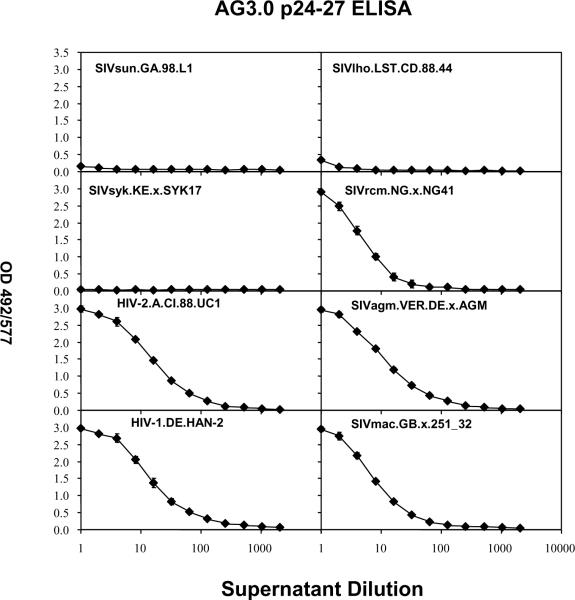
Third, the reactivity of AG3.0 to HIVs and SIVs from five primate lentivirus lineages was tested by antigen capture ELISA. As shown in Fig. 4, the AG3.0-based antigen capture assay was able to detect Gag capsid from HIV-1, HIV-2, SIVmac, SIVagm, and SIVrcm, but not from SIVsun, SIVlho, and SIVsyk. For this experiment, two-fold dilutions of virus supernatants were assayed, and the optical densities decreased proportionally to the virus dilutions. At about a 100-fold dilution of supernatant, the signal became undetectable. Overall, the cross-reactivity of the AG3.0 antibody was consistent with the amino acid conservation of the Gag epitope (see Table 2A). As described above and shown in Table 2A, the epitope SPRTLNA is mostly conserved among the HIV-1/SIVcpz, HIV-2/SIVsmm, and SIVagm lineages, and SIVrcm. In SIVsyk, SIVsun, and SIVlho, it differs by 3–4 amino acids, which prohibit binding to the monoclonal antibody. Viruses from the SIVdeb, SIVgsn/mon/mus, and SIVtal lineages were not tested, but the epitope differs by 2–5 amino acids from SPRTLNA, and it would be unlikely that the AG3.0 antibody cross-reacts with Gag from these viruses. In contrast, the conservation of the epitope in SIVdrl and SIVmnd-2, which are phylogenetically related to SIVrcm. likely predicts cross-reactivity.
Figure 4. Antigen capture ELISA using various isolates.
Supernatants from cells infected with various isolates of SIV and HIV were tested at different dilutions for reactivity in the AG3.0 based antigen capture assay as described in the material and methods.
Affinity determination of the interaction between AG3.0 and Gag proteins from SIVagm and SIVmac
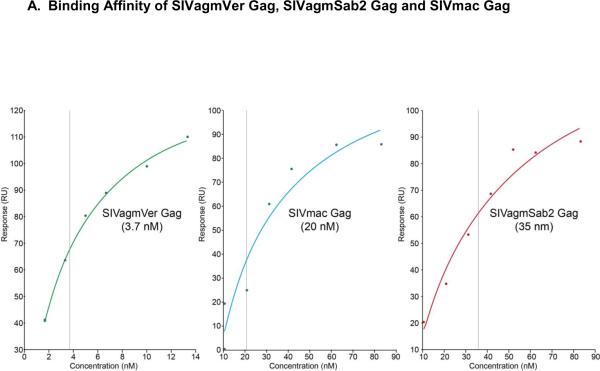
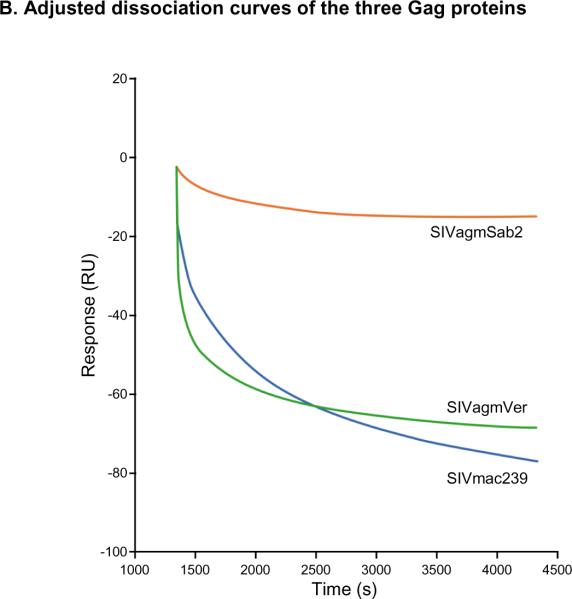
Surface plasmon resonance (SPR) technology was used to evaluate the binding affinity of the monoclonal antibody AG3.0 to different SIV Gag proteins. Although the calculated maximal response (Rmax) was 100 response units (RU), the observed Rmax was more than double, likely due to multimerization of the Gag protein (data not shown). The highest affinity was obtained using SVagmVer (3.7 nM) while the affinities for SIVmac Gag and SIVagmSab Gag were lower but similar with 20 nM and 35 nM, respectively (Fig. 5A). Interestingly, while the dissociation of SIVagmVer Gag and SIVmac Gag was fairly rapid, the binding between the AG3.0 mAb and SIVagmSab Gag was found to be very stable, with very little dissociation detectable (Fig. 5B).
Figure 5. Surface plasmon resonance (SPR) analysis.
A: Binding affinity profiles of Gag proteins derived from different SIVs to AG3.0 measured by BIAcore as described in the materials and methods. Sensor chips coated in AG3.0 were exposed to purified preparations of the different Gag proteins at various concentrations. Binding was measured as a change in RU at equilibrium. The apparent kD values were determined using the BiaEvaluation software (GE Healthcare). B: Dissociation curves for the different Gag proteins (1.5 μg/ml) using an injection period of 1,080 sec, a dissociation time of 3,000 sec, and a flow rate of 5 μl/min.
Sequencing of the AG3.0 antigen binding site
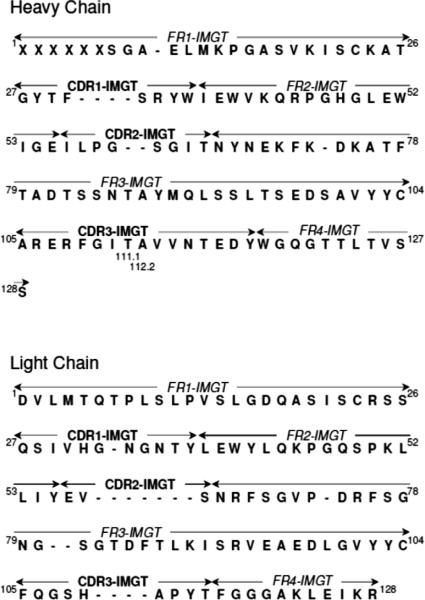
cDNAs generated from RNA coding for the variable fragments of the AG3.0 heavy and light chains were sequenced and analysed using the Vbase2 application (http://www.vbase2.org) to identify the conserved framework regions (FR1–FR4) and variable complementarity determining regions (CDR1–CDR3) according to the International ImMunoGeneTics (IMGT) nomenclature (Fig 6). The three variable heavy chain regions were found to consist of 8, 8 and 16 amino acids for CDR1, CDR2 and CDR3, respectively, whereas the light chain had corresponding CDR lengths of 11, 3, and 9.
Figure 6. Amino acid sequences of the heavy and light chains of the AG3.0 monoclonal antibody.
The four conserved framework regions (FR1–FR4) and the three hypervariable complementarity determining regions (CDR1–CDR3) are displayed according to the IMGT nomenclature (Lefranc, 2007). Unoccupied amino acid positions are marked with a dash (−). X = positions in the conserved FR1 region of the heavy chain, at which the identity of the amino acid could not be unequivocally determined from the sequence.
DISCUSSION
Monoclonal antibodies against SIVagm Gag p27 were developed to allow the Gag capsid expression in African green monkeys and in pathogenic macaque models of SIV infection to be compared. Gag is an important structural protein and is often referred to as a precursor because it is subject to cleavage by the viral protease, which yields the internal structural proteins of the mature virion (Freed, 1998; Swanstrom and Wills, 1997; Vogt, 1997; Wills and Craven, 1991). Three of the Gag cleavage products, matrix, capsid, and nucleocapsid, are common to all retroviruses and are always arranged in this order within the Gag precursor, with matrix being at the N-terminus. Additionally, the Gag precursors of HIV and SIV possess a C-terminal domain termed p6 that is unique to primate lentiviruses, as well as two “spacer” regions that separate capsid from nucleocapsid, and nucleocapsid from p6 (Henderson et al., 1992; Mervis et al., 1988).
Capsid, which directly follows matrix in the Gag precursor, has crucial roles in particle assembly and after entry into a new target cell. However, the function of capsid in the early phase of the replication cycle is not well understood. In the mature virion, capsid forms the shell of the core, which is occasionally tubular but most often conical, a feature that distinguishes lentiviruses, such as HIV-1, from most other retroviruses (Gelderblom, 1991). Capsid has two predominantly α-helical domains that are connected through a flexible linker region (Gamble et al., 1997; Gitti et al., 1996; Momany et al., 1996) and have different roles in virus morphogenesis. The N-terminal domain, which comprises two thirds of HIV-1 capsid, is required for the formation of the mature core but is dispensable for the assembly of immature virus particles (Borsetti, Ohagen, and Gottlinger, 1998; Dorfman et al., 1994; Reicin et al., 1996; Reicin et al., 1995; Srinivasakumar, Hammarskjold, and Rekosh, 1995; Wang and Barklis, 1993). In contrast, the C-terminal capsid domain is crucial both for particle assembly and for core formation (Dorfman et al., 1994; Mammano et al., 1994; McDermott et al., 1996; Reicin et al., 1995). The N-terminal domain of HIV-1 capsid interacts with the human peptidylprolyl cis-trans isomerase cyclophilin A (CyPA), which leads to the specific incorporation of this ubiquitous cytosolic host protein into virions (Franke, Yuan, and Luban, 1994; Luban et al., 1993; Thali et al., 1994). This feature is unique for HIV-1 and absent in other primate lentiviruses and HIV-2. Based on the regular appearance of the synthetic cones, Sundquist et al. propose that retroviral cores are composed of hexagonal lattices that are closed through the incorporation of a total of 12 pentagons (Ganser et al., 2003).
In the study presented here, three monoclonal antibodies against SIVagm from the vervet subspecies (SIVagmTYO-1) were generated and characterized. AG6.0 cross-reacted with two subspecies of African green monkeys (vervet and tantalus), whereas AG5.0 showed a broader cross-reactivity, reacting with SIVagm Gag p26 from vervet, grivet, sabaeus, and tantalus, HIV-1 and HIV-2. Interestingly, although HIV-2 and SIVmac have similar ancestries, having both arisen by transmission from sooty mangabeys (SIVsm), only the p26 from HIV-2ST was recognized. AG3.0 showed the broadest cross-reactivity and recognized not only the SIVagm lineage, but also members of the HIV-2 lineage (HIV-2, SIVmac), HIV-1 lineage, and SIVrcm. In addition, AG3.0 reacted with the Gag precursors p55 and p38 and the Gag-Pol precursor in infected cell lysates. This broadly cross-reactive antibody (AG3.0) is available from the NIH ARRRP as “Monoclonal antibody to HIV-1 p24” under catalog no. 4121 (Simm et al., 1995). In addition, the AG3.0 producing hybridoma cells are also available. A number of monoclonal antibodies specific for HIV and SIV Gag are deposited in the NIH ARRRP but, with the exception of AG3.0, none react with SIVagm or is broadly cross-reactive with SIV and HIV. So far, only Otteken et al. (1992) successfully produced monoclonal antibodies to SIVagm Gag p27 (strain TYO-7). However, these monoclonal antibodies (1.17.3, 1A7 and 1F6) are primarily type-specific, reacting with only certain types of SIVagm and the HIV-2 lineage, but not with HIV-1 or SIVmndGB1. These monoclonal antibodies mapped to a peptide spanning amino acids 152 to 172 in SIVmac251, which is conserved in the HIV-2 lineage, but not in SIVagmTYO-1, HIV-1 IIIB, and SIVmndGB1. One particular monoclonal antibody, FM18, generated against SIVmac, also reacted with SIVagm, but not with HIV-1 (Lairmore et al., 1993). Since this antibody has not been mapped, the full extent of cross-reactivity is unknown. However, our broadest reactive monoclonal antibody, AG3.0, is able to detect p24–27 from all of the main primate lentivirus lineages including HIV-1, SIVmac/HIV-2, SIVagm, and SIVrcm.
Rough and fine mapping revealed the minimal epitope recognized by AG3.0 to be SPRTLNA (Table 1, Fig. 3). This epitope is conserved throughout the HIV-1 (including SIVrcm), HIV-2 and SIVagm lineages, and is partially conserved throughout the other SIV lineages (SIVlho, SIVsyk, SIVcol, SIVdeb, SIVtal, SIVgsn), although it is completely absent in non-primate lentiviruses. However, despite this partial conservation, AG3.0 does not recognize SIVlho, SIVsun, or SIVsyk Gag, indirectly confirming the minimal epitope needed for binding (Fig. 4). Von Schwedler et al. showed that viruses with a mutated arginine and asparagine in the SPRTLNA motif failed to form a cone-shaped core and were not infectious, indirectly demonstrating the importance of this epitope for viral assembly. The Los Alamos Immunology database contains 21 HIV-1 cytotoxic T-lymphocyte (CTL) epitopes restricted by HLA-A, -B, and -C haplotypes spanning the SPRTLNA peptide (http://www.hiv.lanl.gov/content/immunology/ctl_search) suggesting immunogenic dominance of the region. For example, a dominant CTL epitope ISPRTLNAW has been detected in HIV-1 positive individuals having the protective haplotype HLA B*5701 (Bailey et al., 2006), which is found in a very high frequency in HIV-1 infected non-progressors (11/13 or 85% versus 19/200 or 9.5% of progressors). Non-progressors tended to have an immune response that was highly focused on four p24 epitopes that were presented by B*5701, ISPRTLNAW, KAFSPEVIPMF, TSTLQEQIGW, and QASQEVKNW (Migueles and Connors, 2001). THE SPRTLNA epitope is also part of three human CD4+ T helper cell HIV-1 epitopes and two mouse monoclonal antibody HIV-1 epitopes (AISPRTLNAW for the F5-2 antibody and VHQAISPRTLNAWVK for the ID8F6 antibody) (http://www.hiv.lanl.gov/content/immunology/ctl_search).
After mapping the AG3.0 epitope, we sought to develop an antigen capture ELISA using the AG3.0 monoclonal antibody since it showed the broadest cross-reactivity against HIV/SIV (Binninger-Schinzel et al., 2008). Currently, there is only one commercially available antigen capture assay that cross-reacts with SIVmac, SIVagm, SIVdrl, and SIVmnd-2 (from ZeptoMetrix Corp.) (Hu et al., 2003; Pandrea et al., 2006). However this assay is expensive and the epitope recognized by the monoclonal antibody that forms the basis of the test is not public knowledge. Therefore, the degree of cross-reactivity with different HIV/SIV isolates can only be determined empirically. Most in-house antigen capture assays using monoclonal antibodies developed to date are either optimized for the HIV-1/SIVcpz or HIV-2/SIVmac/SIVsm lineage (Lohman et al., 1991; Thorstensson et al., 1991; Wehrly and Chesebro, 1997). Only one assay, based on a polyclonal sandwich system, was also able to detect SIVmnd to an appreciable degree (Beirnaert et al., 1998). Our AG3.0 in-house antigen capture assay had a lower limit of detection of 100 pg/ml, comparable to that of Zeptometrix's SIV p27 antigen ELISA (62.5 pg/ml). Optimization of the reagents used in the AG3.0 antigen capture assay should further enhance its sensitivity.
The binding affinity of AG3.0 to SIVagm and SIVmac Gag purified from native virus was determined using SPR technology. SIVagmSab2 Gag was found to bind slowly to AG3.0, whereas the overall binding affinity was more similar to SIVmac than to SIVagmVer. In addition, the dissociation curve was markedly different from both SIVmac and SIVagmVer and indicated a very stable complex between SIVagmSab2 and AG3.0. The same results were obtained with two different Gag concentrations (data not shown). Although all three Gag proteins share the same minimal epitope, the binding structures could be influenced by residues flanking the SPRTLNA epitope, resulting in different tertiary structures of the whole Gag protein and therefore different binding affinities to AG3.0 (Table 2B). To our knowledge, crystallization of the SIV capsid protein, needed to address this hypothesis, has not yet been achieved. Finally, the AG3.0 monoclonal antibody RNA was sequenced and annotated to facilitate creating a human- or monkey-adapted monoclonal antibody specific for the SPRTLNA epitope. This would allow anti-capsid antibodies for modulation of SIV/HIV infection to be evaluated.
Table 2B.
Alignment of amino acid sequence adjacent to SPRTLNA
In summary, we generated three mouse monoclonal antibodies against SIVagmTYO-1 (Ver) Gag capsid, each with varying degrees of cross-reactivity to SIV/HIV. The broadest cross-reactivity was found with AG3.0, which recognized p24–27 from the three major primate lentivirus lineages (HIV-1/SIVcpz/SIVrcm, HIV-2/SIVsm/SIVmac, and SIVagm) and mapped to a conserved 7 amino acid motif in Gag, SPRTLNA (aa 156–162 of SIVagmTYO-1 Gag). An antigen capture assay was developed, which could detect as little as 100 pg/ml of p24 Gag.
MATERIALS AND METHODS
Monoclonal antibody production
BALB/c mice were immunized by intraperitoneal injection of 10 μg sucrose gradient purified SIVagm(TYO-1) in phosphate buffered saline (days 0 and 21) in addition to 10 μg of 1% Triton X-100-treated SIVagm(TYO-1) (day 14). The mice were sacrificed at day 24 and the spleen removed for fusion of splenic cells with SP-2 mouse myeloma cells. The hybridomas were grown in 96-well plates using DMEM (Life Technologies) supplemented with 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 20% fetal bovine serum, with 10 mM hypoxanthine and 2 μg/ml of azaserine (Sigma-Aldrich) for selection. Supernatant fluid (100 μl) from each well showing visible hybridoma growth was evaluated for reactivity to SIVagm(TYO-1) by whole virus ELISA and positive samples screened by western blotting. Clones of interest were subcloned twice. The isotype of each monoclonal antibody was determined with the Sigma ImmunoType™ Kit according to the manufacturer's instructions.
Virus purification
Culture supernatant fluid (5–10 liters) from SIVagm(TYO-1) infected MOLT4clone8 cells was concentrated 50–100 fold with a Minitan ultrafiltration system (Millipore) containing 5 filters with a 300,000 molecular weight pore size. The retentate (50–100 ml) was layered onto 20/70% discontinuous sucrose gradients, and the virus was recovered from the interface after centrifugation for 2 hours at 71,500 × g. The virus fraction was diluted 2× with TEN (0.05 M Tris-HCl pH 7.2, 0.15 M NaCl, and 1mM Ethylenediaminetetraacetic acid (EDTA)), layered over a 20–70% continuous sucrose gradient and centrifuged overnight at 71,500 × g. One milliliter fractions were assessed with a refractometer, the virus fractions (1.16–1.18 g/cm3) diluted 5-fold with TEN buffer, and the virus pelleted by centrifugation at 71,500 × g for 2 hours at 4°C. The virus pellets were resuspended in a volume of 1.0 ml phosphate buffered saline (PBS), and the protein concentration determined using the Bio-Rad Protein Assay according to the manufacturer's instructions (Bio-Rad Life Sciences). The virus was further analyzed for purity by SDS-PAGE with the protein bands visualized by immunoblotting.
Whole virus ELISA
Purified SIVagm(TYO-1) was diluted to 2 μg/ml in borate buffered saline (BBS) and used to coat Immulon II plates (Dynex Technologies Inc., Chantilly, VA) overnight at 4°C. Wells were blocked with 10% normal goat serum in BBS for 30 min. at 37°C, and the plates washed 3 times with 1% Tween20 in phosphate buffered saline. One hundred microliters of hybridoma supernatant fluid were then added, the plates incubated for 60 min. at 37°C, and washed three times prior to addition of biotinylated anti-mouse antibody. Binding was determined colorimetrically by incubation with streptavidinhorseradish peroxidase (Amersham) before washing and developing with 2,2'-Azinobis(3-ethylbenzthiazoline-sulfonic acid) (ABTS).
Cells and viruses
All human CD4+ T cell lines were maintained in complete medium consisting of RPMI 1640 supplemented with 15% fetal bovine serum (Life Technologies), HEPES, 2 mM L-glutamine, 50 μg/ml streptomycin and 50 U/ml of penicillin. SIVagm strains VER.KE.x.TYO1, GRI_677, SAB.SN.x.SAB1C, SAB.SN.x.SAB2, SAB.SN.x.SAB4, TAN.UG.x.TAN1, and HIV-2ST (Broussard et al., 2001; Fomsgaard et al., 1991; Jin et al., 1994; Kumar et al., 1990; Shibata et al., 1990; Soares et al., 1997) were grown in Molt4clone8 cells, SIVagm(Ver-1) (Jin et al., 1994) in SupT1 cells, SIVmac251 in Hut78 cells, and HIV-1/IIIB in Molt3 cells.
Western blot analysis
Western blotting was performed as previously described (Allan et al., 1991). Virus in culture supernatants taken from infected cell lines at 48–72 hours was purified through a 20% sucrose cushion, separated by SDS-polyacrylamide gel electrophoresis, and then blotted onto nitrocellulose sheets. The nitrocellulose paper was blocked with 3% bovine serum albumin and subsequently incubated with a 1:8 dilution of monoclonal antibody supernatant fluid or a 1:80 dilution of seropositive and seronegative controls. Viral proteins were detected using the streptavidin-biotin system (Amersham) with diaminobenzidine as the substrate for color development.
Radioimmunoprecipitation analysis
Radioimmunoprecipitation analysis (RIPA) of virus lysates was conducted as described previously (Allan et al., 1992). Infected cells were metabolically labeled for 16 hours with 35-S cysteine and methionine (Translabel, ICN Biomedicals, Inc., Costa Mesa, CA) at 1 mCi/107 cells in cysteine-methionine free RPMI 1640 with 15% FBS. The cells were resuspended in RIPA buffer (0.08 M Tris-HCl pH 7.4, 0.15 M NaCl) containing 0.1% SDS, 1% Triton X-100, and 1% deoxycholate with protease inhibitors (1.0 mM phenylmethyl sulfonyl fluoride (Pierce) and 2 μg/ml of leupeptin (Sigma-Aldrich)) and centrifuged at 100,000 × g to remove protein aggregates. Proteins were then immunoprecipitated with monoclonal antibodies bound to protein A sepharose. The complexes were washed 5 times with complete RIPA lysing buffer, the viral proteins solubilized by resuspension in electrophoresis sample buffer (0.08 M Tris-HCl pH 6.8, 2% SDS, 0.1 M dithiothreitol, 10% glycerol, and 1 mg/ml of bromophenol blue), and the viral proteins separated by 11.3% SDS-PAGE. Proteins were visualized by impregnating the gel with EnHance (DuPont NEN, Boston, MA) before drying and exposure to Kodak SB5 radiographic film (Eastman Kodak).
Epitope mapping
A panel of overlapping SIVmac Gag synthetic peptides (20-mers, 10 overlap; kindly provided by the European AIDS Reagent Programme, reference ARP714.1-22) were used in a standard ELISA for rough epitope mapping of the AG3.0 monoclonal antibody. For fine epitope mapping, a series of 13-mer synthetic peptides with an overlap of 12 aa covering the sequence GGNYVHLPLSPRTLNAWVKLIEEKK (SIVmac251 Gag 141–165) were synthesized directly onto a cellulose membrane (Jerini Bio Tools GmbH, Berlin, Germany). Briefly, the membrane was washed 3 times in Tris-buffered saline containing 0.05% Tween20 (T-TBS) and blocked overnight at 4°C with T-TBS plus 2% milk powder and 10% sucrose (blocking buffer). After washing 3 times with T-TBS, the membrane was incubated overnight at 4°C with a 1:50 dilution of purified AG3.0 mAb in blocking buffer. The membrane was then washed and incubated for one hour at room temperature with a 1:10,000 dilution of goat anti-mouse IgG peroxidase conjugate (Sigma-Aldrich). After three final washes, spots were visualized using the enhanced chemiluminescence kit (Amersham), followed by exposure to photographic film (RPN 2103H Hyperfilm, Amersham).
Antigen capture ELISA
Protein G sepharose column (Pharmacia) 4 Fast Flow (25 ml) was used to purify IgG from cell culture supernatants according to the manufacturer's instructions. For the antigen capture ELISA, 50 μl of a 13.5 μg/ml AG3.0 solution in distilled water were dried onto 96-well flat-bottom polystyrene assay plates (PRO BIND, BD Biosciences) overnight at 37°C. The next day, the monoclonal antibody layer was blocked with 100 μl PBS/2% nonfat milk powder for 1 hour and subsequently washed 3 times with PBS/0.05% Tween20. SIV/HIV supernatant or SIV/HIV infected cells were lysed with 0.2 % Tween20 for 30 min., 100 μl lysate plus 50 μl PBS/2% nonfat milk powder/0.05% Tween20 added to the AG3.0 coated wells, and the plates incubated overnight at 4°C and the next day for two hours at 37°C. The plates were washed 3 times with PBS/0.05% Tween20, 50 μl of specific anti-SIV plasma from an infected monkey or plasma pool from HIV-1 infected individuals, diluted 1:1,000 in PBS/2% nonfat milk powder/0.05% Tween20, were added to the wells, and the plates incubated at 37°C for 1 hour. The plates were then washed 5 times with PBS/0.05% Tween20 and under the stream of tab water for five seconds. Finally, 10 μg o-phenylenediaminedihydrochloride (Sigma-Aldrich) in 50 μl PBS pH 6.0 and 0.012 μl H2O2 were added as substrate, and the reaction stopped with 20 μl 2.5 M H2SO4. The optical density of each well was read at 492 nm using a microplate reader.
Immunostaining of cells
Flat bottom 96-well plates were incubated with 5 μg of poly-L-lysine (Sigma) in 100 μl of distilled water overnight at 4°C. Plates were washed twice with PBS, and 200 μl of cell suspension were added to the wells. Plates were incubated for 1 h at 37°C to allow cells to settle, and then the supernatant was removed. Plates were submerged in −20°C methanol and fixed for 30 min. at −20°C before carefully washing three times with PBS and blocking with 100 μl of PBS/2% nonfat milk powder for 1 h at room temperature. The blocking solution was then removed, and 50 μl of the appropriate anti-SIV/HIV plasma diluted in PBS/2% nonfat milk powder added for 1 h at 37°C. Wells were washed five times with PBS and incubated with 50 μl of 1:500 anti-human IgG (Sigma-Aldrich) in PBS/2% nonfat milk powder for 1 h at 37°C. Wells were then washed again five times with PBS and finally the substrate added (10 μg 3-amino-9-ethylcarbazol (Sigma-Aldrich), 2.5 μl dimethylformamide, 47.5 μl 20 mM sodium acetate, pH 5.0, and 0.025 μl H2O2 per well). After 10 to 20 min of incubation with the substrate, SIV/HIV infected cells were stained red. The wells were washed once with PBS, and the substrate reaction stopped with 20% glycerol.
Virus titrations
To determine the TCID50 of virus stocks, supernatants were initially diluted 1:10 and then 11 dilutions steps with 1:3 dilutions were performed. Row 12 served as negative control (medium only). Fifty microliters of virus dilutions or media were added to 2×103 C8166 or Molt4clone8 cells in 8 replicates. The plates were incubated for 7 days, and the SIV/HIV replication was monitored by antigen capture assay or cellular immunostaining.
Surface plasmon resonance (SPR) analysis
To determine the binding affinity of Gag proteins derived from the different SIVs to the monoclonal AG3.0 mAb, samples were analyzed in duplicate with a BIAcore ×100 instrument (GE Healthcare) at 25°C in HBS-EP+ buffer (10 mM Hepes, 150 mM NaCl, 3 mM EDTA, 0.05% Surfactant P20, pH 7.4) using the mouse antibody capture kit (GE Healthcare). The final immobilization level of anti-mouse IgG antibody was 8,500 RU on both flow cells (Fc1 and Fc2) as recommended by the manufacturer. The antibody was captured at a final concentration of 20 μg/ml for 20 sec at a flow rate of 10 μl/min and a constant level of 500 RU and 120 RU for measuring SIV Gag binding. The different analytes were injected under the following conditions: The association/dissociation times for SIVagmVer Gag and SIVmac Gag were 720/1,200 sec and for SIVagmSab Gag 1,080/5,400 sec. Furthermore, association and dissociation curves were estimated for all 3 Gag proteins using an injection period of 1,080 sec and a dissociation time of 3,000 sec at a flow rate of 5 μl/min. The regeneration buffer consisted of 10 mM glycine/HCl, pH 1.7. The apparent kD values were determined using the BiaEvaluation software (GE Healthcare). All three Gag preparations (SIVagmVer, SIVagmSab, and SIVmac) were produced from native purified virus.
Sequencing of the AG3.0 heavy and light chain variable regions
RNA was isolated from 1×107 AG3.0 hybridoma cells using the RNeasy plus kit (Qiagen) according to the manufacturer's instructions and eluted in 30 μl DEPC-treated H2O. cDNA was generated with the Cloned AMV cDNA synthesis kit (Invitrogen) using oligodT primers and a temperature of 50°C for 1h. Amplification of variable region fragments was performed with 1.25U Pfu DNA polymerase (Fermentas) in the supplied PCR buffer containing 2 mM MgSO4, 0.2 mM dNTPs, and 25 pmol of each primer per 50 μl reaction. Forward and reverse primers from a mouse IgG library primer set (Progen) were used in all possible combinations for light and heavy chain fragments. Amplification cycles were as follows: initial denaturation for 5 min at 95°C, followed by 35 cycles of 30 sec/95°C, 30sec/55°C, 60sec/72°C and a final elongation of 10 min at 72°C. Amplified PCR products of approximately 400kb were purified using a PCR purification kit (Fermentas) or, if more than one product was generated, separated on a 1.5% agarose gel and subsequently extracted using a gel extraction kit (Qiagen). Sequencing was performed on an ABI 3500DX sequencer. Sequences obtained were analysed using Vbase2 (http://www.vbase2.org) to identify the complementarity determining regions of the heavy and light chain variable fragments.
ACKNOWLEDGMENTS
This paper is dedicated to the memory of Jonathan S. Allan who generated and initially characterized the AG3.0 monoclonal antibody in his laboratory. We thank Evelyn M. Whitehead, Manuela Schütze, and Nicole Norley for their excellent technical assistance. We would also like to thank Phyllis Kanki and Soulemayne M'Boup for the HIV-2 serum, and the U.S. Air Force Wilford Hall AIDS program for the HIV-1 serum. This work was funded in part from NIH grants AI-28273 and AI-41396 to JSA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aghokeng AF, Ayouba A, Mpoudi-Ngole E, Loul S, Liegeois F, Delaporte E, Peeters M. Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect Genet Evol. 2010;10(3):386–96. doi: 10.1016/j.meegid.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghokeng AF, Bailes E, Loul S, Courgnaud V, Mpoudi-Ngolle E, Sharp PM, Delaporte E, Peeters M. Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages. Virology. 2007;360(2):407–18. doi: 10.1016/j.virol.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan JS, Short M, Taylor ME, Su S, Hirsch VM, Johnson PR, Shaw GM, Hahn BH. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65(6):2816–28. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan JS, Whitehead EM, Strout K, Short M, Kanda P, Hart TK, Bugelski PJ. Strong association of simian immunodeficiency virus (SIVagm) envelope glycoprotein heterodimers: possible role in receptor-mediated activation. AIDS Res Hum Retroviruses. 1992;8(12):2011–20. doi: 10.1089/aid.1992.8.2011. [DOI] [PubMed] [Google Scholar]
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300(5626):1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203(5):1357–69. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–71. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- Beer BE, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley SG, Kurth R, Gautier JP, Gautier-Hion A, Vallet D, Sharp PM, Hirsch VM. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73(9):7734–44. doi: 10.1128/jvi.73.9.7734-7744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer BE, Foley BT, Kuiken CL, Tooze Z, Goeken RM, Brown CR, Hu J, St Claire M, Korber BT, Hirsch VM. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411) J Virol. 2001;75(24):12014–27. doi: 10.1128/JVI.75.24.12014-12027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirnaert E, Willems B, Peeters M, Bouckaert A, Heyndrickx L, Zhong P, Vereecken K, Coppens S, Davis D, Ndumbe P, Janssens W, van der Groen G. Design and evaluation of an in-house HIV-1 (group M and O), SIVmnd and SIVcpz antigen capture assay. J Virol Methods. 1998;73(1):65–70. doi: 10.1016/s0166-0934(98)00044-5. [DOI] [PubMed] [Google Scholar]
- Bibollet-Ruche F, Bailes E, Gao F, Pourrut X, Barlow KL, Clewley JP, Mwenda JM, Langat DK, Chege GK, McClure HM, Mpoudi-Ngole E, Delaporte E, Peeters M, Shaw GM, Sharp PM, Hahn BH. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade. J Virol. 2004;78(14):7748–62. doi: 10.1128/JVI.78.14.7748-7762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binninger-Schinzel D, Muller D, Wolf T, Krause B, Meye B, Winskowsky G, Raupp S, Norley S, Brodt R, Werner A. Characterization of a chemokine receptor CCR5-negative T cell line and its use in determining human immunodeficiency virus type 1 phenotype. J Med Virol. 2008;80(2):192–200. doi: 10.1002/jmv.21064. [DOI] [PubMed] [Google Scholar]
- Borsetti A, Ohagen A, Gottlinger HG. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72(11):9313–7. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11(7):672–5. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- Broussard SR, Staprans SI, White R, Whitehead EM, Feinberg MB, Allan JS. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J Virol. 2001;75(5):2262–75. doi: 10.1128/JVI.75.5.2262-2275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey MA, Santos-Ferreira MO, Laurent AG, Dauguet C, Katlama C, Rouzioux C, et al. Isolation of a new human retrovirus from West African patients with AIDS. Science. 1986;233(4761):343–6. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- Courgnaud V, Abela B, Pourrut X, Mpoudi-Ngole E, Loul S, Delaporte E, Peeters M. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different Cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J Virol. 2003a;77(23):12523–34. doi: 10.1128/JVI.77.23.12523-12534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Formenty P, Akoua-Koffi C, Noe R, Boesch C, Delaporte E, Peeters M. Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African colobids: SIVwrc from Western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus) J Virol. 2003b;77(1):744–8. doi: 10.1128/JVI.77.1.744-748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Pourrut X, Bibollet-Ruche F, Mpoudi-Ngole E, Bourgeois A, Delaporte E, Peeters M. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J Virol. 2001;75(2):857–66. doi: 10.1128/JVI.75.2.857-866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazza MC, Ekwalanga M, Nende M, Shamamba KB, Bitshi P, Paraskevis D, Saragosti S. Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent's Mona monkey (Cercopithecus mona denti) J Virol. 2005;79(13):8560–71. doi: 10.1128/JVI.79.13.8560-8571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger HG. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68(12):8180–7. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomsgaard A, Hirsch VM, Allan JS, Johnson PR. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology. 1991;182(1):397–402. doi: 10.1016/0042-6822(91)90689-9. [DOI] [PubMed] [Google Scholar]
- Franchini G, Gurgo C, Guo HG, Gallo RC, Collalti E, Fargnoli KA, Hall LF, Wong-Staal F, Reitz MS., Jr. Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987;328(6130):539–43. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372(6504):359–62. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251(1):1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, Kalyanaraman VS, Mann D, Sidhu GD, Stahl RE, Zolla-Pazner S, Leibowitch J, Popovic M. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):865–7. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278(5339):849–53. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- Ganser BK, Cheng A, Sundquist WI, Yeager M. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. Embo J. 2003;22(12):2886–92. doi: 10.1093/emboj/cdg276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelderblom HR. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5(6):617–37. [PubMed] [Google Scholar]
- Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273(5272):231–5. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- Henderson LE, Bowers MA, Sowder RC, 2nd, Serabyn SA, Johnson DG, Bess JW, Jr., Arthur LO, Bryant DK, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66(4):1856–65. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Campbell BJ, Bailes E, Goeken R, Brown C, Elkins WR, Axthelm M, Murphey-Corb M, Sharp PM. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J Virol. 1999;73(2):1036–45. doi: 10.1128/jvi.73.2.1036-1045.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Dapolito GA, Goldstein S, McClure H, Emau P, Fultz PN, Isahakia M, Lenroot R, Myers G, Johnson PR. A distinct African lentivirus from Sykes' monkeys. J Virol. 1993a;67(3):1517–28. doi: 10.1128/jvi.67.3.1517-1528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, McGann C, Dapolito G, Goldstein S, Ogen-Odoi A, Biryawaho B, Lakwo T, Johnson PR. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology. 1993b;197(1):426–30. doi: 10.1006/viro.1993.1606. [DOI] [PubMed] [Google Scholar]
- Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339(6223):389–92. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- Hu J, Switzer WM, Foley BT, Robertson DL, Goeken RM, Korber BT, Hirsch VM, Beer BE. Characterization and comparison of recombinant simian immunodeficiency virus from drill (Mandrillus leucophaeus) and mandrill (Mandrillus sphinx) isolates. J Virol. 2003;77(8):4867–80. doi: 10.1128/JVI.77.8.4867-4880.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MJ, Hui H, Robertson DL, Muller MC, Barre-Sinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 1994;13(12):2935–47. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Fomsgaard A, Allan J, Gravell M, London WT, Olmsted RA, Hirsch VM. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J Virol. 1990;64(3):1086–92. doi: 10.1128/jvi.64.3.1086-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Hui HX, Kappes JC, Haggarty BS, Hoxie JA, Arya SK, Shaw GM, Hahn BH. Molecular characterization of an attenuated human immunodeficiency virus type 2 isolate. J Virol. 1990;64(2):890–901. doi: 10.1128/jvi.64.2.890-901.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairmore MD, Hofheinz DE, Letvin NL, Stoner CS, Pearlman S, Toedter GP. Detection of simian immunodeficiency virus and human immunodeficiency virus type 2 capsid antigens by a monoclonal antibody-based antigen capture assay. AIDS Res Hum Retroviruses. 1993;9(6):565–71. doi: 10.1089/aid.1993.9.565. [DOI] [PubMed] [Google Scholar]
- Leblanc JJ, Perez O, Hope TJ. Probing the structural states of human immunodeficiency virus type 1 pr55gag by using monoclonal antibodies. J Virol. 2008;82(5):2570–4. doi: 10.1128/JVI.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP. WHO-IUIS Nomenclature Subcommittee for immunoglobulins and T cell receptors report. Immunogenetics. 2007;59(12):899–902. doi: 10.1007/s00251-007-0260-4. [DOI] [PubMed] [Google Scholar]
- Liegeois F, Courgnaud V, Switzer WM, Murphy HW, Loul S, Aghokeng A, Pourrut X, Mpoudi-Ngole E, Delaporte E, Peeters M. Molecular characterization of a novel simian immunodeficiency virus lineage (SIVtal) from northern talapoins (Miopithecus ogouensis) Virology. 2006;349(1):55–65. doi: 10.1016/j.virol.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Liegeois F, Lafay B, Formenty P, Locatelli S, Courgnaud V, Delaporte E, Peeters M. Full-length genome characterization of a novel simian immunodeficiency virus lineage (SIVolc) from olive Colobus (Procolobus verus) and new SIVwrcPbb strains from Western Red Colobus (Piliocolobus badius badius) from the Tai Forest in Ivory Coast. J Virol. 2009;83(1):428–39. doi: 10.1128/JVI.01725-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli S, Lafay B, Liegeois F, Ting N, Delaporte E, Peeters M. Full molecular characterization of a simian immunodeficiency virus, SIVwrcpbt from Temminck's red colobus (Piliocolobus badius temminckii) from Abuko Nature Reserve, The Gambia. Virology. 2008a;376(1):90–100. doi: 10.1016/j.virol.2008.01.049. [DOI] [PubMed] [Google Scholar]
- Locatelli S, Liegeois F, Lafay B, Roeder AD, Bruford MW, Formenty P, Noe R, Delaporte E, Peeters M. Prevalence and genetic diversity of simian immunodeficiency virus infection in wild-living red colobus monkeys (Piliocolobus badius badius) from the Tai forest, Cote d'Ivoire SIVwrc in wild-living western red colobus monkeys. Infect Genet Evol. 2008b;8(1):1–14. doi: 10.1016/j.meegid.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Lohman BL, Higgins J, Marthas ML, Marx PA, Pedersen NC. Development of simian immunodeficiency virus isolation, titration, and neutralization assays which use whole blood from rhesus monkeys and an antigen capture enzyme-linked immunosorbent assay. J Clin Microbiol. 1991;29(10):2187–92. doi: 10.1128/jcm.29.10.2187-2192.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73(6):1067–78. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Mammano F, Ohagen A, Hoglund S, Gottlinger HG. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68(8):4927–36. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J, Farrell L, Ross R, Barklis E. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J Virol. 1996;70(8):5106–14. doi: 10.1128/jvi.70.8.5106-5114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mervis RJ, Ahmad N, Lillehoj EP, Raum MG, Salazar FH, Chan HW, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62(11):3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Connors M. Frequency and function of HIV-specific CD8(+) T cells. Immunol Lett. 2001;79(1–2):141–50. doi: 10.1016/s0165-2478(01)00276-0. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3(9):763–70. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, Jacquelin B, Bohm R, Marx PA, Barre-Sinoussi F, Hirsch VM, Muller-Trutwin MC, Lackner AA, Veazey RS. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J Virol. 2006;80(10):4858–67. doi: 10.1128/JVI.80.10.4858-4867.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potash MJ, Bentsman G, Muir T, Krachmarov C, Sova P, Volsky DJ. Peptide inhibitors of HIV-1 protease and viral infection of peripheral blood lymphocytes based on HIV-1 Vif. Proc Natl Acad Sci U S A. 1998;95(23):13865–8. doi: 10.1073/pnas.95.23.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reicin AS, Ohagen A, Yin L, Hoglund S, Goff SP. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70(12):8645–52. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reicin AS, Paik S, Berkowitz RD, Luban J, Lowy I, Goff SP. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69(2):642–50. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma R, Noser JA, Ohmine S, Ikeda Y. Rhesus monkey TRIM5alpha restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat Med. 2007;13(5):631–5. doi: 10.1038/nm1562. [DOI] [PubMed] [Google Scholar]
- Santiago ML, Lukasik M, Kamenya S, Li Y, Bibollet-Ruche F, Bailes E, Muller MN, Emery M, Goldenberg DA, Lwanga JS, Ayouba A, Nerrienet E, McClure HM, Heeney JL, Watts DP, Pusey AE, Collins DA, Wrangham RW, Goodall J, Brookfield JF, Sharp PM, Shaw GM, Hahn BH. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii) J Virol. 2003;77(13):7545–62. doi: 10.1128/JVI.77.13.7545-7562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni I, Trapp S, Santarcangelo AC, Piacentini V, Pugliese K, Baur A, Federico M. HIV-1 Nef enhances both membrane expression and virion incorporation of Env products. A model for the Nef-dependent increase of HIV-1 infectivity. J Biol Chem. 2004;279(22):22996–3006. doi: 10.1074/jbc.M312453200. [DOI] [PubMed] [Google Scholar]
- Shibata R, Miura T, Hayami M, Sakai H, Ogawa K, Kiyomasu T, Ishimoto A, Adachi A. Construction and characterization of an infectious DNA clone and of mutants of simian immunodeficiency virus isolated from the African green monkey. J Virol. 1990;64(1):307–12. doi: 10.1128/jvi.64.1.307-312.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm M, Shahabuddin M, Chao W, Allan JS, Volsky DJ. Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J Virol. 1995;69(7):4582–6. doi: 10.1128/jvi.69.7.4582-4586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MA, Robertson DL, Hui H, Allan JS, Shaw GM, Hahn BH. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology. 1997;228(2):394–9. doi: 10.1006/viro.1996.8387. [DOI] [PubMed] [Google Scholar]
- Srinivasakumar N, Hammarskjold ML, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69(10):6106–14. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. 1997 [PubMed] [Google Scholar]
- Takemura T, Ekwalanga M, Bikandou B, Ido E, Yamaguchi-Kabata Y, Ohkura S, Harada H, Takehisa J, Ichimura H, Parra HJ, Nende M, Mubwo E, Sepole M, Hayami M, Miura T. A novel simian immunodeficiency virus from black mangabey (Lophocebus aterrimus) in the Democratic Republic of Congo. J Gen Virol. 2005;86(Pt 7):1967–71. doi: 10.1099/vir.0.80697-0. [DOI] [PubMed] [Google Scholar]
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372(6504):363–5. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Thorstensson R, Walther L, Putkonen P, Albert J, Biberfeld G. A capture enzyme immunoassay for detection of HIV-2/SIV antigen. J Acquir Immune Defic Syndr. 1991;4(4):374–9. [PubMed] [Google Scholar]
- Tsujimoto H, Hasegawa A, Maki N, Fukasawa M, Miura T, Speidel S, Cooper RW, Moriyama EN, Gojobori T, Hayami M. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature. 1989;341(6242):539–41. doi: 10.1038/341539a0. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S, Butel C, Liegeois F, Yangda B, Sharp PM, Mpoudi-Ngole E, Delaporte E, Hahn BH, Peeters M. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368(1):155–71. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444(7116):164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn F, Peeters M. The origins of HIV and implications for the global epidemic. Curr Infect Dis Rep. 2007;9(4):338–46. doi: 10.1007/s11908-007-0052-x. [DOI] [PubMed] [Google Scholar]
- VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev. 2006;19(4):728–62. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese FD, Copeland TD, Oroszlan S, Gallo RC, Sarngadharan MG. Biochemical and immunological analysis of human immunodeficiency virus gag gene products p17 and p24. J Virol. 1988;62(3):795–801. doi: 10.1128/jvi.62.3.795-801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschoor EJ, Fagrouch Z, Bontjer I, Niphuis H, Heeney JL. A novel simian immunodeficiency virus isolated from a Schmidt's guenon (Cercopithecus ascanius schmidti) J Gen Virol. 2004;85(Pt 1):21–4. doi: 10.1099/vir.0.19427-0. [DOI] [PubMed] [Google Scholar]
- Vogt VM. Retroviral virions and genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 1997. 1997. [PubMed] [Google Scholar]
- Von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77(9):5439–50. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CT, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J Virol. 1993;67(7):4264–73. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly K, Chesebro B. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12(4):288–93. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- Wills JW, Craven RC. Form, function, and use of retroviral gag proteins. AIDS. 1991;5(6):639–54. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]