Abstract
In this study we report that Streptomyces asterosporus DSM 41452 is a producer of new molecules related to the non-ribosomal cyclodepsipeptide WS9326A and the polyketide Annimycin. S. asterosporus DSM 41452 is shown to produce six cyclodepsipeptides and peptides, WS9326A to G. Notably, the compounds WS9326F and WS9326G have not been described before. The genome of S. asterosporus DSM 41452 was sequenced and a putative WS9326A gene cluster was identified. Gene deletion experiments confirmed that this cluster is responsible for the biosynthesis of WS9326A to G. Additionally, a gene deletion experiment demonstrated that sas16 encoding a cytochrome P450 monooxygenase is involved in the synthesis of the novel E-2,3-dehydrotyrosine residue found in WS9326A and its derivatives. An insertion mutation within the putative Annimycin gene cluster led to the production of a new Annimycin derivative, Annimycin B, which exhibited modest inhibitory activity against Plasmodium falciparum.
The WS9326A biosynthetic gene cluster in Streptomyces asterosporus DSM 41452 was identified through bioinformatic analysis and mutagenesis. Analysis of the cluster discovered gene sas16 encoding a cytochrome P450 monooxygenase which is involved in the biosynthesis of the E-2,3-dehydrotyrosine residue in WS9326As. A mutant lacking sas16 produced a new WS9326A derivative.
Introduction.
Nonribosomal peptide synthetases (NRPSs) represents one of the most important classes of enzymes involved in natural product biosynthesis[1]. Many pharmaceutically important antibiotics are biosynthesized by NRPSs[2]. Important NRPS derived antibiotics have been discovered from various kinds of microorganisms[3], including vancomycin, produced by Amycolatopsis orientalis[4], and daptomycin, produced by Streptomyces roseosporus[5]. NRPS natural products display a wide range of chemical modifications that deviate from standard peptides, including halogenation, hydroxylation, N-methylation, α/β-dehydrogenation.
The cyclodepsipeptide WS9326A is notable for its unusual spectrum of bioactivities and its production by a number of Streptomyces strains. The molecule was first isolated from Streptomyces violaceusniger sp. 9326 by researchers at Fujisawa Pharmaceutical Co. on the basis of its novel activity as a tachykinin receptor agonist (IC50 = 3.6 μM)[6]. A total chemical synthesis confirmed the structure of WS9326A and probed the relationship of the N-acyl group to bioactivity[7]. Subsequently, WS9326A and series of congeners were isolated from Streptomyces sp. 9078 based on inhibition of an asparaginyl-tRNA synthetase from the filarial nematode parasite Brugia malayl[8]. More recently, WS9326A was found to be a transcriptional inhibitor in Clostridium perfringens, while WS9326B was observed to reduce the toxicity of Staphylococcus aureus to human corneal epithelial cells[9]. WS9326A has also been identified in cultures of Streptomyces calvus ATCC 13382[10]. While the gene cluster encoding WS9326A was identified in this study[10], the specific functions of individual genes were not examined.
WS9326A exhibits an interesting chemical structure. The core NRPS backbone structure consists of four standard amino acids (L-Thr, L-Leu, L-Asn, L-Ser) and three non-proteinogenic amino acids (E-2,3-dehydrotyrosine, D-Phe, and L-allo-threonine). The E-2,3-dehydrotyrosine (ΔTyr) residue is notable for not having been observed previously in NRPS derived peptides. Amino acid residues with α,β-dehydrogenation have been observed in many other important NRPS compound like Telomycin from Streptomyces canus ATCC 12646[11], calcium-dependent antibiotic (CDA) from Streptomyces coelicolor A3(2)[12], and Splenocin from Streptomyces sp. CNQ431[13], along with different mechanisms to form the α,β-alkene. The enzymatic mechanism to form the E-2,3-dehydrotyrosine in WS9326A is still unknown.
Another prominent feature of WS9326A is the N-acyl group, consisting of a Z-pentenyl-cinnamoyl moiety Similar side chains are also found in depsipeptide Skyllamycin[14], pepticinnamins[15], and the dimeric peptides Mohangamide A and B[16]. Structure and bioactivity relationships have shown that the Z-pentenyl-cinnamoyl moiety is essential for activity[17]. Previous biosynthetic studies indicated that this polyketide chain may arise from a complex biosynthetic pathway[14], but the exact mechanism has not been deciphered to date.
We were inspired to study S. asterosporus DSM 41452 in detail upon observing a surprising biosynthetic kinship with Streptomyces calvus ATCC 13382. The genome sequence of S. asterosporus DSM 41452 revealed gene clusters with very high sequence identity to the WS9326A[10] and Annimycin clusters[18] found in S. calvus. Indeed, analysis of culture extracts of S. asterosporus DSM 41452 revealed the presence of WS9326A. It has also been observed that the production of WS9326A in S. calvus is inversely correlated to the production of Annimycin[18]. Therefore, we decided to disrupt the Annimycin gene cluster in S. asterosporus DSM 41452. While titres of WS9326A in the resulting S. asterosporus mutant (S. asterosporus DSM 41452::pUC19Δ3100spec) was slightly improved, we were surprised to observe the production of a new polyketide.
In this study we report that the isolation and elucidation of six WS9326A derivatives from S. asterosporus DSM 41452, among them there are two new truncated derivatives WS9326F and WS9326G. Furthermore, we isolated a new polyketide from the mutant S. asterosporus DSM 41452::pUC19Δ3100spec, a polyene derivative Annimycin B, which shares structural features with annimycin[18] and the antibiotic Sch25424 previously isolated from Kitasatospora sp[19]. Gene inactivation experiments were performed to confirm the identity of the WS9326A cluster. Finally, we were able to show that a P450 cytochrome monooxygenase encoded by sas16 is involved in the biosynthesis of the E-2,3-dehydrotyrosine residue in WS9326A.
Isolation and characterization of WS9326A and derivatives from S. asterosporus DSM 41452.
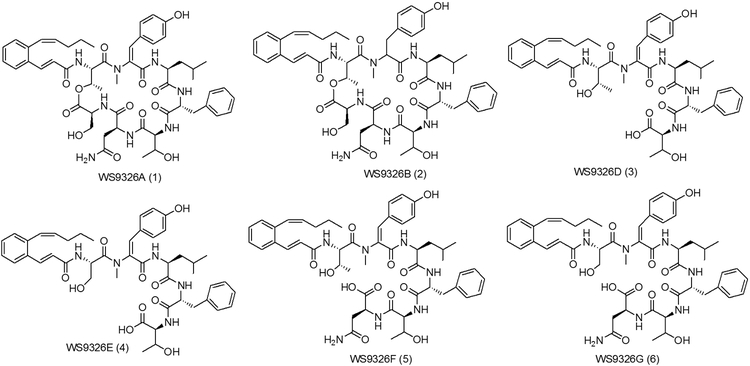
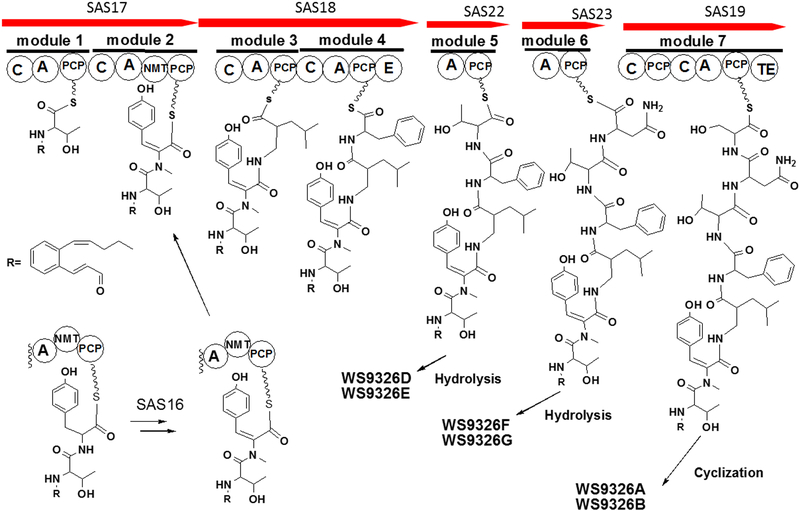
From 10L fermentation of S. asterosporus DSM 41452, four known despipeptides WS9326A (1), WS9326B (2), WS9326D (3), WS9326E (4) were isolated (Figure 1). The NMR and MS/MS spectroscopic data for WS9326A (1) was consistent with previously reported spectra (Supporting information, Table S7)[8], and the amino acid stereochemical configuration was confirmed by Marfey’s method[20] (Supporting Information). The 2E and 10Z alkene configurations of the N-acyl moiety was confirmed by 1H-NMR coupling constants for H2/H3 (J = 15.5 Hz) and H10/H11 (J =11.4 Hz). Likewise, the ortho configured aromatic ring was distinguished by a multiplet arising from four hydrogens (δ 7.25 ppm). Compounds WS9326B (2), WS9326D (3) and WS9326E (4) were assigned based on their MS/MS spectra (Supporting information, Figure S9-S12). Furthermore, we observed two additional compounds by LC-ESI-MS with m/z values of 966.4 (compound 5) and 952.4 (compound 6) (Figure S2). Both compounds were isolated for subsequent NMR and MS/MS spectroscopic analysis to reveal two new WS9326A analogues: WS9326F (5) and WS9326G (6).
Figure 1.
Chemical structures of WS9326A and derivatives produced by Streptomyces asterosporus DSM 41452.
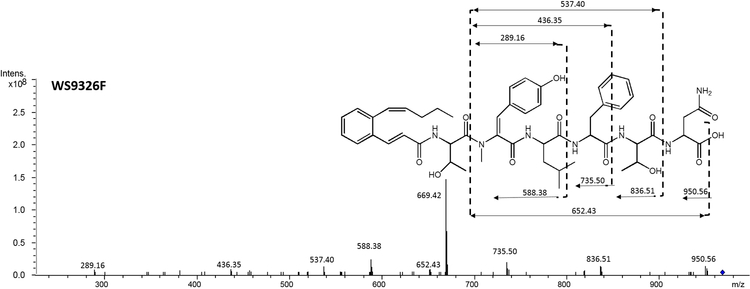
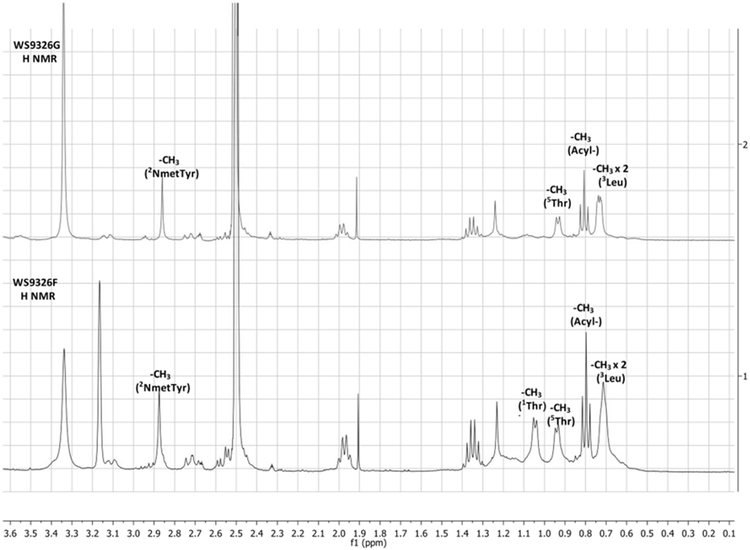
WS9326F (5) (Figure 1) was obtained as white amorphous powder. The molecular ion observed for 5 by HRESI-MS corresponded to a molecule with the formula C51H65N7O12 and 23 degrees of unsaturation (obsvd: m/z = 968.4750 [M+H]+, calcd for C51H66N7O12, m/z = 968.4764) The structure of 5 was determined by careful comparison with the NMR spectroscopic data obtained from WS9326A[8]. The 1H NMR (400 MHz, DMSO-d6) spectrum of 5 (Table S7) exhibited significant signal characteristics of WS9326A, including the presence of five α-amino methines protons at δ 4.29 (1H, m), 4.49 (1H, m), 4.62 (1H, m), 4.23 (1H, m) and 3.17 (1H, m) ppm, which correlated with five sp3 α-amino methine carbons at δ 52.4 (1Thr), 49.6 (3Leu), 54.6 (4Phe), 58.7 (5Thr) and 48.8 (6Asn) ppm, respectively, in the HSQC spectrum. Based on the HMQC spectrum, six sets of methyl protons (δ 0.79, 1.05, 2.87, 0.70, 0.63 and 0.92 ppm) were assigned to the corresponding carbon atoms at δ 14.2 (Acyl-C14), 20.3 (1Thr), 34.8 (2N-methyl-ΔTyr), 22.6 (3Leu), 23.3 (3Leu) and 22.7 (5Thr), respectively. In addition, signals corresponding to 8 carbonyl carbons at δ 165.6, 170.8, 165.2, 171.8, 170.6, 170.2 and 172.0 ppm were observed in the 13C NMR spectrum (100 MHz, DMSO-d6) which further indicate the presence of 6 amino acid residues in 5. Shared MS/MS fragmentation patterns (Figure 2) were observed in the spectra of WS9326A, WS9326D and 5, corresponding to the peptides 2N-methyl-ΔTyr-3Leu-4Phe (−436.35 Da), 2N-methyl-ΔTyr-3Leu-4Phe-5Thr (−537.40 Da), and Acyl-1Thr-2N-methyl-ΔTyr-3Leu-4Phe (−735.50 Da). In contrast, unique m/z values in the MS/MS spectrum of 5 were identified: 588.38 Da (2N-methyl-ΔTyr −3Leu) and 836.51 Da (Acyl-1Thr-2N-methyl-ΔTyr −3Leu-4Phe-5Thr). Accordingly, 5 is a new WS9326A analog with the structure Acyl-1Thr-2N-methyl-ΔTyr-3Leu-4Phe-5Thr-6Asn (Figure 2). As 5 is a truncated analog of WS9326A, it is predicted to have the same amino acid configuration of the ones in WS9326A.
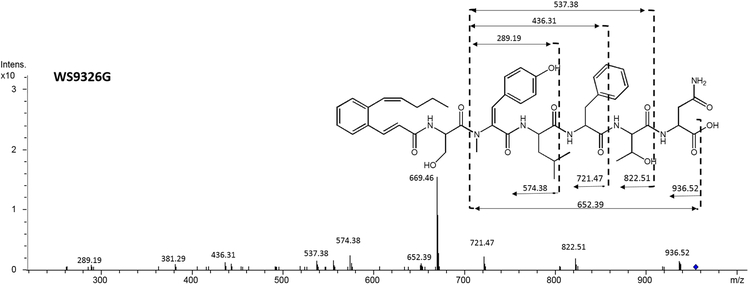
Figure 2.
MS/MS spectra of WS9326F(A) and WS9326G (B) produced by S. asterosporus DSM41452; (C) Partial H NMR Spectrum comparison between WS9326F and WS9326G
WS9326G (6) was also obtained as white amorphous powder. The molecular ion observed by HRESI-MS for 6 corresponded to a molecular formula of C50H63N7O12 (obsvd: m/z = 954.4644 [M+H]+ (calcd for C50H64N7O12, 954.4607). This reveals that 6 is 14 Da smaller than 5 which corresponds to the absence of a methyl group.
The chemical structure of 6 (Figure 1) was elucidated by comparing the MS/MS fragmentation spectrum with that obtained from 5 (Figure 2A and 3B). In addition to the mutual MS/MS mass fragments of 289.19 Da, 436.31 Da, 537.38 Da, three unique masses are observed for 6: 822.51 Da (Acyl-1Ser-2N-methyl-ΔTyr-3Leu-4Phe-5Thr), 721.47 Da (Acyl-1Ser-2N-methyl-ΔTyr-3Leu-4Phe), and 574.38 Da (Acyl-1Ser-2N-methyl-ΔTyr-3Leu). These results demonstrate that 6 shares a similar chemical structure with 5, exception of a methyl group located within the N-terminus. Furthermore, compared to 5, the 1H NMR spectrum of 6 (Figure 2C) reveals only five methyl proton signals (δ 0.79, 1.04, 2.87, 0.70, and 0.63 ppm). This agrees with the presence of an N-terminal Ser in 6 versus Thr in 5. Therefore, 6 was proven to be a new WS9326A analog with the chemical backbone: Acyl-1Ser-2NmetTyr-3Leu-4Phe-5Thr-6Asn, and its absolute configuration also was assigned as that resembling WS9326A.
Figure 3.
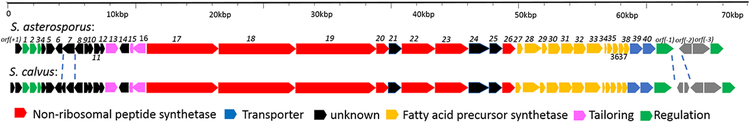
(A) Organization comparison of the WS9326A gene clusters in S. calvus ATCC 13382 and S. asterosporus DSM 41452; (B) NRPS domain organization are shown in the order for the WS9326A biosynthetic assembly line. Domain notation: C, condensation; A, adenylation; PCP, peptidyl carrier protein; E, epimerization; NMT, N-methyltransferase; TE, thioesterase.
Identification and Analysis of the WS9326A gene cluster in S. asterosporus DSM 41452.
The chemical structure of WS9326A predicts that a modular mixed PKS/NRPS gene cluster is involved in its biosynthesis[2]. To find the corresponding biosynthesis gene cluster, the genome of S. asterosporus DSM 41452 was sequenced. Through bioinformatic analysis and gene disruption experiment, one candidate NRPS gene cluster matching our expectations was identified. The cluster is very similar to the predicted WS9326A cluster identified in the S. calvus ATCC 13382 genome[10].
The WS9326A gene cluster is 60.3 kb long and consists of 40 open reading frames (Table S6). To attempt to define the boundaries of the WS9326A gene cluster in S. asterosporus DSM 41452, gene disruption was performed by single crossover. The boundary gene sas1 encodes a protein which shares 48% amino acid identity with a two-component sensor histidine kinase from S. olivaceus (Accession number WP_052410686.1). After deletion of sas1 a mutant strain was obtained. Analysis of the culture extract by HPLC-ESI/MS showed that this mutant did not produce WS9326A and its derivatives, suggesting the sas1 is involved in the biosynthesis of WS9326A in this strain (Figure S4). In contrast, disruption of orf(−1) encoding a putative regulatory protein led to a producing strain, indicating that orf(−1) is not involved in WS9326A biosynthesis (Figure S4). Taken together, our results strongly supported the hypothesis that genes sas1-sas40 are responsible for the biosynthesis of WS9326A and its analogs.
Within the putative SAS gene cluster 18 genes are predicted to be involved in the biosynthesis of the PKS-derived acyl side chain. Very similar genes are found in the Skyllmycin gene cluster[14] (Table S5). In contrast to the Skyllmycin gene cluster, sky11 encoding a putative carboxyltransferase, is absent in our cluster. Thus malonyl-CoA as the starting building block is most likely derived from the primary metabolism in our strain. Further bioinformatic analysis of the genes within this locus revealed that six genes (sas7, sas8, sas30, sas31, sas32, and sas33) are predicted to encode 3-oxoacyl-ACP synthases. These might be involved in a series of condensation reactions with acetyl-CoA and malonyl-CoA units to form the N-acyl C14-polyene before aromatic ring formation occurs[14]. The terminal C12-C13 double bond in this C14-polyene intermediate might be further reduced by a reductase encoded by sas21, which show significant homology (96% amino acid identity) to an oxidoreductase from S. griseoflavus. The configurations of the C14-acyl group in WS9326A and the C12-acyl group in skyllamycin are identical (2E, 10Z) according to NMR spectroscopic data (Table S7). Moreover, the double bond configuration at C4, C6, and C8 may be important for aromatization during biosynthesis. The required configuration conversion is most likely to be introduced by the gene product of sas27 encoding an isomerase, which shares 62% amino acid identity with Sky27 and 57% amino acid identity with Has16 involved in haoxinamide biosynthesis[14]. Finally, aromatization to form the benzene ring could be catalyzed by either the putative oxidoreductase SAS24 or the phytoene dehydrogenase SAS28.
The WS9326A gene cluster contains 5 genes encoding nonribosomal peptide synthetases (sas17, sas18, sas19, sas22, and sas23). The deduced amino acid sequence of SAS17 consists of two modules. The first module might be responsible for loading the first amino acid, Ser or Thr. The second module contains a N-methyltransferase domain, which is involved in the formation of N-methyl-ΔTyr as component of WS9326A. A mutant (ΔNmet) in which the N-methyltransferase encoding gene was deleted by an in-frame gene deletion did not produce any WS9326A derivative (Figure S4). This implies that N-methylation of tyrosine is important for downstream substrate recognition by the condensation domain.
SAS18 also contains two modules. The first module is predicted to be responsible for the incorporation of Leu and the second module for the incorporation of D-Phe (Table S4). Marfey’s analysis of WS9326A confirmed the presence of D-Phe in this molecule (Supporting Information). The epimerization of the second module might catalyze the epimerization of L-Phe into its D-stereoisomer. SAS19 consists of one module with the architecture C-A-PCP-C-A-PCP-TE. The module is responsible for the incorporation of Ser and also for the cyclization and release of the depsipeptides WS9326A and WS9326B. SAS22 and SAS23 contain a Thr-specific A domain and an Asn-specific A domain (Table S4), respectively. Both trans-acting domains are predicted to interact with the condensation domain in module 7 encoded by sas19 to catalyze the incorporation of Thr and Asn in the assembly line. A predicted type II thioesterase is encoded by sas20. Although the function of SAS20 in the biosynthesis of WS9326A is unknown, type II thioesterases are well known to serve editing functions in PKS and NRPS systems[21]. SAS20 may also mediate release of the linear peptides WS9326D, E, F, and G from the trans acting domains SAS22 and SAS23.
Three predicted regulatory genes were found in the 3’-region of the SAS gene cluster. Including the essential biosynthetic gene sas1, encoding a two-component sensor histidine kinase, the gene sas2 shows homology to the LuxR transcriptional regulator found in S. canus [WP_059209681.1], and sas3 is predicted to encode a LysR transcription factor. In addition, 3 transporter genes (sas38, sas39 and sas40) are located at the 5’ end of the SAS cluster. According to the recent report given by Ju et al[22], the putative two-component sensor histidine kinase encoded by sas1 could interact with the isomerase encoded by sas27, thereby enabling the epimerization of the Thr to form L-allo-Thr. The gene sas4 is predicted to encode a MbtH-like protein, which shares 46% identical amino acids with PA2412 involved in pyoverdine biosynthesis in Pseudomonas aeruginosa. We postulate that sas4 is involved in the NRPS assembly by interacting with the corresponding A domain[23].
Genes sas13, sas15 and sas16 were predicted to be involved in the formation of the 2,3-dehydrotyrosine residue. According to a BLAST analysis, SAS16 (407 amino acids) has 97% amino acid identity to the cytochrome P450-SU2 (WP_004931872.1) in the strain of Streptomyces griseoflavus, but its function is unknown. SAS16 also shares 40% amino acid identity with an epothilone b hydroxylase from Streptomycres sp. AA4 (Accession no. EFL04897.1), 38% identity with the P450 hydroxylase from Saccharopolyspora erythraea NRRL 2338 (Accession no. CAM02704.1), and 25% identity with P450sky (CYP163B3), a P450 hydroxylase that introduces β-hydroxyl groups on three amino acids in Skyllamycin[24]. Moreover, SAS13 is predicted to be a 3-hydroxyacyl-ACP dehydratase. Accordingly, we hypothesized that the formation of the double bond in 2,3-dehydrotyrosine is formed by a sequence of hydroxylation and dehydration catalyzed by SAS16 and SAS13, respectively.
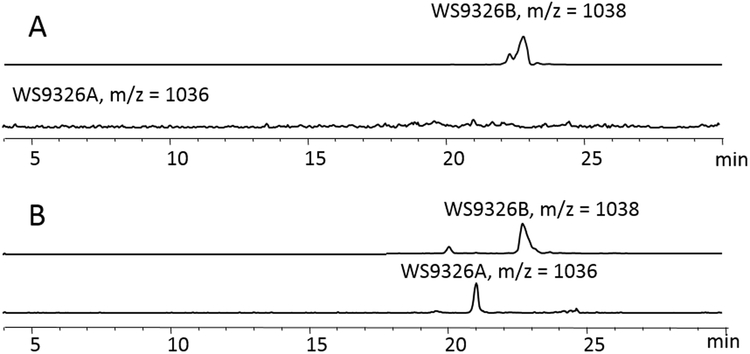
Both genes, sas13 and sas16, were disrupted in S. asterosporus DSM 41452 by single crossover (Supporting information). Crude extracts from both mutants were analyzed by HPLC/ESI-MS (Figure S4). The sas16 mutant did not produce WS9326A, while the sas13 mutant retained the ability to produce WS9326A. As a single crossover mutations may lead to a deleterious polar effect, sas16 was also deleted by replacing sas16 with an antibiotic resistance marker followed by removal of the marker following the REDirect protocol[25]. In this way, an in-frame deletion mutant (S. asterosporus DSM 41452Δsas16) was obtained. Screening the wild-type strain for WS9326A and WS9326B confirmed that both compounds were produced. However, the Δsas16 mutant only produced WS9326B. Complementation of the mutant with sas16 restored WS9326A production (Figure 4), indicating that the encoded P450 monoxygenase is involved in the formation of the double bond of 2,3-dehydrotyrosine.
Figure 4.
(A) The LC/MS extracted ion chromatogram (EICs) for [M-H]- ions corresponding to WS9326A, WS9326B in organic extracts of Streptomyces asterosporus DSM 41452Δsas16; (B) The LC/MS extracted ion chromatogram (EICs) for [M-H]- ions corresponding to WS9326A, WS9326B in organic extracts of S. asterosporus DSM 41452Δsas16::pTESa-SAS16;
Characterization a new polyketide from a S. asterosporus mutant and its possible biosynthesis mechanism.
Analogous to the mutagenesis performed in S. calvus ATCC 13382[18] a single crossover mutation was used to disrupt the predicted Annimycin cluster, yielding the mutant S. asterosporus DSM 41452 :: pUC19Δ3100spec. As observed in S. calvus, we expected to see an increase in production of WS9326A. Instead we detected a new product, Annimycin B (7), in the mutant by LCMS analysis. Annimycin B was isolated as described in the supporting information, and the structure determined by NMR spectroscopy (Table S8).
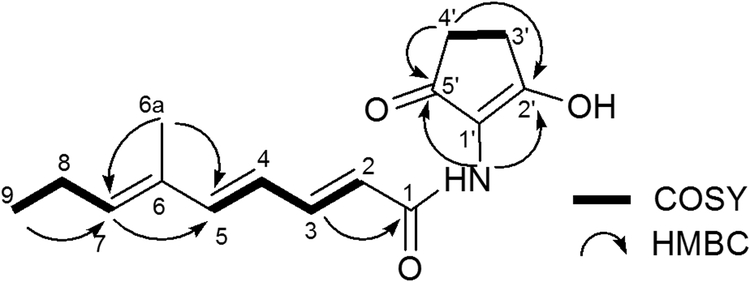
The polyene structure of 7 was elucidated by 1D and 2D NMR spectroscopy. The 1H NMR spectrum indicated the presence of the 4 coupled trans-olefinic protons at δ 5.97 (1H, d, J = 14.9 Hz), 7.38 (1H, dd, J = 14.9 Hz, 11.3 Hz), δ 6.24 (1H, dd, J = 15.3 Hz, 11.3 Hz) and 6.62 ppm (1H, d, J = 15.3 Hz), along with one stand-alone olefinic H-atom at δ 5.76 ppm (1H, t, J = 7.4 Hz). The 13C NMR spectrum shows 15 carbons signals. From correlations in the HSQC spectrum, those carbons were assigned to be 5 olefinic CH carbons at δ 118.7 (C-2), 145.3 (C-3), 123.3 (C-4), 147.5 (C-5) and 141.4 ppm (C-7). Also present are signals for 3 quaternary carbons at δ 133.3 (C-6), 115.2 (C-1ˈ) and 174.5 (C-2ˈ) and two carbonyl carbons at δ 165.4 (C-1) and 197.7 ppm (C-5ˈ).
The overall structure of 7 was assigned by 2D-NMR spectroscopy. The 1H-1H COSY spectrum revealed the presence of three coupled systems. HMBC correlations were observed between the proton at δ 1.79 (H-6a) and carbons at δ 141.4 (C-7) and 147.5 (C-5), consistent with the structure (2E,4E,6E)-6-methylnona-2,4,6-trienamide. Furthermore, the correlation between amino proton NH δ(H) 7.49 and carbons at δ 115.2 (C-1ˈ), 197.7 (C-5ˈ) and 165.4 (C-1) indicated the presence of a C5N ring structure that is connected to the polyene through an amide bond. Taken together, the chemical structure of 7 is consistent with that shown in Figure 5A.
Figure 5.
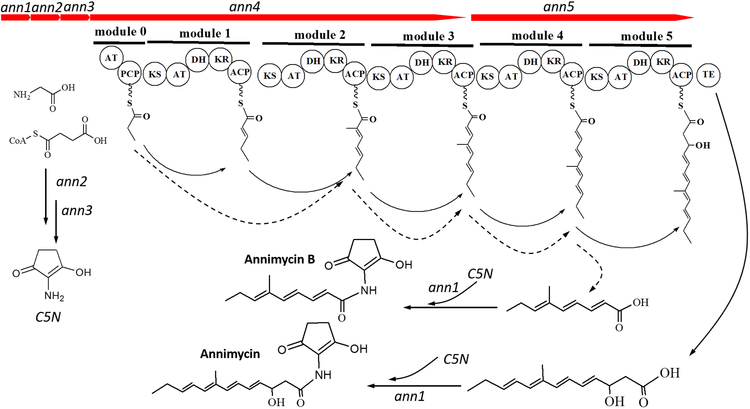
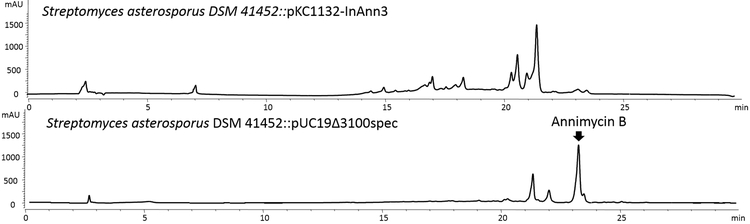
A) Annimycin B chemical structure; B) The proposed biosynthesis of Annimycin B. The arrows depict the sequential synthesis of Annimycin[18]; the dotted arrows depict the module-skipping biosynthesis pathway leading to Annimycin B; C) HPLC chromatograms of the metabolites from S. asterosporus DSM 41452::pKC1132-InAnn3 and S. asterosporus DSM 41452::pUC19Δ3100spec at 340 nm. Annimycin B is marked by the red line.
The significant structural similarity between 7 and Annimycin suggested that 7 is a truncated product, and might originate from the same PKS assembly line. To confirm our hypothesis, we performed gene inactivation experiments of ann3 by single crossover in S. asterosporus DSM 41452. Production of both, 7 and Annimycin was completely abolished in the corresponding mutant strain S. asterosporus DSM 41452::pKC1132-InAnn3.
Biosynthesis of a polyketide by type I PKS, such as that encoded by the annimycin cluster, is typically co-linear[26]. A comparison of the structure of 7 with annimycin reveals that co-linear biosynthesis is no longer operative in the mutant S. asterosporus DSM 41452::pUC19Δ3100spec. Biosynthesis of 7 requires condensation of a propionyl starter unit followed by one methyl-malonyl and two malonyl extender units. 7 appears to be assembled by skipping module 1 of Ann4 (Figure 5). Because the mutation in S. asterosporus DSM 41452::pUC19Δ3100spec disrupts module 5 of Ann5, biosynthesis of 7 also lacks a final extension with a malonyl unit as found in Annimycin biosynthesis[27] (Figure 5).
Biological activity of Annimycin B and WS9326A-G.
WS9326A and its congeners display a surprising range of bioactivities, from tachykinin antagonism[6] to antifilarial activity[8].This prompted us to further investigate the activities of the newly identified WS9326A analogues 5 and 6, along with the annimycin derivative 7. In an antimalarial assay, we evaluated in three Plasmodium falciparum cell-lines (Dd2, HB3 and 3D7), which possess variant drug-resistance phenotypes. Artemether (ATM) was used as positive control. To our surprise 7 had notable activity at the highest concentration tested (2.5 µM) with approximately 30% inhibitory activity against the three cell-lines tested, including the multidrug resistant Dd2 isolate. In contrast, WS9326A and its derivatives 5 and 6 did not demonstrate significant antimalarial activity at the concentrations tested (Figure S3). This suggests 7 would be a useful starting point for the synthesis of derivatives with improved antimalarial activities. Notably, WS9326A and all its derivatives did not show antibiotic activity towards E. coli or Bacillus subtilis.
Conclusions.
In summary, from a new WS9326A producing strain Streptomyces asterosporus DSM 41452, two new WS9326A analogs WS9326F (5) and WS9326G (6) were isolated and structurally characterized. Both compounds are most probably released from the assembly line following an alternative hydrolysis strategy. By means of genome sequencing, bioinformatics analysis and gene inactivation, we identified and validated the corresponding WS9326A biosynthetic gene cluster in Streptomyces asterosporus DSM 41452. Gene deletion studies demonstrated a critical role of N-methylation in WS9326A biosynthesis, and the role of a P450 monoxygenase in forming a 2,3-dehydrotyrosine group. In addition, a new Annimycin analog Annimycin B (7) was discovered from a mutant strain S. asterosporus DSM 41452::pUC19Δ3100spec. Through bioinformatic analysis and gene inactivation experiments, we proposed that Annimycin B is biosynthesized in a module-skipping mechanism, unlike the collinear mechanism followed by Annimycin. Biological activity tests indicate that Annimycin B has modest antimalarial activity. These results set the stage for engineering S. asterosporus DSM 41452 for the production of additional WS9326A and Annimycin analogs.
Supplementary Material
Acknowledgements:
This work was supported by the Division of Preclinical Innovation, National Center for Advancing Translational Sciences, NIH. We thank Ferlaino Sascha (Freiburg University) for acquiring NMR spectra. We are grateful to Dr. Maksym Myronovskyi from the group of Prof. Dr. Andriy Luzhetskyy (Saarbrücken University) for the HR-ESI-MS measurements. We thank Dr. Fu Yan from the group of Prof. Dr. Rolf Müller (Saarbrücken University) for the ESI-MS/MS measurements. We also thank Dr. Roman Makitrynskyy for guidance in mutagenesis. The scholarship of Songya Zhang was supported by China Scholarship Council.
Footnotes
Supporting information available:
Detailed experimental procedures, NMR data and spectra for compound are available.
REFERENCES:
- [1].Sussmuth RD, Mainz A, Angew Chem Int Ed Engl 2017, 56, 3770–3821. [DOI] [PubMed] [Google Scholar]
- [2].Winn M, Fyans JK, Zhuo Y, Micklefield J, Nat Prod Rep 2016, 33, 317–347. [DOI] [PubMed] [Google Scholar]
- [3].Finking R, Marahiel MA, Annu Rev Microbiol 2004, 58, 453–488. [DOI] [PubMed] [Google Scholar]
- [4].Barna JC, Williams DH, Annu Rev Microbiol 1984, 38, 339–357. [DOI] [PubMed] [Google Scholar]
- [5].Baltz RH, Miao V, Wrigley SK, Nat Prod Rep 2005, 22, 717–741. [DOI] [PubMed] [Google Scholar]
- [6].a) Hayashi K, Hashimoto M, Shigematsu N, Nishikawa M, ezaki M, yamashita M, kiyoto S, okuhara M, kohsaka M, imanaka H, J Antibiot 1992, 45, 1055–1063; [DOI] [PubMed] [Google Scholar]; b) hashimoto M, hayashi K, murai M, fujii T, nishikawa M, kiyoto S, okuhara M, kohsaka M, imanaka H, J Antibiot 1992, 45, 1064–1070; [DOI] [PubMed] [Google Scholar]; c) Shigematsu N, Hayashi K, Kayakiri N, Takase S, Hashimoto M, Tanaka H, J Org Chem 1993, 58, 170–175. [Google Scholar]
- [7].Shigematsu N, Kayakiri N, Okada S, Tanaka H, Chem Pharm Bull 1997, 45, 236–242. [DOI] [PubMed] [Google Scholar]
- [8].Yu Z, Vodanovic-Jankovic S, Kron M, Shen B, Org Lett 2012, 14, 4946–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Desouky SE, Shojima A, Singh RP, Matsufuji T, Igarashi Y, Suzuki T, Yamagaki T, Okubo K.-i., Ohtani K, Sonomoto K, Nakayama J, FEMS Microbiol Lett 2015, 362. [DOI] [PubMed] [Google Scholar]
- [10].Johnston CW, Skinnider MA, Wyatt MA, Li X, Ranieri MR, Yang L, Zechel DL, Ma B, Magarvey NA, Nat Commun 2015, 6, 8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fu C, Keller L, Bauer A, Bronstrup M, Froidbise A, Hammann P, Herrmann J, Mondesert G, Kurz M, Schiell M, Schummer D, Toti L, Wink J, Muller R, J Am Chem Soc 2015, 137, 7692–7705. [DOI] [PubMed] [Google Scholar]
- [12].Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BA, Hayes MA, Chem Biol 2002, 9, 1175–1187. [DOI] [PubMed] [Google Scholar]
- [13].Chang C, Huang R, Yan Y, Ma H, Dai Z, Zhang B, Deng Z, Liu W, Qu X, J Am Chem Soc 2015, 137, 4183–4190. [DOI] [PubMed] [Google Scholar]
- [14].a) Pohle S, Appelt C, Roux M, Fiedler HP, Sussmuth RD, J Am Chem Soc 2011, 133, 6194–6205; [DOI] [PubMed] [Google Scholar]; b) Schubert V, Di Meo F, Saaidi PL, Bartoschek S, Fiedler HP, Trouillas P, Süssmuth RD, Chem Eur J 2014, 20, 4948–4955. [DOI] [PubMed] [Google Scholar]
- [15].Omura S, Van der Pyl D, Inokoshi J, Takahashi Y, Takeshima H, J Antibiot 1993, 46, 222–228. [DOI] [PubMed] [Google Scholar]
- [16].Bae M, Kim H, Moon K, Nam SJ, Shin J, Oh KB, Oh DC, Org Lett 2015, 17, 712–715. [DOI] [PubMed] [Google Scholar]
- [17].Shigematsu N, Kayakiri N, Okada S, Tanaka H, Chem Pharm Bull 1997, 45, 236–242. [DOI] [PubMed] [Google Scholar]
- [18].Kalan L, Gessner A, Thaker MN, Waglechner N, Zhu X, Szawiola A, Bechthold A, Wright GD, Zechel DL, Chem Biol 2013, 20, 1214–1224. [DOI] [PubMed] [Google Scholar]
- [19].Yang S-W, Chan T-M, Terracciano J, Patel R, Loebenberg D, Chen G, Patel M, Gullo V, Pramanik B, Chu M, J Antibiot 2005, 58, 192–195. [DOI] [PubMed] [Google Scholar]
- [20].Marfey P, Carlsberg Res Commun 1984, 49, 591–596. [Google Scholar]
- [21].Kotowska M, Pawlik K, Appl Microbiol Biotechnol 2014, 98, 7735–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li Q, Qin X, Liu J, Gui C, Wang B, Li J, Ju J, J Am Chem Soc 2016, 138, 408–415. [DOI] [PubMed] [Google Scholar]
- [23].Zhang W, Heemstra JR Jr., Walsh CT, Imker HJ, Biochemistry 2010, 49, 9946–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Uhlmann S, Süssmuth RD, Cryle MJ, ACS Chem Biol 2013, 8, 2586–2596. [DOI] [PubMed] [Google Scholar]
- [25].Gust B, Challis GL, Fowler K, Kieser T, Chater KF, Proc Natl Acad Sci 2003, 100, 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hertweck C, Angew Chem Int Ed Engl 2009, 48, 4688–4716. [DOI] [PubMed] [Google Scholar]
- [27].Awakawa T, Crüsemann M, Munguia J, Ziemert N, Nizet V, Fenical W, Moore BS, Chembiochem 2015, 16, 1443–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.