Abstract
The accuracy of genotype imputation depends upon two factors: the sample size of the reference panel and the genetic similarity between the reference panel and the target samples. When multiple reference panels are not consented to combine together, it is unclear how to combine the imputation results to optimize the power of genetic association studies. We compared the accuracy of 9,265 Norwegian genomes imputed from three reference panels – 1000 Genomes Phase 3 (1000G), Haplotype Reference Consortium (HRC), and a reference panel containing 2,201 Norwegian participants from the population-based Nord Trøndelag Health Study (HUNT) from low-pass genome sequencing. We observed that the population-matched reference panel allowed for imputation of more population-specific variants with lower frequency (minor allele frequency (MAF) between 0.05% and 0.5%). The overall imputation accuracy from the population-specific panel was substantially higher than 1000G and was comparable with HRC, despite HRC being 15-fold larger. These results recapitulate the value of the population-specific reference panels for genotype imputation. We also evaluated different strategies to utilize multiple sets of imputed genotypes to increase the power of association studies. We observed that testing association for all variants imputed from any panel results in higher power to detect association than the alternative strategy of including only one version of each genetic variant, selected for having the highest imputation quality metric. This was particularly true for lower-frequency variants (MAF < 1%), even after adjusting for the additional multiple testing burden.
Keywords: genotype imputation, GWAS, multiple reference panels, population-specific, study power
Introduction
Many novel disease-associated signals for a wide variety of diseases and traits have been successfully identified using imputation-based meta-analyses(Cheng & Thompson, 2016; Cooper et al., 2008; De Jager et al., 2009; Ge et al., 2016; Horikoshi et al., 2015; Houlston et al., 2008; Jin et al., 2016; Loos et al., 2008; Ruth et al., 2015; Zeggini et al., 2008; Zeggini et al., 2007). Genotype imputation is the process of inferring missing genotypes in study samples using a reference panel of high-density haplotypes(Li, Willer, Sanna, & Abecasis, 2009). Imputation allows variants that are not directly genotyped to be studied without other costs than computation. Previous simulations showed that imputation substantially increases the power of association studies to detect causal loci(Marchini & Howie, 2010; Spencer, Su, Donnelly, & Marchini, 2009). Imputation-based genome-wide association studies (GWAS) have successfully identified novel signals that were undetected in chip-based studies. For example, two disease-associated signals were detected in the 1000G-based imputation(Auton et al., 2015) for the Wellcome Trust Case Control Consortium phase 1 Data (WTCCC), which were missed in the original WTCCC GWAS study that was performed four years before(Burton et al., 2007; J. Huang, Ellinghaus, Franke, Howie, & Li, 2012). Imputation also facilitates fine-mapping studies by allowing most polymorphic variants, including causative ones, to be tested in known disease associated loci. For example, the strongest association signal, observed at the imputed variant rs7903146 of the TCF7L2 locus in the WTCCC type 2 diabetes scan, is suggested to be causal association in the locus(Mahajan et al., 2014; Marchini, Howie, Myers, McVean, & Donnelly, 2007). Furthermore, imputation allows for meta-analysis between samples that have been genotyped using different arrays, increasing power.
However, for studies that have access to population-matched genome sequenced individuals, there is uncertainty in deciding between a smaller, ancestry-matched reference panel and a larger publicly-available cosmopolitan reference panel. An ideal reference panel is expected to have closely matched ancestry to study samples because the genetic similarity increases the accuracy of imputation(Deelen et al., 2014; G. H. Huang & Tseng, 2014; J. Huang et al., 2015; Low-Kam et al., 2016; Mitt et al., 2017; Okada, Momozawa, Ashikawa, Kanai, & Matsuda, 2015; Pistis et al., 2015; Roshyara & Scholz, 2015; Walter et al., 2015). On the other hand, the imputation accuracy increases when larger reference panels are used, especially for lower-frequency variants(Browning & Browning, 2009; B. N. Howie, Donnelly, & Marchini, 2009; L. Huang et al., 2009; Y. Li et al., 2009; Roshyara & Scholz, 2015). Furthermore, different whole-genome reference panels may generate discordant imputed genotypes for the same variants in the same study samples. This brings in challenges for the follow-up association tests. The optimal strategy to perform association tests using genotypes imputed by different reference panels remains unclear. IMPUTE2 provides one possible approach to merge all reference panels to a single larger panel for genotype imputation when multiple reference panels are available (B. N. Howie et al., 2009), which may avoid the problem that different versions of genotypes are imputed for the same variants. The Genome of the Netherlands Consortium and the UK10K study has further shown that the combined reference panel of 1000G and the population-specific reference resulted in better imputation results compared to the two individual panels for rare variants(Deelen et al., 2014; J. Huang et al., 2015). However, this approach is not feasible when individual-level haplotypes within the reference panel are not accessible, as is the case with the Haplotype Reference Consortium (HRC)(McCarthy et al., 2016), primarily due to ethical issues surrounding sharing of individual-level genetic data(McCarthy et al., 2016). Here we genotyped 9,265 Norwegian participants from the HUNT study(Krokstad et al., 2013) for 350,270 polymorphic autosomal variants using the Illumina Human CoreExome array with approximately 240,000 GWAS tagging markers. We created a population-matched reference panel by whole-genome sequencing (WGS) 2,021 individuals from the HUNT study to a mean depth of 5×. We imputed variants from the HUNT WGS reference panel as our ethnically matched panel. We also performed imputation with two additional imputation reference panels: the HRC(McCarthy et al., 2016) and 1000G Phase 3(Auton et al., 2015). First, we systematically evaluated and compared the imputation results from the three reference panels, including the number of successfully imputed variants as well as the imputation accuracy. Next, we evaluated and compared the power of association tests between two approaches to incorporate multiple versions of imputed genotypes. First is the “best Rsq” approach, which retains imputed genotypes only from the panel with highest imputation quality metrics for each variant. Second is the “best p-value” approach that tests association with all imputed genotypes and uses the most significant association p value, adjusting for the additional variants tested.
Materials and Methods
Array-based genotyping
9,265 samples from the HUNT Biobank in Norway were genotyped at 350,270 polymorphism autosomal variants using an Exome + GWAS chip array (HumanCoreExome-12 v1.0, Illumina). Genotype calling was performed using GenTrain version 2.0 in GenomeStudio V2011.1 (Illumina). Samples with <98% genotype calls (N = 37), evidence of gender discrepancy (N = 21), duplicates (N = 66) as well as individuals with non-Norwegian ancestry identified by plotting the first 10 genotype-driven principal components(Springer-Verlag, 1986) (N = 7) were excluded from further analysis (N = 131, 1.19%). As Figure S1 shows, the HUNT GWAS samples have similar ancestry to the samples in the HUNT WGS reference panel. All HUNT research subjects provided informed written consent and IRB approval was obtained for genetic studies.
Relatedness was evaluated based on the estimation of the proportion of identity by descent (IBD) by PLINK(Purcell et al., 2007). We excluded 1,644 samples from the HUNT GWAS sample due to 1st or 2nd degree relatedness to samples in HUNT WGS, defined as IBD ≥ 0.25. We excluded samples that were related to samples within the reference panel to avoid inflating imputation statistics for regions inherited IBD. We performed variant-level quality control by excluding 19,872 variants that met any of the following criteria; variants with a cluster separation score < 0.3 reported by GenomeStudio V2011.1 (Illumina), < 95% genotype call rate, or deviation from Hardy–Weinberg equilibrium (P < 1 × 10−5).
Genotype imputation
Genotype imputation with the 1000G Phase 3(Auton et al., 2015) and the HRC(McCarthy et al., 2016) reference panels was conducted using the Michigan Imputation Server(Das, Forer, Schonherr, Sidore, & Locke, 2016) and imputation with the HUNT WGS reference panel was conducted using a local server. The study samples were phased using SHAPEIT2(v2.r790)(Delaneau, Zagury, & Marchini, 2013) followed by imputation using minimac3(v2.0.1)(Fuchsberger, Abecasis, & Hinds, 2015; B. Howie, Fuchsberger, Stephens, Marchini, & Abecasis, 2012). Two imputation metrics output by minimac3 were used for evaluating the imputation quality: ImpRsq and EmpRsq. ImpRsq is previously known as r̂2 in different versions of the MaCH/minimac(Fuchsberger et al., 2015; B. Howie et al., 2012; Li, Willer, Ding, Scheet, & Abecasis, 2010). ImpRsq is defined for both genotyped and ungenotyped variants in the chip array as an estimate of the squared correlation between imputed dosages and true, unobserved genotypes, calculated as the observed variance over the expected variance. EmpRsq is defined only for genotyped variants in the chip array as the squared correlation between leave-one-out imputed dosages and the true, observed genotypes (See “Estimated Imputation Accuracy” section at http://genome.sph.umich.edu/wiki/Minimac_Diagnostics for details).
Reference panels
The HUNT WGS reference panel contains 1,101 earliest onset cases with myocardial infarction and 1,100 age and sex matched controls that were selected from the HUNT study(Krokstad et al., 2013). Whole genome sequencing to ~5× depth was performed on either Illumina HiSeq 2000 or 2500. We followed the GotCloud SNP calling pipeline to process the whole genome sequencing data(Jun, Wing, Abecasis, & Kang, 2015). The variant sites and genotype likelihood were called using SAMtools(H. Li et al., 2009) and the genotypes for SNPs were refined and phased using Beagle v4(Browning & Browning, 2013). After quality control, 20.2 million single nucleotide variants were retained in 2,201 samples, of which 4 million were unique to our study; not observed in dbSNP 144(Sherry et al., 2001), 1000 Genomes Phase 3(Auton et al., 2015), UK10K(Walter et al., 2015), ESP6500(W. NHLBI GO Exome Sequencing Project (ESP) Seattle, 2013), or ExAC.r0.3(Lek et al., 2016) (Table 1). The individuals in the HUNT WGS panel have similar ancestry to the HUNT study samples (Figure S1) and are from the same geographic region, although we excluded in the genotyped samples any 1st or 2nd degree relatives of the sequenced samples to avoid biased estimates of the accuracy of imputation. Additionally, there were no close relatives within the sequenced samples. The other two reference panels that we used for genotype imputation are the 1000 Genomes Phase 3 (1000G)(Auton et al., 2015) and the HRC release 1(McCarthy et al., 2016) containing 32,488 individuals, both of which are pre-stored in the Michigan Imputation Server(Das et al., 2016) (Table 2). The HUNT cohort contributed an early freeze of whole genome sequencing data consisting of 1,023 samples to the HRC consortium. Thus, the HUNT WGS and the HRC reference panels have 1,023 samples in common. Variants with minor allele counts (MAC) less than or equal to 5 were excluded from HRC(McCarthy et al., 2016).
Table 1.
Summary of the variants in the HUNT whole-genome sequencing reference panel containing 2,201 individuals with average sequencing depth 5×.
| Variant Type | Total number of variants |
Mean number of variants per individual (SD) |
Mean number of unique variants per individual (SD) |
% in 1000 Genomes |
Number of novel variants* |
|---|---|---|---|---|---|
| Splice | 1,265 | 71.5(4.6) | 0.2(0.47) | 36.6 | 355 |
| Nonsense | 2,432 | 71.5(6) | 0.43(0.74) | 36.6 | 585 |
| Missense | 113,576 | 9,480(113) | 13.8(13.6) | 56.3 | 13,927 |
| Synonymous | 77,699 | 10,707(100) | 7.1(7.5) | 68.5 | 5,935 |
| Noncoding | 20,050,237 | 3,342,839(15,415) | 1531(906) | 68.7 | 4,030,199 |
| Total | 20,245,209 | 3,363,168(15,522) | 1,552(919) | 68.6 | 4,051,001 |
Novel: not reported in dbSNP 144(Sherry et al., 2001), 1000 Genomes Phase 3(Auton et al., 2015), UK10K(Walter et al., 2015), ESP6500(W. NHLBI GO Exome Sequencing Project (ESP) Seattle, 2013), or ExAC.r0.3(Lek et al., 2016)
Table 2.
Reference panels used for genotype imputation
| Reference Panels | Variants | Sample Size | Population |
|---|---|---|---|
|
| |||
| Haplotype Reference Consortium(McCarthy et al., 2016) (HRC) | 39 million SNPs (MAC ≥ 5) | 32,488a | Cosmopolitan (mostly European) |
|
| |||
| 1000 Genomes Phase 3 Version 5(Auton et al., 2015) (mean depth < 8×) | 81 million Biallelic SNPs, indels, deletions, complex short substitutions and other structural variant classes (MAC ≥ 2) | 2,504 | Cosmopolitan |
|
| |||
| HUNT Whole Genome Sequencing (HUNT WGS) (mean depth ~ 5×) | 20 million SNPs | 2,201a | Norwegian |
|
| |||
HRC and HUNT whole-genome sequencing data set have 1,023 samples in overlap.
MAC: minor allele count
Permutation test
To determine the genome-wide significance thresholds for association tests using the two approaches to incorporate imputed genotypes, we performed permutation tests. The measurements of the high-density lipoprotein (HDL) cholesterol for the study samples were permuted 1,000 times. Each permutation was followed by a genome-wide association test (GWAS) using the permuted phenotypes. The most significant p-values from each of the 1,000 GWAS were ranked. And the significant threshold with family-wise error rate (FWER) n/1000 equals to the nth smallest p-value. Because the “best p-8 value” approach tests more variants, it will be a more stringent significant threshold than the “best Rsq” approach.
Power estimation
In order to estimate the power to detect association under the two approaches to incorporate imputed genotypes from multiple reference panels, we considered directly genotyped variants as causal variants, and used multiple sets of imputed genotypes to evaluate the power. First, we obtained the leave-one-variant-out imputed dosages for those directly genotyped variants. The official release of minimac3 performs leave-one-out hidden Markov model (HMM) calculation internally to calculate leave-one-out Rsq summary statistics, but does not output individual dosages (Fuchsberger et al., 2015; B. Howie et al., 2012). We modified minimac3 to include the individual leave-out-out dosages in the output VCF for the genotyped variants. Second, we simulated phenotypes based on the genotypes obtained by the chip array. Finally, we evaluated power of the two approaches by performing association tests between the simulated phenotypes and the imputed dosages based on either “best Rsq” or “best p-value” approaches.
The details of simulation follow the steps described below:
Select the non-centrality parameter corresponding to the association test p-value pt. We calculate the non-centrality parameter Nr2 as a chi-square statistics corresponding to the upper-tail probability pt, where N is the total number of study subjects. This ensures that the median p-value is pt when the true phenotypic variance explained by the genotype is r2.
For each variant, we randomly draw ε from the normal distribution with mean 0 and standard deviation . We calculate the effect size β as , where f is the minor allele frequency (MAF) estimated using the chip genotypes of the variant. The phenotype value y is then calculated as Gβ + ε, where the chip genotypes G is 0, 1, or 2. The phenotypic variance explained by G and ε will be r2 and 1−r2, respectively.
We perform the linear regression using the leave-one-variant-out dosages for this variant, which were imputed using the three different reference panels respectively, and the phenotype y.
For the “best p-value” approach, the final association p value equals to most significant one among the three p values associated with the three different versions of imputed dosages. With the “best Rsq” approach, the final p value equals to the one corresponding to the reference panel with the highest imputation quality (ImpRsq), an estimated value for the correlation between imputed genotypes and true, unobserved genotypes.
The power to detect association signals equals to the percentage of final p values exceeding the genome-wide significance threshold determined for each approach by the permutation tests described above.
We performed linkage disequilibrium(LD) based variant pruning for the 289,376 directly genotyped variants that were found by all three reference panels using PLINK(Purcell et al., 2007) and obtained 132,183 variants with LD r2< 0.2 among each other. Then we randomly selected 3,000 variants for each of the MAF categories: MAF ≤ 0.001, MAF > 0.001 and ≤ 0.01, MAF > 0.1 and ≤ 0.05, and MAF > 0.05. We applied ImpRsq > 0.3, 0.5 and 0.8 to remove poorly imputed genotypes and variants that were successfully imputed from at least two references were used for this simulation study. All 5 steps above were repeated given different pt’s ranging from 5×10−8 to 1×10−13. Additionally, the entire process was repeated 5 times across the selected variants to average power.
Partial correlation estimation
To quantify the net gain of imputation accuracy obtained by including another reference panel on top of an existing panel, we estimated the partial correlation between the leave-one-out imputed dosages from the additional panel and the chip genotypes, conditioned on the leave-one-out imputed dosages from the existing panel. The correlation has been estimated for every pair of reference panels among the three on each of the 289,376 genotyped variants that were found in all three panels. For example, to estimate the net gain of including 1000G panel on top of HUNT panel (PartialRsq [1000G,Chip | HUNT]), we first obtained the leave-one-out dosages based on 1000G and HUNT WGS (details described in the Power estimation subsection). Secondly, for each variant, we performed three linear regressions on the chip genotypes: the first one has the imputed dosages from 1000G and HUNT WGS as covariates (model 1), the second one has the imputed dosages from HUNT WGS only as a covariate (model 2), and the third one does not have any other covariate except for the intercept (model 3). Lastly, we obtained sum of squared residuals (SSR) for the three linear regressions and calculated the partial correlation (partial Rsq) as . In a similar notation, the EmpRsq is equivalent to , and their sum should be equivalent to the proportion of explained variance by both sets of imputed dosages. Our intuition is that the more extra information the additional reference panel provides, the higher the partial correlation will be.
Results
Evaluating successfully imputed variants using different reference panels
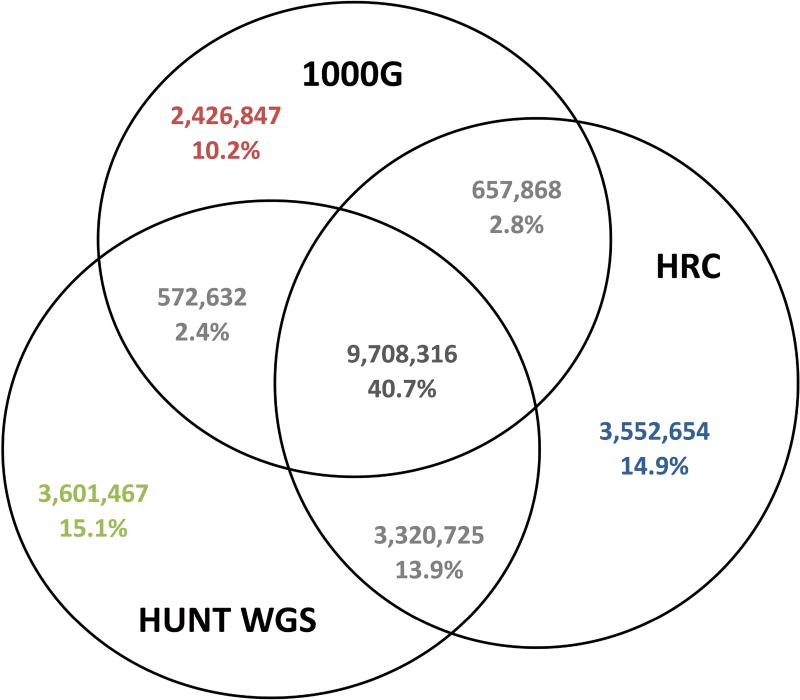
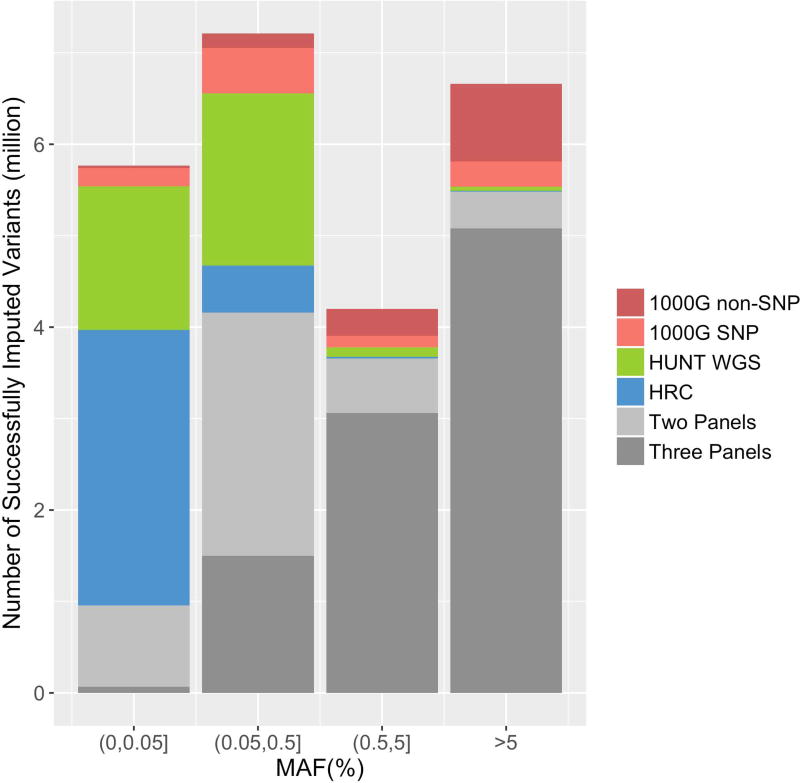
In total, ~23.8 million variants were successfully imputed using minimac 3(Fuchsberger et al., 2015; B. Howie et al., 2012) from at least one of the three reference panels and exceeded the threshold of estimated imputation quality (ImpRsq) ≥ 0.3 (Figure 1). The three reference panels yielded roughly equal number of SNPs with MAF more than 1%, but the 1000G uncovered more unique variants; approximately 75.3% (1,068,228 out of 1,418,417) that were uniquely imputed from 1000G are indels or structural variants, a category of variation that is not available in the other two reference panels. We observed that imputation from the HRC panel resulted in more extremely rare variants (MAF less than 0.05%) than from HUNT WGS and 1000G. Imputation from the HUNT WGS panel uncovered more variants with MAF between 0.05% and 1% than the other two reference panels (Table 3). Approximately 3.6 million variants were uniquely imputed by the HUNT WGS panel (Figure 1) and the majority of them have MAF less than or equal to 0.05%(Figure 2). A threshold ≥ 0.3 for ImpRsq was applied as recommended to remove most of poorly imputed variants while retaining the vast majority of well imputed SNPs(Y. Li et al., 2009). We observed that the average EmpRsq remained above 0.6 for all MAF categories from all three reference panels when the ImpRsq ≥ 0.3 threshold was applied (Figure S2).
Figure 1. Number of variants that are imputed by different reference panels.
The corresponding percentage is the variants number out of all 23.8 million variants that are successfully imputed by any of the three reference panels.
Table 3.
Numbers of imputed variances contributed by each reference panel categorized by MAF. The threshold ImpRsq > 0.3 was applied. Each reference panel contributed uniquely imputed variants.
| HRC Release 1 (39.2M SNPs, 32,488 samples including 1,203 HUNT samples) |
1000G Phase3 v5 (81.2M markers, 2,504 samples) |
HUNT 5× WGS (20.2M SNPs, 2,201 samples) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| MAF | Number of Passed Variants |
Percent of Passed Variants |
Number of Uniquely Imputed Variants |
Number of Passed Variants |
Percent of Passed Variants |
Number of Uniquely Imputed Variants |
Number of Passed Variants |
Percent of Passed Variants |
Number of Uniquely imputed Variants |
|
| |||||||||
| (0, 0.0005) | 4,337,138 | 23.9% | 3,009,729 | 567,481 | 2.4% | 230,186 | 2,291,216 | 50.6% | 1,570,259 |
|
| |||||||||
| (0.0005, 0.001) | 1,339,096 | 91.1% | 373,964 | 501,248 | 11.4% | 176,252 | 1,668,837 | 94.4% | 901,106 |
|
| |||||||||
| (0.001, 0.005) | 2,964,988 | 97.5% | 140,318 | 2,119,956 | 33.6% | 475,376 | 3,917,801 | 98.0% | 982,320 |
|
| |||||||||
| (0.005, 0.01) | 1,125,181 | 99.2% | 7,426 | 1,074,885 | 68.9% | 126,616 | 1,279,200 | 98.6% | 47,426 |
|
| |||||||||
| (0.01, 0.05) | 2,314,490 | 99.6% | 10,525 | 2,554,206 | 89.2% | 295,991 | 2,538,140 | 99.1% | 55,490 |
|
| |||||||||
| > 0.05 | 5,158,670 | 99.8% | 10,692 | 6,547,887 | 98.1% | 1,122,426 | 5,507,946 | 99.6% | 44,866 |
|
| |||||||||
| Total | 17,239,563 | 55.1% | 3,552,654 | 13,365,663 | 29.5% | 2,426,847 | 17,203,140 | 87.4% | 3,601,467 |
|
| |||||||||
The greatest number of the uniquely imputed variants among the three reference panels for variants in each MAF category is highlighted in red.
MAF: minor allele frequency; ImpRsq, imputation quality metric R2
Figure 2. Distribution of numbers of variants that are imputed from only one reference panel or from multiple reference panels in different MAF categories.
Variants that are imputed by 1000G only are categorized as SNPs and non-SNP variants, including indels, deletions, complex short substitutions and other structural variant classes. 1000G, 1000 Genomes Phase 3; WGS, whole-genome sequencing; HRC, Haplotype Reference Consortium; MAF, minor allele frequency
Comparing imputation accuracy from different reference panels
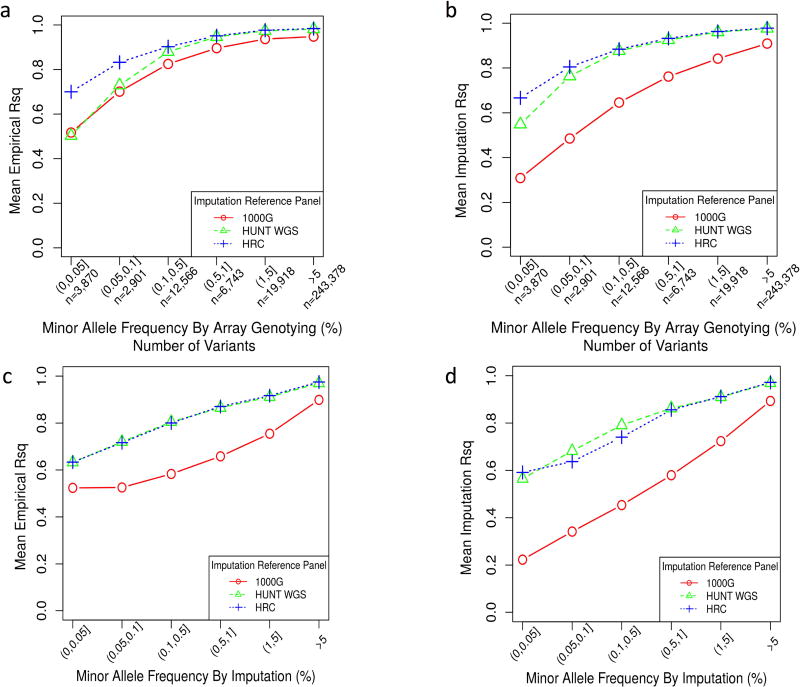
To compare the imputation accuracy across the three reference panels, we examined all 289,376 variants that were directly genotyped by the chip array and available in all three reference panels. “Leave-one-variant-out” imputation results were used for these directly genotyped variants, meaning that one-by-one, each genotyped variant was masked, imputed, and then compared to the directly genotyped calls. The EmpRsq was estimated for each genotyped variant from each panel, which is the squared Pearson correlation between the imputed allele dosages and the genotypes called by direct genotyping. Figure 3a compares the average EmpRsq for all genotyped variants categorized by MAF among different reference panels. The MAF is estimated using the genotypes called by the chip array. Imputation from HRC has higher imputation accuracy for rare variants with MAF < 0.5% than the other two reference panels, which is expected because the number of samples available in HRC is much larger than the other two panels and the imputation accuracy for extremely rare variants depends on the number of copies of alternate alleles(Roshyara & Scholz, 2015). What is unexpected is that for variants with MAF ≥ 0.5%, HRC and HUNT WGS panels show comparable imputation accuracy, even though the size of the HUNT WGS panel is 15 times smaller than HRC. Consistent to previous studies, this result demonstrated the value of whole-genome sequencing for ancestry matched samples as a reference panel for genotype imputation(Deelen et al., 2014; G. H. Huang & Tseng, 2014; J. Huang et al., 2015; Low-Kam et al., 2016; Okada et al., 2015; Pistis et al., 2015; Roshyara & Scholz, 2015; Walter et al., 2015). It is also noticed that imputation from 1000G has lower average ImpRsq than the other two reference panels (Figure 3b–d), which is consistent to the lower proportion of variants passing the various ImpRsq thresholds in 1000G observed in Figure S2.
Figure 3. HRC and HUNT WGS panels show comparable imputation quality.
a. comparing the mean empirical R2 (y axis) reported by different reference panels for variants that are directly genotyped categorized by the MAF (x axis) without any ImpRsq threshold applied.
b. comparing the mean Imputation R2 (y axis) reported by different reference panels for variants that are directly genotyped categorized by the MAF (x axis) without any ImpRsq threshold applied.
c. comparing the mean Imputation R2 (y axis) reported by different reference panels for all imputed variants (ImpRsq > 0.3) by the MAF (x axis).
d. comparing the mean Imputation R2 (y axis) reported by different reference panels for all imputed variants by the MAF (x axis) without any ImpRsq threshold applied.
1000G, 1000 Genomes Phase 3; WGS, whole-genome sequencing; HRC, Haplotype Reference Consortium; MAF, minor allele frequency; ImpRsq, imputation quality metric R2
To further evaluate the impact of the sample size of the HUNT WGS panel on the imputation accuracy, we have randomly drawn 500, 1000, and 1500 samples from the original HUNT reference panel for imputation. Figure S3 shows the comparison of the average EmpRsq for all genotyped variants categorized by MAF among the target samples, across all reference panels. As expected, increases in the sample size of the HUNT WGS reference panels resulted in higher imputation accuracy, particularly for less frequent variants with MAF < 0.5%. Interestingly, we observed that the HUNT WGS with 500 samples outperforms 1000G(Auton et al., 2015) for variants with MAF > 0.5%. These results are consistent with other studies with population specific reference panels(Mitt et al., 2017; Pistis et al., 2015). The subset of 1000 samples provides better imputation accuracy than 1000G(Auton et al., 2015) even for variants with MAF as low as 0.1% and comparable imputation accuracy to HRC(McCarthy et al., 2016) for variants with MAF > 0.5%.
We examined whether our evaluation of imputation accuracy is biased in favor of HUNT WGS due to relatedness. Previous studies have shown that the relatedness between study samples and reference samples increases genotype imputation efficiency since related individuals tends to share longer haplotype stretches than unrelated ones(G. H. Huang & Tseng, 2014). To avoid the bias of imputation accuracy due to the relatedness between our study samples and the samples in the HUNT WGS reference panel, we excluded 1,644 study samples who are up to 2nd degree relatives of HUNT WGS samples. Relatedness was based on the estimation of the proportion of IBD by PLINK(Purcell et al., 2007). We observed that excluding these study samples did not affect the imputation accuracy except causing a slight decrease of the imputation accuracy for those very rare variants with MAF < 0.05% (Figure S4).
Evaluating two possible association test strategies to use multiple sets of imputed genotypes
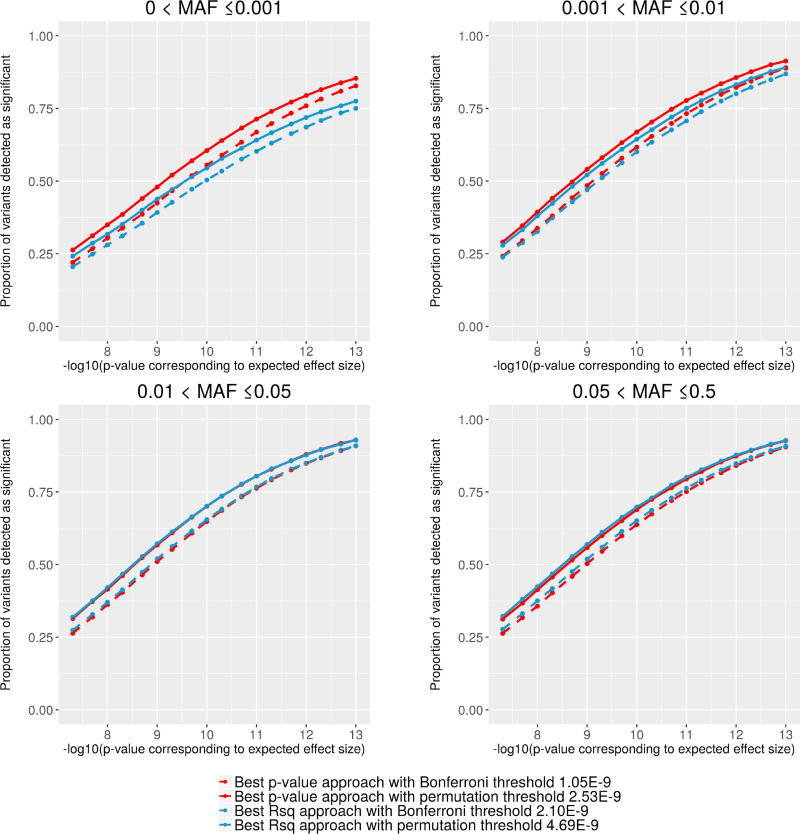
As Figure 1 shows, approximately 60% of all successfully imputed variants were imputed from more than one reference panel, which makes it unclear how to perform downstream association tests. We compared two possible strategies: the “best p-value” and the “best Rsq” approaches. The “best p-value” approach uses each version of imputed genotypes to choose the lowest association p-value, thereby increasing the burden of adjusting for multiple hypothesis testing. The “best Rsq” approach selects the imputed variant with the highest estimated imputation quality ImpRsq, which is expected to be a reasonable approximation of the association between imputed and true genotypes, especially for common variants (Figure S5). We have compared the power of the two approaches to detect association signals accounting for the fact that the “best p-value” approach needs adjusting for the additional variants tested. To determine the significant thresholds for association tests with a family-wise error rate (FWER) 0.05, we estimated the number of independent tests using 1,000 permutations. For the “best Rsq” approach, where fewer ‘variants’ are analyzed, the significance threshold is 4.69×10−9 (2.10×10−9 with a Bonferroni correction) and for the best p-value approach, it is 2.53×10−9 (1.05×10−9 with a Bonferroni correction).
Using the permutation-derived significance thresholds above, we evaluated the power of the two approaches for association tests with quantitative traits through a simulation study (details described in methods). Our results indicated that the “best p-value” approach has more power to detect association signals than the “best Rsq” approach, particularly for rare variants with MAF < 1%, no matter how stringent the ImpRsq threshold was used for filtering out the poorly imputed genotypes (Figure 4, Figure S6 and Table S1). This is probably because the estimated imputation quality ImpRsq does not always agree with empirical imputation quality EmpRsq especially for rare variants (Figure S5), resulting in loss of variants with highest empirical imputation quality when selecting the “best Rsq” strategy. In addition, the distributions of the ImpRsq are quite different from different panels. Notably, from 1000G(Auton et al., 2015), the ImpRsq and EmpRsq were substantially lower for low-frequency variants (0.5% < MAF < 5%), and ImpRsq tends to underestimate EmpRsq(Figure S5). The two approaches have comparable association power for variants with MAF ≥ 1%, where estimated and empirical imputation qualities highly agree with each other (Figure S5). Our observation suggests that the inaccurate prediction of imputation quality have a higher impact than increased burden of multiple testing in association test with rare variants.
Figure 4. Comparison of power to detect true associations between best p-value and best Rsq approaches via simulation studies.
For each MAF category, 3,000 directly genotyped variants were randomly selected based on their MAF estimated with genotypes obtained from the chip array to estimate the power. The power is calculated as the proportion of significantly associated variants across three imputed panels based on each strategy given the corresponding significance threshold. ImpRsq > 0.3 was applied to remove poorly imputed genotypes. The numbers of variants that were successfully imputed from at least two reference panels and used in the simulation studies are: 2,513 with MAF > 0 and ≤ 0.001; 2,989 with MAF > 0.001 and ≤ 0.01; 3,000 with MAF > 0.01 and ≤ 0.05; and 3,000 with MAF > 0.05. MAF, minor allele frequency; ImpRsq, imputation quality metric R2
Evaluating net gain of imputation accuracy by including an additional reference panel
Finally, we quantified the net gain of imputation accuracy by including an additional reference panel as a “partial Rsq” conditioned on the imputed genotypes from an existing reference panel (See Materials and Methods for details). Intuitively, this represents the difference between the “optimal EmpRsq” linearly combined between two sets of imputed genotypes and the EmpRsq from the original imputed genotypes. 289,376 genotyped variants that were found in all three panels were used to evaluate the additional information that were gained from one reference panel given imputed dosages based on another panel. As Figure S7 presents, each reference panel is able to provide additional information to improve imputation accuracy. However, relatively less information could be be gained by including 1000G(Auton et al., 2015) panel on top of HRC across all MAF categories. This is expected since 1000G samples are included in the HRC panel, with the caveat that only single nucleotide variants with minor allele count ≥ 5 were retained. Note that evaluation of indels and structural variants absent in HRC were not included in this experiment. In contrast, given the imputed dosages from 1000G, both HUNT WGS and HRC provide substantial net gain of imputation accuracy, which is consistent to our observations. Furthermore, HUNT WGS and HRC provide additional information conditional on each other. More specifically, more extra information was obtained from HRC given HUNT WGS than those were obtained from HUNT WGS given HRC for these genotyped variants, which is also consistent to our observations in Figure 3.
Discussion
Many studies have performed whole genome sequencing of a subset of samples followed by imputation into samples with GWAS data(Holm et al.; Lane, Vlasac, & Anderson, 2016; Nalls et al., 2014; van Leeuwen et al., 2016). However, the trade-offs between the panel size, imputable variant types, and population specificity across different reference panels make it challenging to decide on the optimal strategy for imputation and downstream association analysis. We evaluated methods for genotype imputation when different reference panels are available. Our findings have demonstrated the benefits of uncovering novel variants with low frequency by using population-specific reference panels as has been reported by previous studies(J. Huang et al., 2015). Since the population-specific HUNT panel shared 1,023 samples with HRC(McCarthy et al., 2016), we expect to see an even bigger advantage in the number of novel low frequency variants imputed by the population-specific panel if there were no overlap between the two reference panels.
We have also observed that large-scale publicly available reference panels, as exemplified by HRC (McCarthy et al., 2016) and 1000G(Auton et al., 2015), contribute a large number of variants that are not captured by population-specific reference panels. More specifically, HRC(McCarthy et al., 2016), which has much larger sample size and contains more general European populations, contributes 3.5 million variants that could not be imputed by the other two panels. Since 1000G(Auton et al., 2015) has additional advantages that indels and structural variants are comprehensively detected and genotyped, 1.3 million non-SNP variants have only been imputed by 1000G(Auton et al., 2015). Furthermore, each reference panel may provide additional information to improve imputation accuracy. Therefore, to increase the variant coverage and imputation accuracy as much as possible, we recommend using all three reference panels for imputation if available. If a single panel has to be chosen, each option will have different advantages and disadvantages. We have shown that imputation from population-specific reference panels provides comparable imputation accuracy for variants with MAF > 0.1%. as using reference panels with 15 times larger sample size with only broad ancestry-matching (i.e. European). Although panel sizes are similar, the population-specific reference panel results in higher imputation accuracy than the mixed-ancestry 1000G panel (Auton et al., 2015) for variants with MAF ≥ 0.05%. This has also been observed by a recently published study on Estonians(Mitt et al., 2017).
To address the issue of imputing different versions of the same variant from different reference panels, we propose the “best p-value” approach, which analyzes all versions of each imputed variant and accounts for the multiple testing. Our simulation study demonstrated that this approach has higher power for detecting association signals than selecting the imputed variant with highest imputation quality given the distributions of the imputation quality metrics from different reference panels may be quite different, even adjusting for additional variants tested.
The UK10K study and the Genome of the Netherlands (GoNL) Consortium suggested that merging multiple reference panels to a larger reference panel would improve imputation performance, especially for less frequent variants(Deelen et al., 2014; J. Huang et al., 2015). Compared to this approach, our “best p-value” approach does not require access to all reference panes and is feasible even if not all reference panel haplotypes are directly accessible. If large imputation reference panels, such as the HRC(McCarthy et al., 2016), are not directly accessible, conducting association tests for all imputed versions of genotype with slightly higher computational cost will be an effective strategy.
In summary, we recommend creating a small size ancestry-matched reference panel using whole genome sequencing to allow for improved imputation of low frequency variants that may be enriched in that ancestral group, performing genotype imputation using the ancestry-matched reference panel and other large publicly available databases, and analyzing all versions of imputed variants in downstream association testing.
Supplementary Material
Acknowledgments
The whole genome sequencing of 2,201 HUNT samples was supported by NHLBI HL109964(CJW). Genotyping services were supported by The Liaison Committee between the Central Norway Regional Health Authority, the Norwegian University of Science and Technology, the Research Council of Norway, and the University of Michigan. We wish to thank all HUNT study participants who contributed to scientific research. We also appreciate the reviewers and editors for their thoughtful and constructive comments that helped improve the manuscript substantially.
Footnotes
Conflicts of Interest
The authors have no conflict of interest to declare.
Description of Supplemental Data
Supplemental data include seven figures and one table.
References
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194(2):459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Samani NJ. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TH, Thompson DJ. Five endometrial cancer risk loci identified through genome-wide association analysis. 2016;48(6):667–674. doi: 10.1038/ng.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Todd JA. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40(12):1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schonherr S, Sidore C, Locke AE. Next-generation genotype imputation service and methods. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, Oksenberg JR. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41(7):776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen P, Menelaou A, van Leeuwen EM, Kanterakis A, van Dijk F, Medina-Gomez C, Kreiner-Moller E. Improved imputation quality of low-frequency and rare variants in European samples using the 'Genome of The Netherlands'. 2014;22(11):1321–1326. doi: 10.1038/ejhg.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31(5):782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Wang Y, Shao W, Jin J, Du M, Ma G, Zhang Z. Rare variants in BRCA2 and CHEK2 are associated with the risk of urinary tract cancers. Sci Rep. 2016;6:33542. doi: 10.1038/srep33542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm H, Saemundsdottir J, Helgadottir HT, Johannsdottir H, Sigfusson G, Thorgeirsson G, Sulem P. [Google Scholar]
- Horikoshi M, Mgi R, van de Bunt M, Surakka I, Sarin AP, Mahajan A, Morris AP. Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation. PLoS Genet. 2015;11(7):e1005230. doi: 10.1371/journal.pgen.1005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Dunlop MG. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40(12):1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GH, Tseng YC. Genotype imputation accuracy with different reference panels in admixed populations. BMC Proc. 2014;8(Suppl 1 Genetic Analysis Workshop 18Vanessa Olmo):S64. doi: 10.1186/1753-6561-8-s1-s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur J Hum Genet. 2012;20(7):801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Howie B, McCarthy S, Memari Y, Walter K, Min JL, Durbin R. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. 2015;6(8111) doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84(2):235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Andersen G, Yorgov D, Ferrara TM, Ben S, Brownson KM, Koks S. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. 2016 doi: 10.1038/ng.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Wing MK, Abecasis GR, Kang HM. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25(6):918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokstad S, Langhammer A, Hveem K, Holmen TL, Midthjell K, Stene TR, Holmen J. Cohort Profile: the HUNT Study, Norway. Int J Epidemiol. 2013;42(4):968–977. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- Lane JM, Vlasac I, Anderson SG. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. 2016;7:10889. doi: 10.1038/ncomms10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Mohlke KL. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Kam C, Rhainds D, Lo KS, Provost S, Mongrain I, Dubois A, Lettre G. Whole-genome sequencing in French Canadians from Quebec. Hum Genet. 2016;135(11):1213–1221. doi: 10.1007/s00439-016-1702-6. [DOI] [PubMed] [Google Scholar]
- Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Morris AP. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Durbin R. A reference panel of 64,976 haplotypes for genotype imputation. Nature Genetics. 2016 doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitt M, Kals M, Parn K, Gabriel SB, Lander ES, Palotie A, Palta P. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur J Hum Genet. 2017;25(7):869–876. doi: 10.1038/ejhg.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, Singleton AB. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014;46(9):989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Momozawa Y, Ashikawa K, Kanai M, Matsuda K. Construction of a population-specific HLA imputation reference panel and its application to Graves' disease risk in Japanese. 2015;47(7):798–802. doi: 10.1038/ng.3310. [DOI] [PubMed] [Google Scholar]
- Pistis G, Porcu E, Vrieze SI, Sidore C, Steri M, Danjou F, Sanna S. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur J Hum Genet. 2015;23(7):975–983. doi: 10.1038/ejhg.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshyara NR, Scholz M. Impact of genetic similarity on imputation accuracy. BMC Genet. 2015;16:90. doi: 10.1186/s12863-015-0248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruth KS, Campbell PJ, Chew S, Lim EM, Hadlow N, Stuckey BG, Perry JR. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5(5):e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer-Verlag JIT. Principal component analysis. NEW YORK: 1986. [Google Scholar]
- van Leeuwen EM, Sabo A, Bis JC, Huffman JE, Manichaikul A, Smith AV, van Duijn CM. Meta-analysis of 49 549 individuals imputed with the 1000 Genomes Project reveals an exonic damaging variant in ANGPTL4 determining fasting TG levels. J Med Genet. 2016;53(7):441–449. doi: 10.1136/jmedgenet-2015-103439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W. NHLBI GO Exome Sequencing Project (ESP) Seattle. 2013 Retrieved from: http://evs.gs.washington.edu/EVS/
- Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, Soranzo N. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.