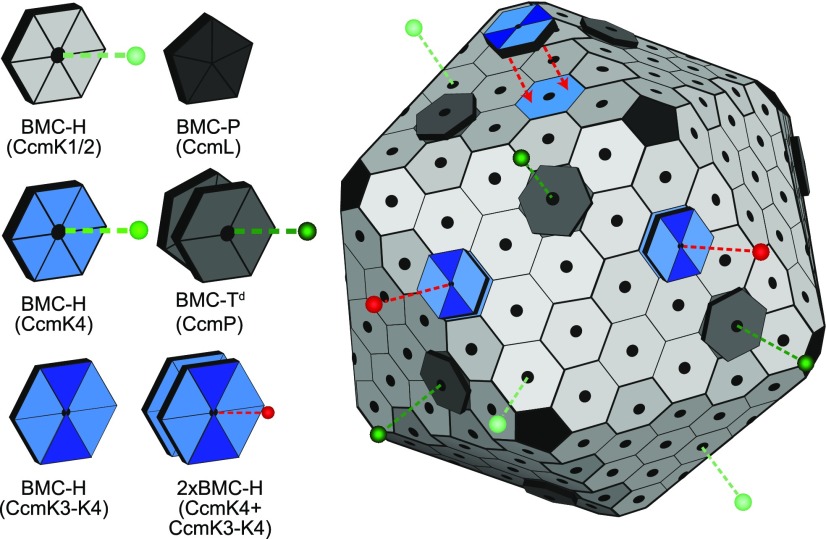
Two shell protein paralogs of the carboxysome, the carbon dioxide fixation microcompartment in β-cyanobacteria, form heterohexamers that stack into dodecamers, which would alter shell permeability.
Abstract
Bacterial microcompartments (BMCs) encapsulate enzymes within a selectively permeable, proteinaceous shell. Carboxysomes are BMCs containing ribulose-1,5-bisphosphate carboxylase oxygenase and carbonic anhydrase that enhance carbon dioxide fixation. The carboxysome shell consists of three structurally characterized protein types, each named after the oligomer they form: BMC-H (hexamer), BMC-P (pentamer), and BMC-T (trimer). These three protein types form cyclic homooligomers with pores at the center of symmetry that enable metabolite transport across the shell. Carboxysome shells contain multiple BMC-H paralogs, each with distinctly conserved residues surrounding the pore, which are assumed to be associated with specific metabolites. We studied the regulation of β-carboxysome shell composition by investigating the BMC-H genes ccmK3 and ccmK4 situated in a locus remote from other carboxysome genes. We made single and double deletion mutants of ccmK3 and ccmK4 in Synechococcus elongatus PCC7942 and show that, unlike CcmK3, CcmK4 is necessary for optimal growth. In contrast to other CcmK proteins, CcmK3 does not form homohexamers; instead CcmK3 forms heterohexamers with CcmK4 with a 1:2 stoichiometry. The CcmK3-CcmK4 heterohexamers form stacked dodecamers in a pH-dependent manner. Our results indicate that CcmK3-CcmK4 heterohexamers potentially expand the range of permeability properties of metabolite channels in carboxysome shells. Moreover, the observed facultative formation of dodecamers in solution suggests that carboxysome shell permeability may be dynamically attenuated by “capping” facet-embedded hexamers with a second hexamer. Because β-carboxysomes are obligately expressed, heterohexamer formation and capping could provide a rapid and reversible means to alter metabolite flux across the shell in response to environmental/growth conditions.
Bacterial microcompartments (BMCs) are polyhedral organelles that were first discovered in cyanobacteria (Drews and Niklowitz, 1957). Similar particles in chemoautotrophs were later shown to contain ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco) (Shively et al., 1973)—the central enzyme of the Calvin–Benson–Bassham Cycle—and the particles were accordingly named “carboxysomes.” All carboxysomes also encapsulate carbonic anhydrase, and the colocalization of the two enzymes enhances CO2 fixation and reduces photorespiration (Price and Badger, 1989; So et al., 2004; Long et al., 2007; Kerfeld and Melnicki, 2016). Due to the abundance of cyanobacteria in the open ocean, carboxysomal Rubisco contributes substantially to global primary carbon production (Garcia-Pichel et al., 2003). The catalytic efficiency of carboxysomes has inspired attempts to transfer the organelle into crop plants to enhance biomass production and yield (Price et al., 2013; Zarzycki et al., 2013; Lin et al., 2014a, 2014b; McGrath and Long, 2014).
Carboxysomes are delimited by a selectively permeable shell made of proteins (Cannon et al., 2001; Kerfeld et al., 2005). There are three classes of constituent proteins that form a shell with icosahedral symmetry: Hexamer-forming proteins containing a single Pfam00936 domain (BMC-H) (Kerfeld et al., 2005); trimer/pseudohexamer-forming proteins consisting of a fusion of two Pfam00936 domains (BMC-T) (Klein et al., 2009; Cai et al., 2013; Larsson et al., 2017), and pentamer-forming proteins containing the Pfam03319 domain (BMC-P) (Tanaka et al., 2008; Sutter et al., 2013). Each oligomer has a concave and convex side. The facets of BMC shells are comprised of a monolayer of these proteins, with their concave face exposed to the cytosol, punctuated by a subset of BMC-T pseudohexamers that form stacked oligomers (dimers of trimers, referred to as BMC-Td) (Sutter et al., 2017) (Fig. 1). Pores, typically at the cyclic axes of symmetry of the oligomers, are the conduits for substrates and products.
Figure 1.
Model of the beta carboxysome shell. Selective permeability of each pore type is represented by dashed lines and spheres in shades of green. Proposed capping of carboxysome facet CcmK4 hexamers by CcmK3-CcmK4 heterohexamers is indicated with red arrows. Altered metabolite flux after capping is represented in red.
There are two types of carboxysomes, distinguished by the form of Rubisco they encapsulate: Form IA in α-carboxysomes and Form IB in β-carboxysomes (Tabita, 1999; Rae et al., 2013; Zarzycki et al., 2013; Kerfeld and Melnicki, 2016). Accordingly, cyanobacterial species encoding each carboxysome type were termed α- and β-cyanobacteria (Badger et al., 2002). α-cyanobacterial strains occupy the open ocean (Prochlorococcus and the marine Synechococcus), and their carboxysomes are encoded in a single genomic locus (Badger et al., 2006; Roberts et al., 2012). In contrast, β-cyanobacteria inhabit a variety of dynamic habitats. Most β-carboxysome genes, including the genes encoding the BMC-H proteins CcmK1 and CcmK2, are situated in the main carboxysome locus and are constitutively expressed across a wide range of conditions (Billis et al., 2014; Hernández-Prieto et al., 2016). But all β-cyanobacterial genomes also contain varying numbers of additional CcmK paralogs (CcmK3-CcmK6) in satellite loci (Sommer et al., 2017). It has been hypothesized that the different CcmK paralogs have differing metabolite selectivities (Kerfeld et al., 2005; Rae et al., 2013; Sommer et al., 2017).
Two of these paralogs, CcmK3 and CcmK4, are present in ∼90% of all sequenced β-cyanobacterial genomes (Sommer et al., 2017). Gene deletion of ccmK3 and ccmK4 has been shown to cause impaired growth, although carboxysomes are still formed (Rae et al., 2012). Whereas the exact function of CcmK3 and CcmK4 remains unknown, Rae et al. (2012) found that the genes of both ccmK3 and ccmK4 must be knocked out in Synechococcus elongatus PCC7942 (Syn7942) to cause a slow-growth phenotype, and they thus concluded that the gene products act redundantly. However, deletion of only ccmK4 was sufficient to produce a slow-growth phenotype in Synechocystis sp. PCC 6803 (Syn6803) (Zhang et al., 2004). A recent comprehensive survey of carboxysome genes in β-cyanobacteria showed that ccmK3 and ccmK4 genes always co-occur; it was suggested that the proteins are not redundant but are functionally linked (Sommer et al., 2017). Because our understanding of BMC hexamers is based on crystal structures of purified BMC-H proteins of a single type, we hypothesized that the functional interdependence may involve heterohexamer formation.
In this study, we investigated the function of the satellite locus-encoded proteins CcmK3 and CcmK4. Growth analysis of deletion mutants shows that CcmK3 and CcmK4 are not redundant and that CcmK3 is a component of the β-carboxysome that is expendable under ideal growth conditions. CcmK3 does not form homohexamers when expressed recombinantly, but CcmK3 and CcmK4 form a heterohexameric complex that can further form dodecamers under certain conditions. Previous studies have provided examples of homohexamer formation among BMC proteins; here we provide an example of heterohexamer formation by a BMC-H protein. We suggest that heterohexamer formation could provide a means of fine-tuning β-carboxysome shell permeability.
RESULTS
In Vivo Characterization of ccmK3 and ccmK4
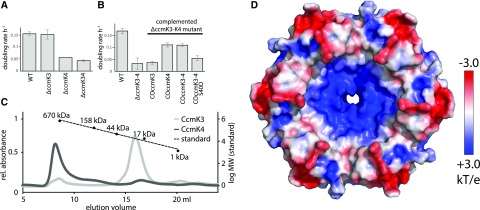
We created ΔccmK3 and ΔccmK4 mutants in Syn7942 and evaluated their growth in air under low light (80 µe). The doubling rate of the wildtype and the ΔccmK3 mutant were similar, with a doubling rate of 0.15 per hour (Fig. 2A). The ΔccmK4 mutant and the ΔccmK3-ΔccmK4 double mutant showed substantial growth impairments with doubling rates of 0.06 and 0.04 per hour, respectively. This is in contrast to a previous study by Rae et al. (2012) who did not detect a growth phenotype for the ΔccmK4 mutant. However, they grew the mutant in pH 8.0, whereas we carried out our growth experiments at pH 7.4. When growth of the strains generated in this study was measured at pH 8.0, we observed a decrease in the difference of growth between the ΔccmK4 mutant and the wild type, which was about 3-fold as long in pH 7.4 and only 2-fold as long in pH 8.0 (Supplemental Fig. S1). This finding can in part explain the differences between the studies.
Figure 2.
Characterization of CcmK3 and CcmK4. A and B, Growth of (A) Syn7942 ΔccmK3 and ΔccmK4 mutants, and (B) lines complementing the ΔccmK3-K4 mutant. Values are means ± sd from three biological replicates. C, Gel filtration of purified Halothece CcmK3-His, CcmK4-Strep and protein standard. D, Concave surface of a hexamer of the CcmK4 crystal structure, colored by electrostatic potential according to the scale on the right.
To verify our growth rate results, the ΔccmK3-ΔccmK4 mutant was complemented with either ccmK3, ccmK4, or the complete ccmK3-ccmK4 locus from Syn7942 (Fig. 2B). When ccmK3 was inserted into the genome of the double deletion mutant, growth remained slow (0.04 h−1). Complementation of the double mutant with ccmK4 or the whole locus, however, resulted in a recovery to a doubling rate to 0.11 h−1. When a mutated version of ccmK4 (that version with pore residue S40 replaced by Asp) was used to rescue the double mutant, growth remained slow with a doubling rate of 0.05 h−1, highlighting the importance of CcmK4 for function. Although CcmK3 is not essential for growth, it is a component of the carboxysome; CcmK3 labeled with yellow fluorescent protein (YFP) colocalizes with carboxysomes labeled with Rubisco-cyan fluorescent protein (CFP) (Supplemental Fig. S2).
Characterization of Purified CcmK3 and CcmK4 Oligomeric States
The oligomeric state of heterologously expressed and purified CcmK3 (12 kD) and CcmK4 (14 kD) from Halothece PCC 7418 was determined by size exclusion chromatography (SEC, Fig. 2C). CcmK4 formed aggregates (>1,000 kD), with a small shoulder at 150 kD that could represent a dodecamer. In contrast, CcmK3 formed only smaller oligomers (29 kD) that could be dimers or trimers but formed no hexamers or higher order oligomers. For example, CcmK2 (10.9 kD), the main carboxysomal BMC-H protein, elutes as a hexamer (64 kD, Supplemental Fig. S3). Also, all structurally characterized BMC-H proteins are symmetrical hexamers that sometimes form higher order assemblies in crystals (Kerfeld et al., 2005; Samborska and Kimber, 2012).
We were not able to crystallize heterohexamers of CcmK3 and CcmK4, but we successfully determined the structure of Halothece CcmK4 at a resolution of 1.8 Å (Fig. 2D, Supplemental Table S2). The protein forms typical BMC-H hexamers with a positive partial charge surrounding the pore (Fig. 2D). The hexamers packed into loosely associated concave-to-concave facing dodecamers in the crystal, consistent with a peak observed in SEC (Fig. 2C). These dodecamers are similar to what has previously been observed for CcmK2 (Samborska and Kimber, 2012). We analyzed the interface between the hexamers with PISA (Krissinel and Henrick, 2007) (Supplemental Table S1); the surface area of each CcmK4 hexamer that is involved in dodecamer formation is ∼2,200 Å2, which is less than half of the area buried in a typical double stacking BMC-Td protein (e.g. CcmP; Fig. 1[Cai et al., 2013]), which are stable in solution.
Interaction of CcmK Paralogs
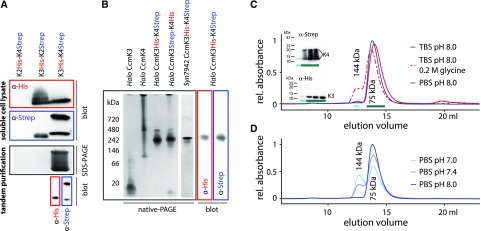
We analyzed interactions between the different CcmK proteins using sequential affinity purification of proteins recombinantely expressed and purified from E. coli. We coexpressed pairs of CcmK paralogs where one protein was His-tagged and the other Strep-tagged. Coexpression of CcmK2-His and CcmK4-Strep yielded mostly insoluble protein (Supplemental Fig. S4 A) with no detection of His- or Strep-tag by western blotting in the soluble fraction (Fig. 3A top). Soluble protein was obtained from coexpression of CcmK3-His and CcmK2-Strep as well as CcmK3-His and CcmK4-Strep (Fig. 3A top). CcmK4-Strep migrates as two bands that show up on both SDS-PAGE (Supplemental Fig. S4) and α-Strep western blot (Fig. 3A bottom). The soluble lysate fractions were loaded on a His-trap column, and the eluate then was applied on a Strep-trap affinity column. No protein was detected from the Strep-trap eluate when CcmK3-His was coexpressed with CcmK2-Strep, indicating they do not stably interact. In contrast, sequential affinity purification of coexpressed CcmK3-His and CcmK4-Strep showed both proteins in the Strep-eluate on SDS-PAGE (Supplemental Fig. S4), and their identity was confirmed by western blotting (Fig. 3A), indicative of a stable interaction between the two types of proteins.
Figure 3.
Characterization of CcmK3-CcmK4 complexes. A, Identification of CcmK proteins by SDS-PAGE and western blots after recombinant expression (soluble lysate) and tandem purification (final elution from second column). B, Native PAGE of recombinantly expressed and purified Syn7942 and Halothece CcmK proteins. Western blots of native gels of copurified CcmK3-CcmK4 protein. C and D, SEC of copurified Halothece CcmK3-CcmK4 complexes in different mobile phases.
Nondenaturing PAGE was performed to compare the copurified proteins to CcmK3 and CcmK4 homocomplexes (Fig. 3B). One major band was visible on the gel for the CcmK3-K4 complex that migrates differently than either CcmK3 or CcmK4 alone (Fig. 3B). Western blots confirmed the presence of CcmK3 and CcmK4 in this complex. Control experiments with the Strep-Tag on CcmK3 and the His-Tag on CcmK4, as well as with Syn7942 orthologs, yielded comparable complexes (Fig. 3B).
The molecular mass of the CcmK3-CcmK4 heterocomplex was determined by SEC. Elution of the mixed complex from the SEC column using a standard buffer (TBS 20/50, pH 7.4) yielded a major peak at 75 kD, corresponding to a hexamer (Fig. 3C) that was preceded by a small peak corresponding to the size of a dodecamer (144 kD). We confirmed the presence of both CcmK3-His as well as CcmK4-Strep in both the main hexamer and small dodecamer peak with western blots (Fig. 3C, inset). When 0.2-m Gly was added to the buffer, the relative amount of this peak increased to about 15%. The amount of the putative dodecamer formed was also affected by pH of the running buffer (Fig. 3D), with significantly more dodecamer observed at pH 7.0 (36%) than at pH 8.0 (3%).
The stoichiometry of CcmK3 and CcmK4 in the heterohexamers was determined using a quantitative dot blot (Supplemental Fig. S5, A, B). The concentration of CcmK3 in an undiluted Halothece CcmK3-CcmK4 sample after copurification was calculated as 78 µm (+/− 8 µm). The concentration of CcmK4 in this sample was calculated to 174 µm. The ratio of CcmK3 and CcmK4 is equivalent to 1.9 CcmK3 monomers per hexamer of the mixed complex. To confirm the results from the dot blot with a further independent method, we performed quantitative analysis of the heterocomplex via liquid chromatography and mass spectrometry (LC-MS/MS). This also resulted in 1.9 CcmK3 monomers per hexamer (Supplemental Table S2).
Characterization of the Interaction between CcmK3-CcmK4 in the Heterohexamers
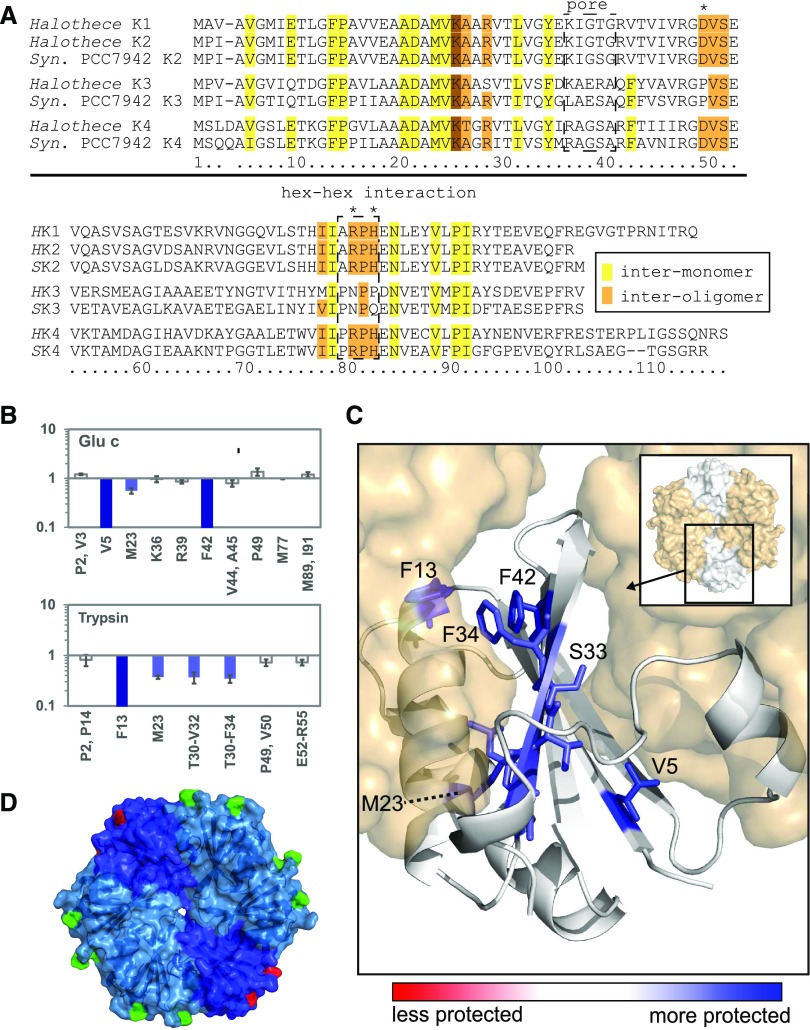
Because we were unable to crystallize the CcmK3-CcmK4 complex, we characterized the interprotein interactions using hydroxyl radical x-ray footprinting coupled to LC-MS (Fig. 4, A–C; Xu and Chance, 2007). The relative oxidation of residues by hydroxyl radicals measured in x-ray footprinting mass spectrometry (XFMS) correlates to solvent accessibility of a residue; profiles were collected for purified CcmK3 and for the CcmK3-CcmK4 complex. In isolated CcmK3, 25 residues were susceptible to oxidation. Of these, nine showed a decreased solvent accessibility (dose response) in the presence of CcmK4. In a model assuming a typical hexameric assembly consisting of CcmK3 and CcmK4 (Fig. 4C), the CcmK3 residues with a decrease in solvent accessibility in the presence of CcmK4 map to the intermonomer surface (V5, F13, L31, F34) or the concave surface (T30, V32, S33, F42), with the exception of M23, which points into the hydrophobic core between the beta sheet and alpha helices of the BMC fold. Notably, solvent accessibility of residues at the convex side surface (V53, E54, R55, M77, M89, I91), the hexamer perimeter (P2, V3, P49, E52), and the central pore (K36, R39) remained unchanged. This finding indicates that CcmK3 only forms the typical intermonomer interactions observed within hexamers in the presence of CcmK4.
Figure 4.
Interactions between CcmK3 and CcmK4 in heterohexamers determined by XFMS. A, Alignment of the sequences of CcmK2, CcmK3, and CcmK4 of Syn7942 and Halothece for reference. Conserved residues involved in interoligomer (lateral tiling) contacts are shaded brown, and those facing the intermonomer interface are yellow. Asterisks indicate substitutions of conserved residues at the interoligomer surface in CcmK3. Numbering below follows Halothece CcmK3. Boxes highlight important regions. B, Ratio of dose response to irradiation of CcmK3 residues, comparing a CcmK3-CcmK4 heterocomplex to isolated CcmK3. C, Concave surface of a CcmK3 model replacing a protomer of a CcmK4 hexamer. Blue coloring shows residues in CcmK3 buried upon interaction with CcmK4 as determined by decreased dose response. D, Structural model of a CcmK3-CcmK4 heterohexamer (assuming 1:2 ratio and symmetrical distribution). Two residues involved in interoligomer contacts in CcmK4 (D50, R81) are colored in green, and the residues replacing them in CcmK3 are in red.
Sequence and Structural Analysis and Molecular Dynamics Simulations of CcmK3, CcmK4, and Their Complexes
An alignment of CcmK3 with the paralogs CcmK2 and CcmK4 reveals conservation of intermonomer residues (Fig. 4A). On the other hand, three residues conserved among CcmK1, CcmK2, and CcmK4 that are required for lateral contacts between adjacent hexamers in shell facets are not conserved in CcmK3 (D49P, R80N, and H42P). Another distinctive feature of CcmK3 is the substitution of a Glu residue for an absolutely conserved Gly in the sequence motif that converges at the pore in all CcmK homohexamers (Fig. 4A; Kerfeld et al., 2005; and figure 2C in Sommer et al., 2017). Given the differences in primary structure features between CcmK3 and its paralogs, we compared energetics of their (putative) oligomeric states by molecular dynamics.
Quantifying the propensity for formation of (hetero)hexamers by Halothece BMC-H proteins would require investigating the process by which six monomers come together to form a hexamer. Molecular dynamics simulations of this self-assembly process is, however, currently intractable. Instead, we chose to study models of hexameric assemblies under physiological conditions and compare their structures during all atom relaxation simulations to gain insights into the propensity of monomers to form hexamers.
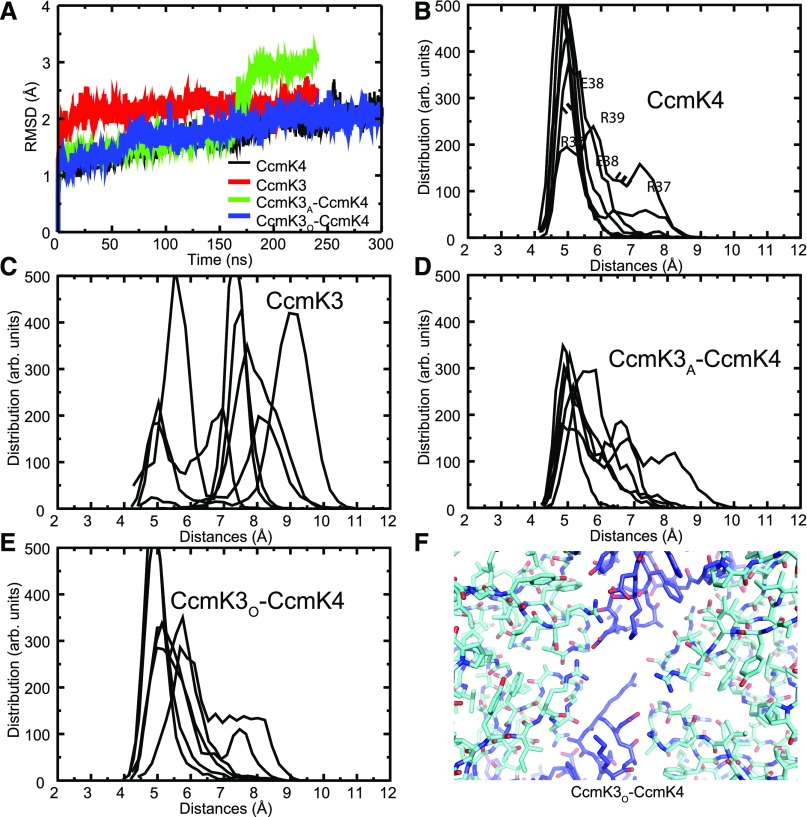
We used the crystal structure of the Halothece CcmK4 hexamer (Fig. 2D), and we constructed a homology model of a CcmK3 hexamer using this structure as a template. We also constructed two different CcmK3-CcmK4 heterohexamers using the experimentally observed 1:2 stoichiometry. In CcmK3A-CcmK4, the two CcmK3 monomers are adjacent to each other, and in CcmK3O-CcmK4 the CcmK3 monomers are opposite one another (separated by two CcmK4 monomers). The simulations were computed over a window of 240 ns for CcmK3, CcmK3A-CcmK4 and a window of 300 ns for CcmK4 and CcmK3O-CcmK4.
We used root mean squared deviations (RMSDs) between the initial and the relaxed state at specific time points to track the course of the simulation. After an initial fast rise, the RMSD of the CcmK3 hexamer reaches a plateau after 50 ns, indicating that the simulation has reached equilibrium (Fig. 5A). In contrast, the RMSDs of the CcmK4, CcmK3O-CcmK4, and CcmK3A-CcmK4 hexamers continue to increase and only reach a plateau after 200 ns in the simulations. A sharp RMSD change takes place in CcmK3A-CcmK4 at 160 ns, at which time the RMSD increases from 1.5 Å to 3 Å within ∼30 ns. This dramatic change is due to a conformational transition of the C-terminal helix of a subunit that dissociates from the compact structure of the monomer and swivels almost 180° to interact with a neighboring subunit. This helix is found disordered in some crystal structures (Tanaka et al., 2009) and interacts with neighboring hexamers in others (Kerfeld et al., 2005; Samborska and Kimber, 2012; Cai et al., 2015). The flexibility could help it sample different conformations, yielding to the double stacking interactions observed previously for CcmK2 (Samborska and Kimber, 2012) as well as here for Halothece CcmK4. Analysis of the RMSD of CcmK4 reveals a peak at around 100 ns, corresponding to a similar structural transition.
Figure 5.
Molecular dynamics simulations of a CcmK3-CcmK4 heterohexamer model. A, RMSDs for of all Cα-atoms. CcmK3 and CcmK3A-CcmK4 were found to reach convergence at 240 ns, whereas for CcmK4 and CcmK3O-CcmK4, 300 ns were needed. B to E, Distances between adjacent residues surrounding the central pore of homohexamers and heterohexamers. Shown are Ser-Ser distances in CcmK4, (B); Arg-Arg distances in CcmK3, (C); Arg-Arg, Arg-Ser, and Ser-Ser distances in CcmK3A-CcmK4, (D); and Arg-Ser and Ser-Ser distances in CcmK3O-CcmK4, (E). F, Structure of the final simulation model of CcmK3O-CcmK4 shows interaction between E38 of CcmK3 (blue) and R37 of CcmK4 (cyan). R39 of CcmK3 is pointing to the center of the pore.
The distinctive feature of CcmK3 is a conserved Glu in the pore region; CcmK2 and CcmK4 have a conserved Gly residue in this position (Fig. 4A). We focused on the behavior of the pore residues by calculating the distance between equivalent positions over the whole simulation time. From this calculation we obtained a distribution of distance values (Fig. 5, B–E; Supplemental Fig. S6, A–D): clearly, the pore residues for the CcmK3 homohexamer are further apart than those of the heterohexamers or the CcmK4 homohexamer. This distance is probably a consequence of each monomer of CcmK3 contributing two bulky residues each (Glu and Arg) to the pore region, which causes steric clashes. Such interactions are ameliorated in the CcmK3-CcmK4 heterohexamers. Moreover, for the heterohexamers, the pore is larger in the CcmK3A-CcmK4 heterohexamer, in which bulky residues are placed in adjacent monomers, than in the CcmK3O-CcmK4 heterohexamer. As monomers converge at the pore, repulsion between the bulky residues likely precludes the formation of a stable CcmK3 homohexamer in solution. For CcmK4, CcmK3A-CcmK4, and CcmK3O-CcmK4, in which there is comparatively less intermonomer repulsion, there is a comparatively smaller energetic penalty when comparing the initial versus the final states of the simulations (Supplemental Fig. S7). These data are consistent with a ratio of two CcmK3 to four CcmK4 as observed experimentally (Supplemental Fig. S5, Supplemental Table S2).
Notably, the final state of the CcmK3O-CcmK4 simulation reveals an interesting interaction between the absolutely conserved Glu-38 in CcmK3 and Arg-38 in CcmK4 (Fig. 5F). The hydrogen bonds between these residues could stabilize and therefore favor a heterohexameric K3-K4 complex. Those interactions are present over the whole course of the simulation (Supplemental Figure S6E). Arg-39 of CcmK3 is pointing to the center of the pore and seems to close it in this conformation.
DISCUSSION
In this study we re-evaluated the function of the satellite locus-encoded β-carboxysome shell proteins CcmK3 and CcmK4 for autotrophic growth of Syn7942. Growth experiments with ccmK3 and ccmK4 deletion mutants indicate that the loss of CcmK4 reduces growth (Fig. 2A), implying that the ccmK4 mutant cannot be functionally compensated by ccmK3. This indication is similar to what was previously observed for a ΔccmK4 mutant of Syn6803 (Zhang et al., 2004), but it stands in contrast to a study reporting that both ccmK3 and ccmK4 need to be deleted to cause retarded growth in Syn7942 (Rae et al., 2012). The apparent functional redundancy observed by Rae et al. (2012) may be partly due to the higher pH of the media used in their phenotypic characterization (pH 8.0 versus pH 7.4); we also see a slightly higher grow rate of the ccmK4 knockout in control experiments at pH 8.0 (Supplemental Fig. S1). Our results indicate that deletion of ccmK3 has little effect on growth, and ccmK3 was unable to rescue the ccmK3-ccmK4 mutant (Fig. 2, A–B). These data and the strict co-occurrence of ccmK3 and ccmK4 (Sommer et al., 2017) support our conclusion that CcmK3 and CcmK4 are not functionally redundant.
Our results also indicate the two paralogs are structurally distinct. In contrast to CcmK4 (Fig. 2D) (Kerfeld et al., 2005; Cai et al., 2015), purified CcmK3 does not form a hexamer; instead it forms a smaller oligomer, either a dimer or trimer (Fig. 2C). Coelution of a complex of CcmK3 and CcmK4 after tandem-purification (Fig. 3B) demonstrates that CcmK3 and CcmK4 form a stable heterocomplex; and the interaction between these two paralogs appears to be specific, because neither of the proteins forms a heterocomplex with CcmK2 (Fig. 3A). To verify that this distinction was not an attribute of CcmK3 and CcmK4 from Halothece specifically, we reproduced complex formation using CcmK3-CcmK4 from Syn7942 (Fig. 3B, far right lane of native PAGE). CcmK4 by itself has previously been shown to form homohexamers when expressed separately (Kerfeld et al., 2005; Cai et al., 2015).
Both quantitative analysis by dot blotting and tandem mass spectrometry indicate a ratio of four CcmK4 to two CcmK3 monomers (Supplemental Fig. S5, Supplemental Table S1) in the CcmK3-CcmK4 complex. In XFMS analysis, the CcmK3 residues that would be at an intermonomer interface within the CcmK3-CcmK4 hexamer were less accessible to solvent in the presence of CcmK4 (Fig. 4, B and C). In conjunction with the solution state data, this result strongly suggests that CcmK3 and CcmK4 assemble into heterohexamers (Fig. 4D).
In Halothece CcmK3, the sequence motif that forms the pore in other paralogs contains two bulky charged residues, a Glu (E38) and an Arg (R39) residue, in which generally small and uncharged residues are found (Sommer et al., 2017). Among structurally characterized hexamers, an exception is the RMM1 BMC-H protein, which has a conserved Asp at the pore (Mallette and Kimber, 2017); however, this Asp is in the structurally equivalent position of the R39 residue and is flanked by two Gly residues that might provide additional space that would alleviate steric clashes. The larger, charged sidechains in CcmK3 may sterically and/or electrostatically preclude formation of stable homohexamers. The physical properties of the pore formed by CcmK3-CcmK4 heterohexamers when modeled are substantially different from those of a CcmK4 homohexamer pore (Fig. 5F) and, accordingly, these properties are predicted to alter its permeability. The CcmK4 S40D mutant was created to simulate the effect that insertion of CcmK3 into a mixed hexamer with CcmK4 may have on the pores. The mutant protein was unable to rescue the ΔccmK3-ccmK4 mutant. Expression and purification of CcmK4 S40D in E. coli confirmed that it was able to form hexamers (Supplemental Fig. S8). It is thus likely that CcmK4 homohexamers function as specific metabolite transporters and that the observed phenotype is due to altered biochemical properties of the pore. In CcmK3-CcmK4 heterohexamers, the bulky and charged sidechains of the CcmK3 loop converging at the pore would alter the permeability characteristics. Our results suggest that CcmK4 can form both homohexamers and heterohexamers with CcmK3, and the two types of hexamers have distinct permeability properties.
Formation of two functionally distinct complex types by CcmK4 may increase the flexibility of carboxysome shell composition and permeability. It also provides an explanation for the positioning of ccmK3 and ccmK4 in a satellite locus. The β-carboxysome is one of the few BMCs that contain its constituents in satellite loci, distal to essential organelle genes (Axen et al., 2014), which enables their differential expression (Sommer et al., 2017). This distinction is particularly important for the β-carboxysome, as it is to date the only known constitutively expressed BMC type, and the permeability of its shell may require rapid adjustments to changing environmental conditions.
We confirmed that CcmK3 is carboxysome associated (Supplemental Fig. S1), although it was not identified in purified Syn7942 carboxysomes, implying that it is a minor component (Faulkner et al., 2017). Prediction of interhexamer interactions based on sequence analysis indicates that the edges formed by CcmK3 are atypical (Fig. 4A) and would likely reduce the affinity of the heterohexamer for embedding in facets. For example, residues highly conserved across BMC-H proteins (e.g. R81, H83 in Halothece CcmK4) involved in forming the interhexamer interface (Sinha et al., 2014; Sutter et al., 2017) are replaced by Asn and Pro in Halothece CcmK3 (Fig. 4A). CcmK3 also lacks an acidic residue (D50 in Halothece CcmK4 is a Pro in CcmK3) that is conserved in other CcmK proteins and is involved in establishing hexamer-pentamer contacts (Sommer et al., 2017; Sutter et al., 2017). This supports our hypothesis that heterohexamers containing CcmK3 are less likely to be integrated into the shell.
In addition to hexamers, CcmK3-CcmK4 complexes can also form dodecamers (Fig. 3, C and D). Interaction occurs at the concave surfaces of two hexamers, as indicated by the decreased solvent accessibility of the concave surface of CcmK3 measured in heterocomplexes during XFMS analysis in conditions promoting dodecamer formation (Fig. 4B). This finding would also be consistent with the dodecamer formation observed previously for CcmK2 (Samborska and Kimber, 2012) as well as for our CcmK4 crystal structure. However, both of those cases are interactions between homohexamers. The CcmK3-K4 heterohexamers provide the possibility of a heterotypic interface that could potentially have more favorable interactions. Interestingly, the sequence predicted to be the C-terminal helix in CcmK3 (E98-R101) is quite conserved (Sommer et al., 2017), supporting such a hypothesis.
The percentage of CcmK3-CcmK4 heterohexamers forming dodecamers varied with buffer conditions (Fig. 3, C and D), implying that formation of dodecamers could be dynamic and could potentially be triggered by molecular cues. For example, glycine, which is the precursor of a cyanobacterial osmoregulator (Hagemann, 2011), caused an increase in dodecamerization (Fig. 3C). Dodecamer formation also correlated with pH (Fig. 3D). A structural response to changes of the pH is not unusual for large protein assemblies; the virus capsid, a proteinaceous shell with many parallels to the BMC shell, can be assembled, reshaped, and gated through pH changes (Allison et al., 1995; Speir et al., 1995). A dodecamer has previously been characterized for the Thermosynechococcus elongatus CcmK2 (Samborska and Kimber, 2012). Key residues for this interaction are conserved glutamates on the C-terminal helix, which interestingly are also present in CcmK3 (E98) and CcmK4 (E99 and E103). Their charge state could play a role in the pH-dependent dodecamer formation. Whereas it is impossible to predict all the effects of pH on such a complex system, the pH-dependent dimerization of BMC-H hexamers could represent a means to dynamically modify proteinaceous compartments through metabolic signals.
Given that the carboxysome shell is a monolayer, what are the possible roles for transient dodecamers? Because the concave surface of the hexamers is exposed to the cytosol, dodecamers could form by the appression of two carboxysomes. This scenario seems unlikely, because evidence suggests that carboxysomes are spatially distributed in the cell (Savage et al., 2010), and because CcmK3-CcmK4 heterohexamers may not be embedded in the facets. Alternatively, free hexamers (expressed but not integrated into the shell) could interact with a facet-embedded hexamer, effectively “capping” it. We propose a model in which CcmK3 and CcmK4 form heterohexamers under specific conditions, and these are recruited to cap hexamers embedded in an extant carboxysome shell (Fig. 1). The dodecameric assemblies observed recapitulate this interaction in isolation, and the looseness of the association is consistent with the proposal that this interaction provides a means for dynamic regulation of shell permeability, i.e. under some conditions the cap will be removed.
A concave-concave interaction of CcmK3-CcmK4 heterohexamers with facet-embedded shell proteins could, thus, be formed on the outside surface of the organelle. An alternative mechanism to regulate shell permeability would be replacing facet-embedded hexamers in extant carboxysomes. But this replacement has been shown to be an infrequent event even in simple monolayers of BMC-H hexamers (Sutter et al., 2016). Moreover, removal of a hexamer would compromise the diffusive barrier provided by the shell and, based on the lack of residue conservation on their perimeters noted previously, CcmK3-CcmK4 heterohexamers seem less likely to integrate into the shell. This possibility is consistent with the absence of any indication of the heterohexamers to form higher order assemblies beyond the stacked dodecamers (Fig. 3, C and D) and the lack of detectable CcmK3 protein in purified β-carboxysomes (Faulkner et al., 2017). Capping hexamers embedded in the facet, however, could provide a means to quickly alter the permeability of the β-carboxysome shell under fluctuating environmental conditions. For example, in light-limited conditions, capping of selected shell protein pores may be used to decrease shell permeability for certain metabolites. This capping may enhance carbon dioxide fixation by Rubisco when metabolism is slow due to light-energy limitation, but the method may not be efficient under ideal growth conditions. Alternatively, certain metabolites could be actively retained in, or precluded from, the carboxysome to control cytosolic metabolite pools and regulate metabolism both rapidly and transiently. The cytosolic pH decreases from 8.4 in light to 7.3 in darkness in cyanobacteria (Mangan et al., 2016); such a change could trigger the formation of dodecamers with CcmK3-CcmK4 heterohexamers in light-limited conditions.
In conclusion, we show that two β-carboxysome shell proteins, CcmK3 and CcmK4, are able to form heterohexamers. The long-standing assumption that all shell proteins form homooligomers has arisen from their structural characterization by x-ray crystallography, which takes as its starting point a single purified BMC-H paralog. The ability to form either CcmK4 homohexamers or CcmK3-CcmK4 heterohexamers, and their expression from a satellite locus, increases the flexibility of carboxysome shell assembly and permeability. This increased flexibility may be advantageous, because the β-carboxysome is an organelle constitutively expressed and subject to environmental changes that alter metabolite levels. To maintain this flexibility, it is essential to express CcmK3 and CcmK4 as separate proteins, which may explain why their genes have not fused to produce a BMC-T gene. Considering that carboxysomes, and BMCs in general, are attracting researchers’ intense efforts to repurpose them for biotechnological applications (Frank et al., 2013; Chessher et al., 2015), shell capping as proposed here for the β-carboxysome offers a unique strategy to altering permeability in a synthetic BMC shell.
MATERIALS AND METHODS
Purification of Recombinant Halothece and Syn7942 CcmK Proteins
For copurification of proteins, ccmK sequences were amplified by PCR and inserted into pCDFDuet-1, fusing an affinity tag sequence to the C terminus of each gene. For the expression of single Halothece proteins, the gene of the other protein was then deleted from the plasmid. Plasmids were transformed into E. coli BL21 cells. After expression (4 h at 37°C) and lysis, soluble proteins were purified by affinity purification. For coexpressed CcmK proteins, lysate was purified with a 5-mL HIS-trap (GE) and subsequently the eluate was applied on a 5-mL STREP-trap (GE). Bio-Rad gels (Mini-PROTEAN 4% to 20%) and Tris-Gly buffers were used for the native and SDS-PAGE of protein samples. Full-size images of the native PAGE gels in Figure 3B are shown in Supplemental Figure S9.
Syn7942 Mutants
Genetic deletions were produced in the Syn7942 wild type through gene replacement with a kanamycin cassette using homologous recombination. Segregation of the strains was confirmed through PCR. For complementation, genes were introduced into neutral site 1, driven by a Ptrc promoter and an Apca ribosome binding site, followed by an aadA gene. Cultures were grown in two different setups, one configuration, using baffled flasks, shaking at 110 rpm in air, BG11 medium buffered with 20-mm Tris at pH 7.4, with a light-emitting diode (LED) lamp (Supplemental Fig. S10) as the light source; 1-mm isopropyl β-D-1-thiogalactopyranoside (IPTG) was added for complementation lines. An LED lamp (Supplemental Fig. S10) was used for illumination. Growth was determined at 150-μe sunlight equivalent, from three replicates at a starting OD730 around 0.3 over the course of 1 h, 4 h after starting the cultures from exponentially growing precultures. The second setup used the same conditions as in Rae et al. (2012))): in air, BG11 medium buffered with 20-mm Tris at pH 7.4 or 25-mm HEPES pH 8.0 and 80 μE of cool-white fluorescent light. Growth rates were determined for three replicates at a starting OD750 of about 0.08 over the course of 2 d, about 4 h after starting the cultures from exponentially growing precultures.
Colocalization by Fluorescent Labeling
pSL175a contains a CcmK3-YFP translational fusion with a GS linker. The fusion gene was cloned downstream of the LacUV promoter in a derivative of pNS3, in which the antibiotic resistance gene has been replaced to Spectinomycin/Streptomycin. pSL175a was introduced into the genome of the wild-type Syn7942 and of the mutant JC007, in which RbcL-CFP fusion is expressed in NS2 from ccmK2 promoter as described in Cameron et al. (2013). Plasmid introduction was carried out by conjugation using the conjugative plasmid pRL443 (Elhai et al., 1997) and the colK derivative pDS4101 (Twigg and Sherratt, 1980).
Western Blots
For western blots of protein gels, proteins were electroblotted to a nitrocellulose membrane. For the dot blot, 50 µL of each protein sample were spotted in technical triplicates on nitrocellulose membrane resting on a glass vacuum filter to minimize bleeding of dots. Immunolabeling of His-tags and Strep-tags was done with 1:5,000 dilutions of 6x-HIS tag antibody-horseradish peroxidase (HRP) Conjugate (Thermo) and StrepTactin-HRP Conjugate (Bio-Rad), respectively. SuperSignal Chemiluminescence Substrate (Thermo) was used to visualize HRP. Signal intensity was determined densitometrically, using ImageJ. Linear regression of the chemiluminescence intensity of CcmK3 standards was fitted with a slope of 7,977 µm−1 and 4.2% inaccuracy (±335 µm−1). The derived concentrations of CcmK3 in two diluted CcmK3-CcmK4 samples were used to fit a second regression line (inaccuracy: 6.0%), from which the CcmK3 concentration in the undiluted CcmK3-CcmK4 sample was calculated.
Size Exclusion Chromatography
Purified proteins were transferred to different buffers using PD10 columns. Two mg of protein were subjected to gel filtration, using a Superdex 200 Increase 10/300 GL column. TBS = 20-mm Tris-HCl, 50-mm NaCl; PBS = 10-mm sodiumphosphate, 50-mm NaCl. For plots, the A280 was normalized to the area under each curve, and the highest overall value was set to 1. Sizes were determined from comparison to the Bio-Rad gel filtration standard, run in TBS pH 8.0.
Sequence Alignment
Sequences of CcmK2/3/4 from Syn7942 and Halo7418 were aligned using Clustalx (Larkin et al., 2007). Residues were marked intermonomer based on the structure described in Sutter et al. (2017). Conservation was assessed based on the sequence conservation logos of CcmK2/3/4 published in Sommer et al. (2017).
Crystallization
Halothece CcmK4 was crystallized at 25°C using the sitting drop vapor diffusion method. Protein sample was concentrated to 25 mg/mL in TBS 20/50, pH 7.4; and 0.2 µL were mixed with 0.2 µL of reservoir solution (40% [w/v] MPD, 0.1-m HEPES, pH 7.0). The crystal was mounted in nylon loops, cryo-stabilized by adding ethylene glycol and flash frozen in liquid nitrogen. Diffraction data were integrated with XDS (Kabsch, 2010) and scaled with SCALA (www.ccp4.ac.uk) (Winn et al., 2011). The structure was solved using molecular replacement with CcmK4 of Syn6803 (PDB ID 2A10). Manual rebuilding/refinement cycles using Coot (Emsley and Cowtan, 2004) and phenix.refine (Afonine et al., 2012) were then applied to improve the model. Statistics for diffraction data collection, structure determination, and refinement are summarized in Supplemental Table S3. Figures of crystal structures and homology models were prepared using pymol (www.pymol.org). To create the CcmK3-CcmK4 heterohexamer model, two opposing monomers of the CcmK4 hexamer were replaced by CcmK3 models using the “align” function. The Protein Data Bank accession ID for Halothece CcmK4 is 5VGU.
XFMS
During XFMS, the microsecond x-ray pulse ionizes both bulk and bound water rapidly and isotropically to generate hydroxyl radicals in situ, which then react with proximal residues to yield stable modification products (Xu and Chance, 2007; Gupta et al., 2014). Purified CcmK3 and CcmK3-CcmK4 sample was exchanged into 10-mm sodium phosphate, pH 7.0, 50-mm NaCl on a PD10 column and diluted to 10 μM. The samples were subjected to a series of increasing exposure, from 0-μs to 400-μs time range as described previously (Gupta et al., 2014, 2015). High-resolution quantitative bottom-up liquid chromatography–tandem mass spectrometry (LCMS) provided a residue-specific dose response for each site of modification and covered ∼80% of sequence of the protein (Supplemental Figure S11, Supplemental Table S4). Modifications were detected at 25 CcmK3 residues. The pseudo-first-order fit of the dose-response gives the hydroxyl radical reactivity rate constant (k s−1), which is a measure of both intrinsic reactivity and solvent accessibility of the modified side chain (Supplemental Figure S11). In a comparative structural analysis, the R of rate constants between CcmK3 and CcmK3-CcmK4 sample determines the relative solvent accessibility changes, which are independent of any sequence context (Gupta et al., 2015). Complex formation decrease (R = kCcmK3-CcmK4/kCcmK3 < 1) or (R = kCcmK3-CcmK4/kCcmK3 > 1) in solvent accessibility of certain residues is indicative of binding and conformational change. In our study, we considered more than ∼2.5-fold change in accessibility as significant change (Supplemental Table S4).
Simulation of All Atom Relaxation
A homology model of Halothece CcmK1, K2, and K3 was built with SWISS-MODEL based on the pdb entry 4OX7, which is CcmK2 of Syn7942. During simulation, delta-protonated His parameters were used, simulating buffer conditions of pH ∼8. The Cys residue of CcmK4 was treated as fully protonated. Hydrogen atoms were added using the program VMD (Humphrey et al., 1996), plugin psfgen. Each hexamer was solvated in a 150-nm water cube with 100-mm NaCl, using TIP3P water molecules. All molecular dynamics simulations were performed with the NAMD program (Phillips et al., 2005) with the CHARMM22 force fields. The short-range cutoff distance for nonbonded interactions was set to 12 Å. Long-range electrostatic interactions were calculated through the particle mesh Ewald method using a grid spacing of 1.0 Å. Langevin dynamics and a Langevin piston algorithm were both used, keeping temperature at 310 K and pressure at 1 atm. Time steps of 2 fs were employed. Each system was treated with two equilibration steps: 1,000 steps of minimization and 240 (300) ns equilibration for CcmK3, CcmK3A-CcmK4 (CcmK4, CcmK3O-CcmK4).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EMBL ABB57451.1 (ccmK2), ABB56316.1 (ccmK3), and ABB56317.1 (ccmK4) for Syn7942, and EMBL AFZ45643.1 (ccmK1), AFZ45642.1 (ccmK2), AFZ42582.1 (ccmK3), and AFZ42581.1 (ccmK4) for Halo.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Growth rates of Syn7942 ΔccmK3 and ΔccmK4 mutants.
Supplemental Figure S2. Colocalization of labeled CcmK3 with carboxysomes.
Supplemental Figure S3. Gel filtration of purified Syn7942 CcmK2.
Supplemental Figure S4. Copurification of CcmK proteins with a HIS-trap and a STREP-trap column.
Supplemental Figure S5. Densitometric analysis of CcmK3:CcmK4 R.
Supplemental Figure S6. Analysis of the pore residue distances in the molecular dynamics simulations of CcmK3-4 complexes.
Supplemental Figure S7. Initial and equilibrated arrangement of the Cα atoms surrounding the pore in all the hexamers studied.
Supplemental Figure S8. Complex formation of Syn7942 CcmK complexes.
Supplemental Figure S9. Full-size images of the native PAGE gels in Figure 3B.
Supplemental Figure S10. Light spectrum of the LED lamp used for growth of Syn7942 cultures.
Supplemental Figure S11. Dose response plots for the comparative XFMS analysis of CcmK3 (black) and CcmK3-CcmK4 (red) complexes.
Supplemental Table S1. CcmK4 interhexamer complex interaction statistics.
Supplemental Table S2. Quantification of CcmK3 and CcmK4 by LC/MS-MS.
Supplemental Table S3. Data collection and refinement statistics of Halo CcmK4 crystal structure analysis.
Supplemental Table S4. R of hydroxyl radical reactivity between CcmK3-4 complex and CcmK3.
Acknowledgments
Thank you to the Center for Nanophase Materials Sciences (a DOE Office of Science User Facility), where molecular dynamics simulations were conducted.
Footnotes
Senior author.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (to M.So.) and the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy (DOE) (award number DE–FG02–91ER20021) with infrastructure support from Michigan State University AgBioResearch (to C.A.K.). This work used resources of the Advanced Light Source, the Joint BioEnergy Institute, and the National Energy Research Scientific Computing Center, all supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy (contract no. DE–AC02–05CH11231).
Articles can be viewed without a subscription.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX (1995) Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J Virol 69: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axen SD, Erbilgin O, Kerfeld CA (2014) A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLOS Comput Biol 10: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Hanson DT, Price GD (2002) Evolution and diversity of CCMs in cyanobacteria. Funct Plant Biol 29: 161–173 [DOI] [PubMed] [Google Scholar]
- Badger MR, Price GD, Long BM, Woodger FJ (2006) The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J Exp Bot 57: 249–265 [DOI] [PubMed] [Google Scholar]
- Billis K, Billini M, Tripp HJ, Kyrpides NC, Mavromatis K (2014) Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation. PLoS One 9: e109738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA (2013) The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem 288: 16055–16063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA (2015) Engineering bacterial microcompartment shells: Chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol 4: 444–453 [DOI] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA (2013) Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell 155: 1131–1140 [DOI] [PubMed] [Google Scholar]
- Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM (2001) Microcompartments in prokaryotes: Carboxysomes and related polyhedra. Appl Environ Microbiol 67: 5351–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessher A, Breitling R, Takano E (2015) Bacterial microcompartments: biomaterials for synthetic biology-based compartmentalization strategies. ACS Biomater Sci Eng 1: 345–351 [DOI] [PubMed] [Google Scholar]
- Drews G, Niklowitz W (1957) Beiträge zur Cytologie der Blaualgen. III. Untersuchungen über die granulären Einschlüsse der Hormogonales. Arch Mikrobiol 25: 333–351 [PubMed] [Google Scholar]
- Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP (1997) Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J Bacteriol 179: 1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12): 2126–2132. [DOI] [PubMed] [Google Scholar]
- Faulkner M, Rodriguez-Ramos J, Dykes GF, Owen SV, Casella S, Simpson DM, Beynon RJ, Liu LN (2017) Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes. Nanoscale 9: 10662–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Lawrence AD, Prentice MB, Warren MJ (2013) Bacterial microcompartments moving into a synthetic biological world. J Biotechnol 163: 273–279 [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F, Belnap J, Neuer S, Schanz F (2003) Estimates of global cyanobacterial biomass and its distribution. Algol Stud 109: 213–227 [Google Scholar]
- Gupta S, Celestre R, Petzold CJ, Chance MR, Ralston C (2014) Development of a microsecond x-ray protein footprinting facility at the Advanced Light Source. J Synchrotron Radiat 21: 690–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Guttman M, Leverenz RL, Zhumadilova K, Pawlowski EG, Petzold CJ, Lee KK, Ralston CY, Kerfeld CA (2015) Local and global structural drivers for the photoactivation of the orange carotenoid protein. Proc Natl Acad Sci USA 112: E5567–E5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M. (2011) Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35: 87–123 [DOI] [PubMed] [Google Scholar]
- Hernández-Prieto MA, Semeniuk TA, Giner-Lamia J, Futschik ME (2016) The transcriptional landscape of the photosynthetic model cyanobacterium Synechocystis sp. PCC6803. Sci Rep 6: 22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: Visual molecular dynamics. J Mol Graph 14: 33–38, 27––28. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010) XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Melnicki MR (2016) Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol 31: 66–75 [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309: 936–938 [DOI] [PubMed] [Google Scholar]
- Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA (2009) Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol 392: 319–333 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, et al. (2007) Clustal W and clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Larsson AM, Hasse D, Valegård K, Andersson I (2017) Crystal structures of β-carboxysome shell protein CcmP: Ligand binding correlates with the closed or open central pore. J Exp Bot 68: 3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR (2014a) β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MA, Hanson MR (2014b) A faster Rubisco with potential to increase photosynthesis in crops. Nature 513: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD (2007) Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem 282: 29323–29335 [DOI] [PubMed] [Google Scholar]
- Mallette E, Kimber MS (2017) A complete structural inventory of the mycobacterial microcompartment shell proteins constrains models of global architecture and transport. J Biol Chem 292: 1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan NM, Flamholz A, Hood RD, Milo R, Savage DF (2016) pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. Proc Natl Acad Sci USA 113: E5354–E5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JM, Long SP (2014) Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol 164: 2247–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR (1989) Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO(2)-requiring phenotype: evidence for a central role for carboxysomes in the CO(2) concentrating mechanism. Plant Physiol 91: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Pengelly JJ, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR (2013) The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J Exp Bot 64: 753–768 [DOI] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD (2012) Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: Roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 7: e43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD (2013) Functions, compositions, and evolution of the two types of carboxysomes: Polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev 77: 357–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EW, Cai F, Kerfeld CA, Cannon GC, Heinhorst S (2012) Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J Bacteriol 194: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samborska B, Kimber MS (2012) A dodecameric CcmK2 structure suggests β-carboxysomal shell facets have a double-layered organization. Structure 20: 1353–1362 [DOI] [PubMed] [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA (2010) Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327: 1258–1261 [DOI] [PubMed] [Google Scholar]
- Shively JM, Ball FL, Kline BW (1973) Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J Bacteriol 116: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Cheng S, Sung YW, McNamara DE, Sawaya MR, Yeates TO, Bobik TA (2014) Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol 426: 2328–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AK, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC (2004) A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J Bacteriol 186: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M, Cai F, Melnicki M, Kerfeld CA (2017) β-Carboxysome bioinformatics: Identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J Exp Bot 68: 3841–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir JA, Munshi S, Wang G, Baker TS, Johnson JE (1995) Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by x-ray crystallography and cryo-electron microscopy. Structure 3: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Wilson SC, Deutsch S, Kerfeld CA (2013) Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species. Photosynth Res 118: 9–16 [DOI] [PubMed] [Google Scholar]
- Sutter M, Faulkner M, Aussignargues C, Paasch BC, Barrett S, Kerfeld CA, Liu LN (2016) Visualization of bacterial microcompartment facet assembly using high-speed atomic force microscopy. Nano Lett 16: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Greber B, Aussignargues C, Kerfeld CA (2017) Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356: 1293–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR. (1999) Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 60: 1–28 [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO (2008) Atomic-level models of the bacterial carboxysome shell. Science 319: 1083–1086 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Sawaya MR, Phillips M, Yeates TO (2009) Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci 18: 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg AJ, Sherratt D (1980) Trans-complementable copy-number mutants of plasmid ColE1. Nature 283: 216–218 [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Chance MR (2007) Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev 107: 3514–3543 [DOI] [PubMed] [Google Scholar]
- Zarzycki J, Axen SD, Kinney JN, Kerfeld CA (2013) Cyanobacterial-based approaches to improving photosynthesis in plants. J Exp Bot 64: 787–798 [DOI] [PubMed] [Google Scholar]
- Zhang S, Laborde SM, Frankel LK, Bricker TM (2004) Four novel genes required for optimal photoautotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803 identified by in vitro transposon mutagenesis. J Bacteriol 186: 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]