Proteomics analysis reveals the mechanisms underlying root architectural changes in response to N deficiency.
Abstract
Rapeseed (Brassica napus) is an important oil crop worldwide. However, severe inhibition of rapeseed production often occurs in the field due to nitrogen (N) deficiency. The root system is the main organ to acquire N for plant growth, but little is known about the mechanisms underlying rapeseed root adaptions to N deficiency. Here, dynamic changes in root architectural traits of N-deficient rapeseed plants were evaluated by 3D in situ quantification. Root proteome responses to N deficiency were analyzed by the tandem mass tag-based proteomics method, and related proteins were characterized further. Under N deficiency, rapeseed roots become longer, with denser cells in the meristematic zone and larger cells in the elongation zone of root tips, and also become softer with reduced solidity. A total of 171 and 755 differentially expressed proteins were identified in short- and long-term N-deficient roots, respectively. The abundance of proteins involved in cell wall organization or biogenesis was highly enhanced, but most identified peroxidases were reduced in the N-deficient roots. Notably, peroxidase activities also were decreased, which might promote root elongation while lowering the solidity of N-deficient roots. These results were consistent with the cell wall components measured in the N-deficient roots. Further functional analysis using transgenic Arabidopsis (Arabidopsis thaliana) plants demonstrated that the two root-related differentially expressed proteins contribute to the enhanced root growth under N deficiency conditions. These results provide insights into the global changes of rapeseed root responses to N deficiency and may facilitate the development of rapeseed cultivars with high N use efficiency through root-based genetic improvements.
Nitrogen (N) is an essential macronutrient for the synthesis of proteins, nucleic acids, and many secondary metabolites and is a key factor in ensuring high crop yields. N deficiency affects crop growth and development, with symptoms including leaf chlorosis, stunted plants, and decreased yields (Coruzzi and Bush, 2001; Kusano et al., 2011). In many agricultural regions, increases in crop production rely on chemical N fertilization. The consumption of N fertilizers worldwide has increased more than 9-fold since 1960 (Hinsinger et al., 2011). However, excessive use of chemical N fertilizers has caused severe environmental problems, such as eutrophication and global warming (Gruber and Galloway, 2008; Zheng et al., 2015). Therefore, it is increasingly important to find widely applicable solutions for reducing chemical N fertilization without decreasing crop production.
Understanding the mechanisms of plant adaptation to N deficiency is crucial for improving crop N efficiency and reducing chemical N fertilization. It has been well documented that plants have evolved a number of mechanisms to adapt to N-deficient soils (Scheible et al., 2004; Hermans et al., 2006), with known responses including enhancement of N uptake and translocation, remobilization of N from source organs to newly growing tissues, and alteration of carbohydrate partitioning (Paul and Driscoll, 1997; Fan et al., 2009; Krapp et al., 2011). Changes in root architecture are important for enhancing plant N acquisition under N deficiency conditions (Liu et al., 2008; Gao et al., 2015; Li et al., 2016). In recent years, several studies have identified genes involved in the regulation of root architecture under low-N conditions, with most of the focus on the model plant Arabidopsis (Arabidopsis thaliana). One gene, NRT1.1, a dual-affinity nitrate transporter, also is an auxin transporter. The activity of NRT1.1 can affect auxin distribution and thereby modify root architecture (Mounier et al., 2014). Another gene, auxin signaling F-box3 (AFB3), encodes an auxin receptor targeted by a nitrate-inducible microRNA, miR393, that has been shown to integrate internal and external N availability signals to control root architecture. Knockout of AFB3 and overexpression of miR393 has been shown to alter the nitrate-controlled growth of primary and lateral roots (Vidal et al., 2010). In maize (Zea mays), rootless with undetectable meristems1, which encodes an auxin/indole-3-acetic acid protein, is required for the initiation of embryonic seminal and postembryonic lateral roots (von Behrens et al., 2011). Thus, specific roles have been identified for a number of genes with regard to root responses to low N. However, the global mechanisms underlying rapeseed (Brassica napus) root adaption to N deficiency remain largely unknown.
Since cellular proteins perform most of the catalytic work required by biological processes, understanding relevant protein dynamics is likely to yield insights into how plants respond to environmental stimuli. To this end, proteomic techniques can be important for exploring molecular mechanisms underlying plant adaptations to abiotic stress. As such, there are numerous publications available detailing proteomics studies of plants experiencing nutrient deficiency stresses, primarily N, phosphorus, and iron (Rellán-Alvarez et al., 2010; Chen et al., 2011, 2015; Wang et al., 2012). Although several stress-responsive proteins have been identified in these studies, they have been limited nonetheless by the utilization of 2D gel electrophoresis, a technique that is insufficient for the separation of large and small proteins as well as highly acidic or hydrophobic proteins (Zieske, 2006). Moreover, low-abundance proteins, such as membrane proteins, are difficult to detect using this method. Developed shotgun proteomic techniques, such as tandem mass tag (TMT) labeling-based methods, have overcome most of these limitations and have enabled protein identification and the quantification of relative changes in complex samples across multiple experimental conditions (Zieske, 2006; Turek et al., 2015; Wang et al., 2016; Hao et al., 2017). Therefore, modern proteomic techniques promise to contribute novel insights into plant adaptation to environmental stresses.
Rapeseed is the third most widely grown oil crop. Due to the high protein content in seeds, large amounts of N are required to increase rapeseed yield. Root architecture determines the ability of plants to acquire N from soils. Therefore, an understanding of root architectural responses to low N availability is essential for determining the potential of rapeseed to improve its tolerance to N deficiency. Although the response to N deficiency at the protein level has been studied in some crops (Møller et al., 2011; Wang et al., 2012), little proteomic-based information is available in rapeseed, especially for roots. In this work, dynamic changes of root architectural traits in N-deficient rapeseed plants were evaluated by 3D in situ quantification. Root proteomes from N-deficient and N-sufficient rapeseed were compared through TMT-based proteomic techniques, and proteins with significant alterations in expression were characterized further. The objective of this research is to provide the fundamental information needed to identify the strategies employed by rapeseed roots to cope with N deficiency. This information could be used for research focused on improving N use efficiency and the production of rapeseed.
RESULTS
Architectural Responses of Rapeseed Roots to N Deficiency
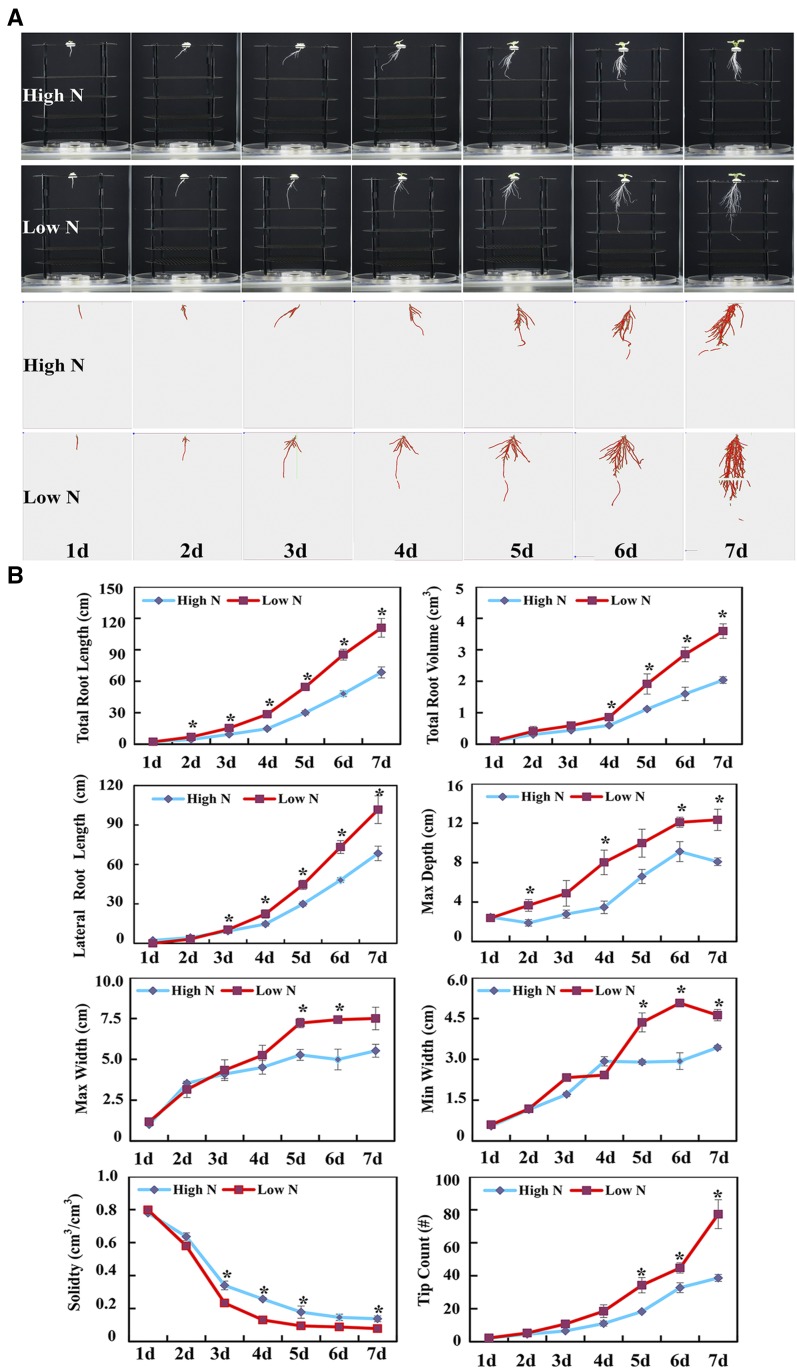
To investigate the dynamic responses of rapeseed root architecture to N deficiency, 3D in situ root reconstructions were generated daily and analyzed in the RootReader3D platform (Clark et al., 2011). As illustrated in Figure 1A, rapeseed root architecture was altered dramatically by N availability, namely high N (7,500 μm N) or low N (190 μm N in hydroponics). Among all the quantified root architectural traits, total root length and maximum depth (namely main root length) of roots were most sensitive to N availability, with significant effects observed after 2 d of N treatment (Fig. 1B). Changes in lateral tip count, minimum width, and convex hull volume all reached significant levels after 3 d and reached maximum differences after 5 d of differential N treatment (Fig. 1B). These observations support the notion that deeper and wider roots are needed to expand the soil volume explored when N is limiting. Low N availability also reduced root solidity starting after 3 d of differential N treatment (Fig. 1B). This suggests that N deficiency not only affects root elongation and lateral root branching but also changes root stiffness.
Figure 1.
Architectural and anatomical changes of rapeseed roots as affected by N availability. A, Dynamic changes of rapeseed root system architecture in response to N deficiency. Rapeseed plants were grown in the growth chamber under a 14-h/10-h light/dark photoperiod in hydroponic solution containing 7,500 μm N (High N) or 190 μm N (Low N) at pH 5.8. 3D reconstructions of root systems were generated from daily imaging of roots over a 7-d period using the RootReader3D hardware and software (Clark et al., 2011). The skeletonized root systems generated with the RootReader3D software are shown in red below. B, Root architectural traits (3D) of rapeseed plants as affected by N availability. Total root length (total lengths of main and lateral roots), total root volume, lateral root length, maximum depth, maximum width, minimum width, solidity, and tip count data were collected after 7 d of N treatments. Max Depth, Maximum vertical depth of the whole root system, namely main root length; Max (or Min) Width, maximum (or minimum) horizontal width of the whole root system. Plots show means ± sd (n = 4). Asterisks indicate significant differences in root architectural traits between the two N treatments (Student’s t test, *, P < 0.05).
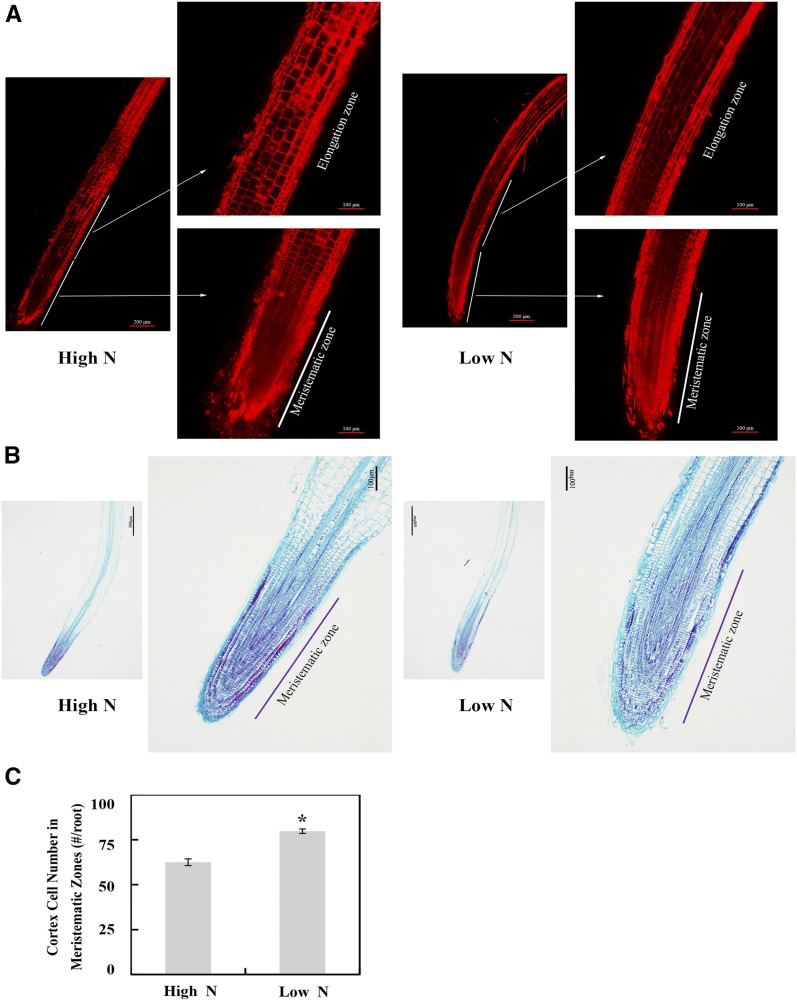
To further determine whether root growth in N-deficient rapeseed is correlated with cell division or cell elongation, the number and size of cells in root tips were examined with a confocal microscope with propidium iodide (PI) staining (Fig. 2A) and with an optical microscope with Fast Green/safranin staining (Fig. 2B), respectively. As shown in Figure 2A, cells in the meristematic zone were denser and cells in the root elongation zone were larger in the N-deficient rapeseed roots than in N-sufficient plants. Additionally, the number of cells in the meristematic zone of root tips was significantly different between two N treatments, with a 27.6% increase in the N-deficient plants (Fig. 2, B and C). This result indicates that both cell division and cell expansion might contribute to the increased root length of rapeseed in response to N deficiency.
Figure 2.
Effects of N availability on cell number and cell size in the tips of rapeseed main roots. A, Images of root tips stained with PI using a confocal laser scanning microscope. B, Images of root tips stained with Fast Green/safranin using an optical microscope. C, Cortex cell number in the meristematic zone. Root tip (0.5-cm root segment starting from the root tip) samples were examined from two N treatments: high N, 7,500 μm N; low N, 190 μm N. The number of cortex cells in the meristematic zone was counted with images from Fast Green/safranin paraffin staining. Data shown are means ± sd (n = 8). The asterisk indicates a significant difference between the two N treatments (Student’s t test, *, P < 0.05).
Overview of Rapeseed Root Proteome Changes in Response to N Deficiency
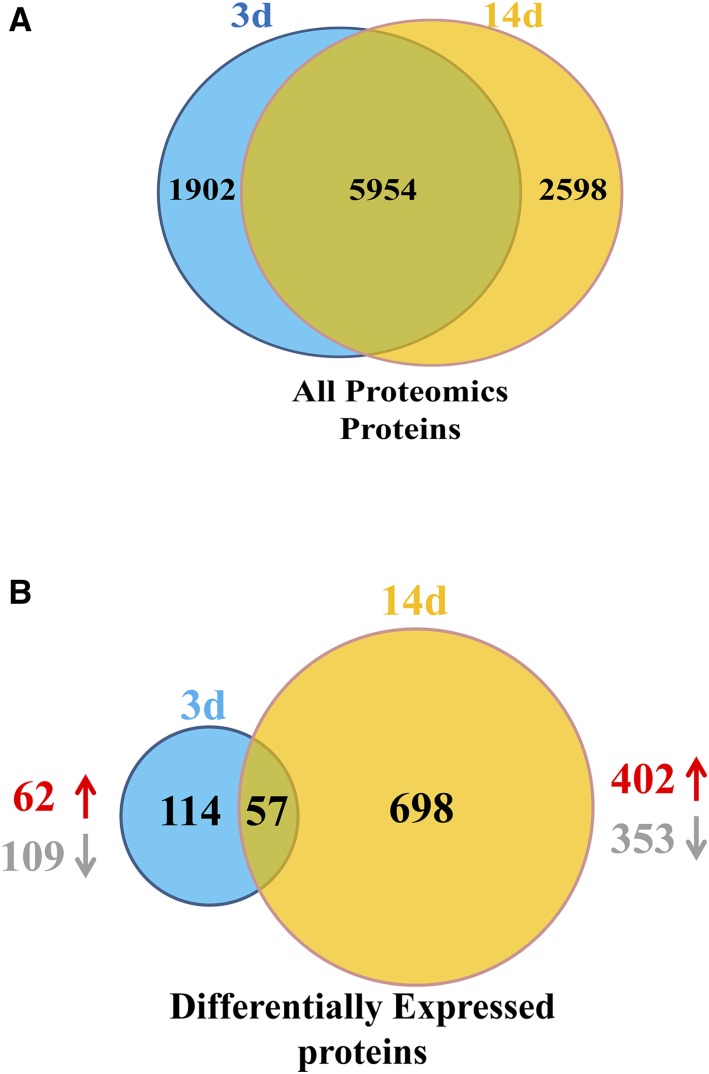
A total of 7,856 and 8,552 rapeseed root proteins were identified through TMT-based proteomic analysis on days 3 and 14, respectively (Fig. 3A). The number of differentially expressed proteins (DEP) increased with increasing duration of N treatment. On day 3, there were 171 root DEPs, of which 109 and 62 were decreased and increased in abundance, respectively, in low N compared with high N (Fig. 3B). Then, 755 DEPs were observed on day 14, of which 353 were decreased in abundance and 402 exhibited increased expression (Fig. 3B). These results illustrate that, as N deficiency progresses, the changes in root architecture and biochemistry become more significant and a larger number of stress-responsive proteins are produced.
Figure 3.
Patterns of proteins and differentially expressed proteins (DEPs) identified in rapeseed roots after implementing the 3-d and 14-d low-N treatment. A, All proteomics proteins. B, DEPs. Venn diagrams show the number of total proteins identified at 3 d (blue) and 14 d (yellow), with the proteins in common to both sampling times in the overlapping sections. Proteins were identified by TMT-based measurements from three biological replicates of rapeseed roots. DEPs were the proteins expressed at different levels in low-N compared with high-N conditions on the same sample day. Up and down arrows represent up-regulated and down-regulated proteins, respectively.
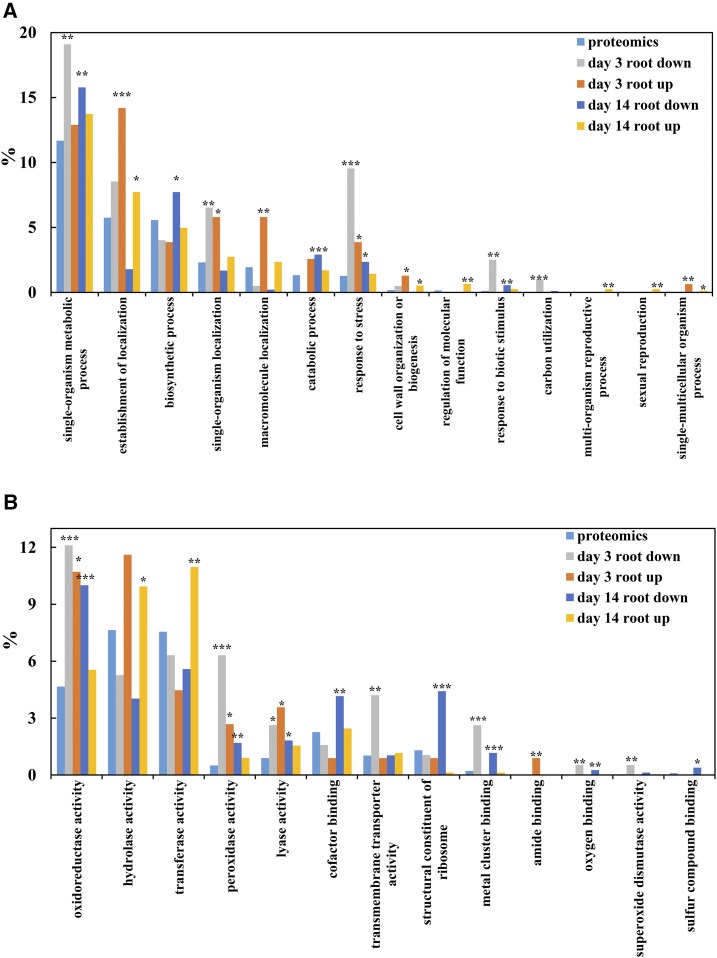
According to Gene Ontology (GO) analysis, all identified proteins and DEPs were classified mainly into 14 biological processes and 13 molecular functions (Fig. 4). For biological processes, the most abundant terms were single-organism metabolism process, followed by establishment of localization, biosynthetic process, and response to stress. In short, terms related to single-organism metabolism process were overrepresented for root down-regulated DEPs on both days 3 and 14 of N deficiency, while establishment of localization was overrepresented for root DEPs with increased abundance (Fig. 4A). Furthermore, oxidoreductase activity, hydrolase activity, transferase activity, and peroxidase (POD) activity represented the most abundant groups in the molecular functions category. It is obvious that oxidoreductase activity was overrepresented significantly for root down-regulated DEPs on both days 3 and 14 of N deficiency and only for root up-regulated DEPs on day 3 (Fig. 4B). Notably, hydrolase activity and transferase activity were overrepresented significantly for root up-regulated DEPs only on day 14 of N deficiency (Fig. 4B).
Figure 4.
Representation of GO categories present among BLAST hits of rapeseed proteins identified 3 and 14 d after implementing the low-N treatments. GO terms were parsed from annotations of the top 10 hits, and parent terms were parsed from the biological process (A) and molecular function (B) ontologies. Overrepresentation was determined using false discovery rate (FDR)-corrected hypergeometric tests at each time point, with comparisons for each set of DEPs made against representation in all proteomics proteins. The significance threshold in FDR-corrected (Benjamini and Hochberg, 1995) hypergeometric tests was q = 0.05. Significantly overrepresented categories are marked by asterisks: *, q < 0.05; **, q < 0.01; and ***, q < 0.001.
DEPs Related to Cell Wall Modifications in Response to N Deficiency
As illustrated above, notable changes in rapeseed root architecture were observed in response to N deficiency (Fig. 1). It is well known that root elongation is based on cell division in the meristematic zone, followed by cell expansion, both of which are tightly associated with cell wall modifications. In this study, terms related to cell wall organization or biosynthesis were overrepresented for root up-regulated DEPs on both days 3 and 14 of N deficiency (Fig. 4A). It is noteworthy that there were more than 4 times as many cell wall-related DEPs detected in day-14 N-deficient roots compared with day-3 roots (Table 1). Specifically, several DEPs were involved in cell wall modification, including xyloglucan endotransglucosylase/hydrolase (XTH; i.e. an enzyme that modulates the synthesis of xylogucans) and expansins (EXPs; i.e. plant cell wall-loosening proteins involved in cell elongation and other developmental processes). The former were up-regulated in both day-3 and day-14 N-deficient roots, and the latter were significantly up-regulated by N starvation in day-14 N-deficient roots but not in day-3 roots (Table 1). Besides XTH and EXP, as mentioned above, other cell wall-modifying enzymes also were significantly up-regulated by N deprivation, including enzymes involved in cellulose synthesis (e.g. endoglucanase, cellulose synthase, and chitinase-like proteins) and enzymes involved in pectin biosynthesis or modification (e.g. galacturonosyl transferase, putative pectate lyase, and pectin methylesterase). The up-regulation of cell wall-modifying proteins and enzymes functioning in cell wall extensibility and plasticity might mediate cell enlargement and expansion during root elongation under N deficiency, as quantified using 3D in situ root analysis systems.
Table 1. Up-regulated DEPs related to root growth of rapeseed plants after 3 and 14 d of high-N and low-N treatment.
| Protein Identifier | Arabidopsis Homolog | Ratio 3 d | Ratio 14 d | Abbreviation in Arabidopsis | Protein | No. of Unique Peptides | No. of Peptides | No. of Peptide Spectrum Matches |
|---|---|---|---|---|---|---|---|---|
| Cell wall organization or biogenesis | ||||||||
| BnaAnng23050D | NP_194756.1 | 1.750 | – | XTH24 | Xyloglucan:xyloglucosyl transferase | 1 | 2 | 6 |
| BnaCnng62600D | NP_178708.1 | 1.659 | 1.816 | XTH4 | Xyloglucan endotransglucosylase/hydrolase4 | 11 | 13 | 28 |
| BnaC03g55190D | NP_190085.1 | – | 2.227 | XTH31 | Xyloglucan:xyloglucosyl transferase | 2 | 8 | 19 |
| BnaA10g05530D | NP_199783.1 | – | 2.765 | GH9A1 | Endoglucanase25 | 1 | 14 | 40 |
| BnaA02g17370D | NP_177697.1 | – | 1.943 | GH9B7 | Endoglucanase10 | 4 | 11 | 42 |
| BnaCnng23250D | NP_192138.1 | – | 1.796 | GH9B13 | Endoglucanase17 | 3 | 3 | 3 |
| BnaA03g52020D | NP_194967.1 | – | 1.813 | CESA1 | Cellulose synthase1 | 9 | 19 | 43 |
| BnaA03g55200D | NP_196136.1 | – | 1.899 | CEV1 | CONSTITUTIVE EXPRESSION OF VSP1 | 1 | 10 | 20 |
| BnaC02g02440D | NP_196136.1 | – | 1.811 | CEV1 | CONSTITUTIVE EXPRESSION OF VSP1 | 1 | 10 | 19 |
| BnaC08g35320D | NP_179803.4 | – | 1.614 | CSI1 | Cellulose synthase-interactive protein1 | 10 | 11 | 14 |
| BnaC01g37020D | NP_188048.1 | – | 1.942 | PME3 | Pectinesterase3 | 2 | 4 | 13 |
| BnaC01g37010D | NP_188048.1 | – | 1.541 | PME3 | Pectinesterase3 | 2 | 2 | 3 |
| BnaUnng03540D | NP_190491.1 | – | 1.983 | AT3G49220 | Pectinesterase | 5 | 6 | 7 |
| BnaC02g17500D | NP_564906.1 | – | 1.513 | AT1G67750 | Putative pectate lyase5 | 1 | 2 | 3 |
| BnaA03g30660D | NP_187552.3 | – | 1.649 | AT3G09410 | Putative pectinacetylesterase | 1 | 10 | 40 |
| BnaA04g24790D | NP_181833.1 | 1.723 | – | ATPMEPCRD | Putative pectinesterase/pectinesterase inhibitor16 | 1 | 3 | 5 |
| BnaA04g24790D | NP_181833.1 | – | 2.865 | ATPMEPCRD | Putative pectinesterase/pectinesterase inhibitor16 | 3 | 3 | 5 |
| BnaC07g45050D | NP_195174.6 | – | 1.617 | AT4G34480 | Glucan endo-1,3-β-glucosidase7 | 2 | 5 | 8 |
| BnaC06g03450D | NP_188616.1 | – | 1.905 | DWF1 | DWARF1 | 6 | 12 | 24 |
| BnaC05g03990D | NP_172076.1 | – | 1.523 | POM1 | Chitinase family protein | 1 | 2 | 4 |
| BnaC06g13400D | NP_851111.1 | – | 2.457 | RWP1 | Reduced levels of wall-bound phenolics1 | 1 | 4 | 4 |
| BnaC07g15930D | NP_569048.1 | – | 1.880 | ARA12 | Subtilisin-like protease | 2 | 21 | 52 |
| BnaC07g45310D | NP_567972.1 | – | 1.935 | M4E13.40 | Subtilisin-like Ser protease2 | 3 | 10 | 32 |
| BnaC01g20780D | NP_567803.1 | – | 3.628 | EXPB3 | Expansin B3 | 2 | 10 | 15 |
| BnaA06g40800D | NP_190183.1 | – | 1.786 | EXLA1 | Expansin-like A1 | 2 | 5 | 11 |
| BnaCnng23400D | NP_190183.1 | – | 1.682 | EXLA1 | Expansin-like A1 | 6 | 6 | 12 |
| BnaA10g24090D | NP_196304.1 | 2.183 | – | PGIP1 | Polygalacturonase inhibitor1 | 3 | 3 | 5 |
| BnaA03g29620D | NP_850525.1 | – | 1.553 | AT3G06770 | Polygalacturonase-like protein | 1 | 4 | 6 |
| BnaA03g34310D | NP_188308.1 | – | 1.917 | AT3G16850 | Glycoside hydrolase family28 protein | 10 | 10 | 15 |
| BnaC09g45990D | NP_196618.1 | – | 1.524 | AT5G10560 | Putative β-d-xylosidase6 | 7 | 16 | 44 |
| BnaA07g06190D | NP_189024.1 | – | 1.576 | GAE6 | UDP-d-glucuronate 4-epimerase6 | 3 | 4 | 5 |
| BnaCnng29190D | NP_186768.1 | 2.397 | – | AT3G01190 | Peroxidase27 | 2 | 13 | 49 |
| BnaC01g31810D | NP_188814.1 | 2.204 | – | AT3G21770 | Peroxidase30 | 3 | 4 | 11 |
| BnaA02g02520D | NP_197022.1 | 1.618 | – | AT5G15180 | Peroxidase56 | 2 | 14 | 52 |
| BnaA01g11800D | NP_567641.1 | – | 2.584 | PRXR1 | Peroxidase42 | 10 | 10 | 25 |
| BnaC01g31810D | NP_188814.1 | – | 1.830 | AT3G21770 | Peroxidase30 | 6 | 12 | 24 |
| BnaA09g08390D | NP_850652.1 | – | 1.806 | AT3G32980 | Peroxidase32 | 1 | 12 | 99 |
| BnaC09g08690D | NP_850652.1 | – | 1.791 | AT3G32980 | Peroxidase32 | 2 | 14 | 112 |
| BnaCnng07140D | NP_188814.1 | – | 1.694 | AT3G21770 | Peroxidase30 | 6 | 12 | 31 |
| BnaC08g35130D | NP_179828.1 | – | 1.607 | AT2G22420 | Peroxidase | 2 | 10 | 18 |
| Cell cytoskeleton organization | ||||||||
| BnaA10g12850D | NP_851227.1 | 1.659 | – | ADF3 | Actin-depolymerizing factor3 | 1 | 4 | 15 |
| BnaC04g48100D | NP_187818.1 | – | 1.521 | ACT11 | Actin11 | 2 | 2 | 3 |
| BnaA10g20420D | NP_196786.1 | – | 1.726 | TUB6 | β-6 tubulin | 1 | 17 | 30 |
| BnaA06g26420D | NP_568437.1 | – | 1.657 | TUB8 | β-8 tubulin | 4 | 4 | 5 |
| BnaC01g24230D | NP_190146.1 | – | 1.727 | TET3 | Tetraspanin3 | 11 | 12 | 17 |
| BnaA09g16950D | NP_199226.1 | – | 1.644 | FLA13 | Fasciclin-like arabinogalactan protein13 protein | 4 | 4 | 8 |
| BnaC05g02150D | NP_563692.1 | – | 2.012 | FLA9 | Fasciclin-like arabinoogalactan9 | 1 | 10 | 20 |
| Auxin related | ||||||||
| BnaAnng17240D | NP_566306.3 | – | 2.159 | AIR12 | Auxin-responsive-like protein | 1 | 11 | 13 |
| BnaC03g05800D | NP_196834.3 | – | 1.651 | SFC | GTPase-activating protein | 2 | 2 | 3 |
| Other root growth related | ||||||||
| BnaC06g32200D | NP_565008.1 | 1.712 | 1.588 | LPR2 | Low phosphate root2 | 2 | 2 | 2 |
| BnaA09g47290D | NP_563901.1 | – | 1.697 | BFRUCT4 | β-Fructofuranosidase | 2 | 5 | 8 |
| BnaA05g35560D | NP_193087.1 | 1.951 | 3.896 | AMT1;1 | Ammonium transporter1;1 | 2 | 2 | 3 |
DEPs Involved in Phenylpropanoid Biosynthesis in Response to N Deficiency
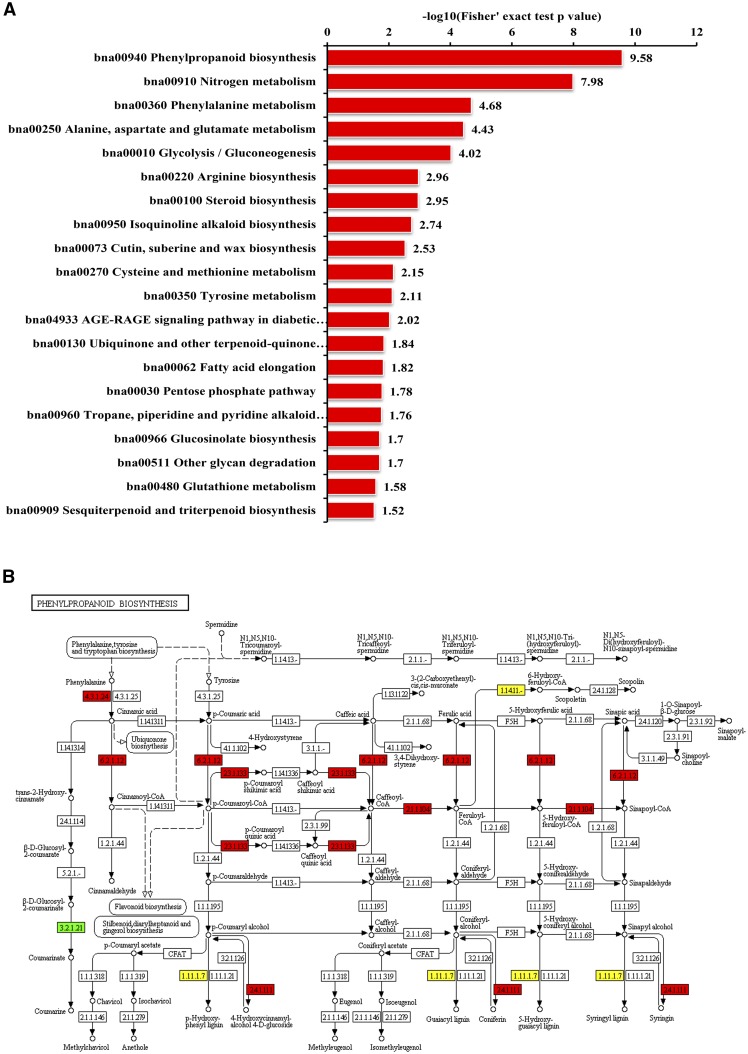
To better understanding the biological functions of the identified DEPs on the biochemical pathways, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed in this study. The top three enriched pathways were phenylpropanoid biosynthesis, N metabolism, and Phe metabolism (Fig. 5A). In short, the abundance of catalytic enzymes involved in phenylpropanoid biosynthesis was increased by N deficiency, especially in the day-14 N-deficient roots, including four phenylalanine ammonia lyase (PAL), three coumarate:coenzyme A ligase (4CL), and one caffeoyl coenzyme A O-methyltransferase (CCoAOMT). Notably, PODs were significantly overrepresented in day-14 roots (Fig. 4B), which putatively participate in phenylpropanoid biosynthesis (Fig. 5B). However, a total of 14 and 20 PODs were identified as DEPs in day-3 and day-14 N-deficient roots, respectively (Tables 1 and 2), and most of the identified PODs were significantly down-regulated in N-deficient roots (Table 2).
Figure 5.
KEGG enrichment analysis of day-14 DEPs in rapeseed roots and DEPs enriched in the phenylpropanoid pathway. A, KEGG enrichment analysis of identified DEPs in day-14 rapeseed roots (P < 0.05). The −log10 values of Fisher’s exact test P values for significant pathways are shown on the x axis. The KEGG database was used to identify enriched pathways by a two-tailed Fisher’s exact test in a comparison of DEPs against all identified proteins. Pathways with a corrected P < 0.05 were considered significant. B, DEPs identified in day-14 N-deficient roots that mapped to the phenylpropanoid pathway. Up- and down-regulated protein expression patterns are boxed in red and green, respectively, and yellow boxes indicate reactions mapped with both up- and down-regulated proteins.
Table 2. Down-regulated DEPs related to root growth of rapeseed plants after 3 and 14 d of high-N and low-N treatment.
| Protein Identifier | Arabidopsis Homolog | Ratio 3 d | Ratio 14 d | Abbreviation in Arabidopsis | Protein | No. of Unique Peptides | No. of Peptides | No. of Peptide Spectrum Matches |
|---|---|---|---|---|---|---|---|---|
| Cell wall organization or biogenesis | ||||||||
| BnaA03g47620D | NP_194312.1 | 0.440 | – | XTH14 | Xyloglucan endotransglucosylase/hydrolase14 | 1 | 3 | 3 |
| BnaA10g05530D | NP_199783.1 | 0.615 | – | GH9A1 | Endoglucanase25 | 2 | 6 | 8 |
| BnaA05g05070D | NP_182099.1 | 0.580 | – | AT2G45750 | Putative methyltransferase PMT16 | 5 | 5 | 10 |
| BnaA03g01580D | NP_196115.1 | 0.605 | – | AT5G04960 | Putative pectinesterase/pectinesterase inhibitor46 | 3 | 4 | 10 |
| BnaC09g11880D | NP_176598.1 | – | 0.560 | DIR5 | Dirigent protein5 | 4 | 5 | 21 |
| BnaC08g46340D | NP_563732.1 | 0.595 | – | AT1G05240 | Peroxidase1/2 | 3 | 6 | 10 |
| BnaAnng05140D | NP_201541.1 | 0.538 | – | RHS19 | Peroxidase73 | 2 | 7 | 29 |
| BnaA03g47730D | NP_567738.1 | 0.612 | – | AT4G26010 | Peroxidase44 | 7 | 9 | 29 |
| BnaA03g47720D | NP_567738.1 | 0.606 | – | AT4G26010 | Peroxidase44 | 6 | 15 | 55 |
| BnaCnng11080D | NP_174372.1 | 0.571 | – | AT1G30870 | Peroxidase7 | 1 | 3 | 3 |
| BnaA06g23250D | NP_201215.1 | 0.605 | – | AT5G64100 | Peroxidase69 | 6 | 10 | 53 |
| BnaA09g25570D | NP_174372.1 | 0.336 | – | AT1G30870 | Peroxidase7 | 1 | 3 | 4 |
| BnaA03g06740D | NP_197284.1 | 0.490 | – | AT5G17820 | Peroxidase57 | 3 | 9 | 20 |
| BnaC09g07300D | NP_201541.1 | 0.428 | – | RHS19 | Peroxidase73 | 10 | 16 | 45 |
| BnaAnng10890D | NP_179488.1 | – | 0.589 | AT2G18980 | Peroxidase16 | 2 | 13 | 59 |
| BnaA09g07380D | NP_201541.1 | – | 0.653 | RHS19 | Peroxidase73 | 2 | 16 | 47 |
| BnaA09g53760D | NP_192617.1 | – | 0.646 | Prx37 | Peroxidase37 | 1 | 16 | 61 |
| BnaC08g46330D | NP_563732.1 | – | 0.626 | AT1G05240 | Peroxidase1/2 | 6 | 15 | 26 |
| BnaA02g34060D | NP_201215.1 | – | 0.293 | AT5G64100 | Peroxidase69 | 1 | 12 | 52 |
| BnaCnng51060D | NP_201217.1 | – | 0.585 | AT5G64120 | Peroxidase71 | 10 | 10 | 17 |
| BnaA05g34020D | NP_186768.1 | – | 0.611 | AT3G01190 | Peroxidase27 | 5 | 16 | 49 |
| BnaA06g23250D | NP_201215.1 | – | 0.459 | AT5G64100 | Peroxidase69 | 8 | 13 | 55 |
| BnaA10g03270D | NP_563732.1 | – | 0.613 | AT1G05240 | Peroxidase1/2 | 5 | 8 | 13 |
| BnaA03g28350D | NP_187017.1 | – | 0.457 | AT3G03670 | Peroxidase | 5 | 5 | 8 |
| BnaC09g25420D | NP_192617.1 | – | 0.657 | Prx37 | Peroxidase37 | 1 | 16 | 61 |
| BnaC01g04790D | NP_567919.1 | – | 0.626 | AT4G33420 | Peroxidase | 5 | 5 | 7 |
| BnaA03g17000D | NP_181250.1 | 0.617 | 0.654 | AT2G37130 | Peroxidase | 4 | 13 | 37 |
| BnaC02g07500D | NP_197284.1 | 0.565 | 0.634 | AT5G17820 | Peroxidase57 | 4 | 14 | 56 |
| Cell cytoskeleton organization | ||||||||
| BnaCnng08860D | NP_171680.1 | 0.407 | – | ADF11 | Actin-depolymerizing factor11 | 2 | 4 | 9 |
| Auxin related | ||||||||
| BnaC01g04630D | NP_199960.1 | – | 0.454 | AT5G51470 | Auxin-responsive GH3 family protein | 2 | 2 | 3 |
| Root hair related | ||||||||
| BnaA02g20520D | NP_192136.1 | 0.294 | – | RHS13 | Root hair specific13 | 3 | 3 | 8 |
| BnaA06g39800D | NP_178467.1 | 0.555 | – | MRH6 | Morphogenesis of root hair6 | 1 | 2 | 3 |
| BnaC06g18560D | NP_175895.1 | 0.318 | – | PRP1 | Pro-rich protein1 | 1 | 3 | 4 |
Interaction Networks of Identified DEPs in N-Deficient Rapeseed Plants
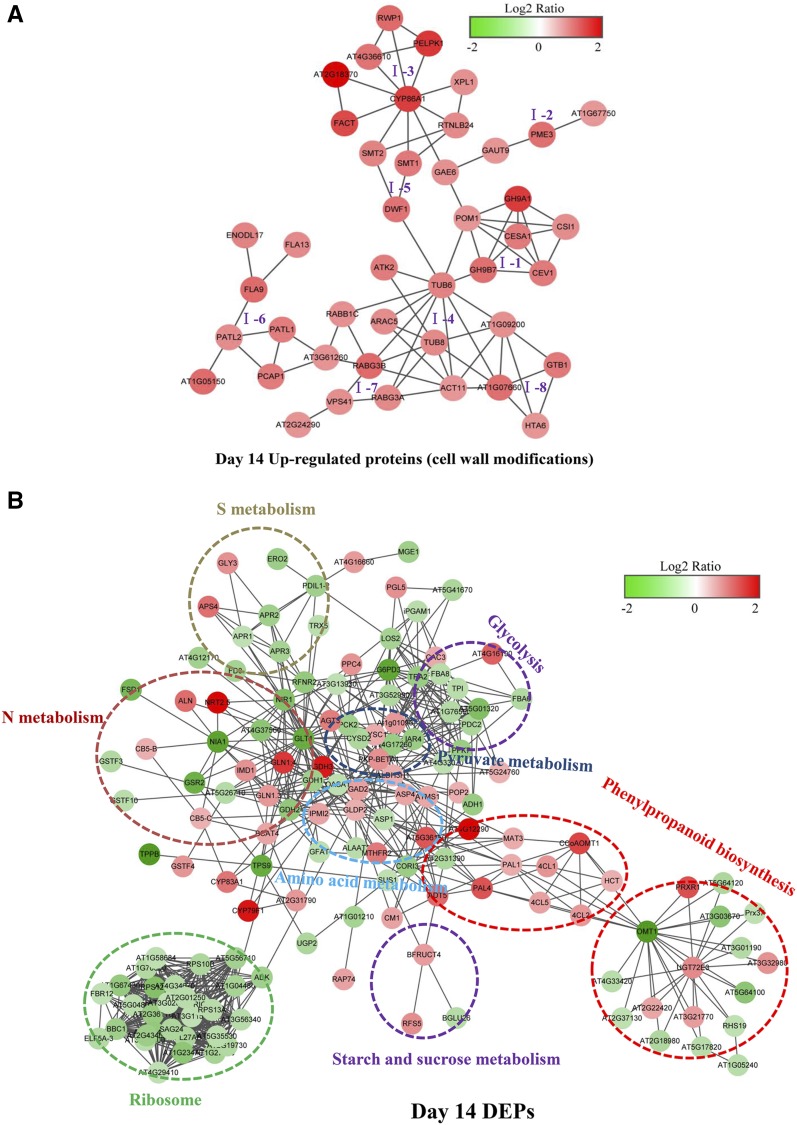
Proteins in living cells often function in complex interrelated networks. The DEPs up-regulated by N deficiency were analyzed using the STRING database of known and predicted protein-protein interactions. Very few DEPs interacted with each other in day-3 N-deficient roots; thus, further analysis was focused on the DEPs from day-14 N-deficient roots. First, a group of DEPs involved in cell wall metabolism were analyzed through STRING analysis, which could be divided into eight subclusters (Fig. 6A). Specifically, the subclusters were involved mainly in the formation of cell wall components and cytoskeleton organization, including subclusters I-1 (cellulose synthesis), I-2 (pectin metabolism), I-3 (cutin, suberin, and wax biosynthesis), and I-4 (cytoskeleton organization; Fig. 6A). The protein interactions for subcluster I-5 (sterol composition) participate in cell elongation and polarity, and subclusters I-6 and I-7 are involved in membrane and vacuolar trafficking, respectively (Fig. 6A). Finally, subcluster I-8 is involved in signal transduction and transcription and also interacted with proteins involved in cytoskeleton organization (Fig. 6A). These results further suggested that cell wall metabolism might be tightly connected with membrane/vacuolar trafficking and signal transduction during N deficiency.
Figure 6.
Predicted protein-protein interaction networks of DEPs in day-14 N-deficient rapeseed roots. A, Predicted protein-protein interaction networks of DEPs (up-regulated proteins) involved in cell wall metabolism in day-14 N-deficient rapeseed roots. Eight subclusters are grouped according to the pathways connected with these proteins: I-1, cellulose synthesis; I-2, pectin metabolism; I-3, cutin, suberin, and wax biosynthesis; I-4, cytoskeleton organization; I-5, sterol composition; I-6, membrane trafficking; I-7, vacuolar trafficking; and I-8, signal transduction and transcription. B, Predicted protein-protein interaction networks of all DEPs, except for those involved in cell wall metabolism, in day-14 N-deficient roots. DEPs within dotted circles share common metabolic pathways. The intensity of color in the circles indicates the degree of up-regulation (red) or down-regulation (green) of the DEPs. This interaction network was constructed using the STRING protein-protein network analysis program (http://string-db.org/) with the Arabidopsis database and confidence scores greater than 0.7. Network files were downloaded from STRING, and network images were made in Cytoscape 3.3.0. The full names for all the DEPs shown in the protein-protein interaction networks are listed in Supplemental Table S3.
Next, the protein-protein interaction networks of other DEPs in day-14 N-deficient roots were analyzed further using STRING. Proteins were grouped into eight large units, including units associated with ribosome biogenesis, glycolysis, phenylpropanoid biosynthesis and N metabolism, sulfur metabolism, amino acid metabolism, pyruvate metabolism, and starch and Suc metabolism. As illustrated in Figure 6B, the up-regulated and down-regulated proteins were integrated into interconnected protein networks. In short, all the DEPs involved in ribosome biogenesis interacted closely, and the abundance of them was reduced in N-deficient rapeseed roots. Most of the DEPs associated with sulfur metabolism and glycolysis also were down-regulated by N deprivation. As expected, the DEPs related to N metabolism and amino acid metabolism were tightly connected and differentially regulated by N deficiency. In this network, nitrate reductase1 and nitrite reductase1 were among the down-regulated proteins and interacted with NRT2.5, which was among the up-regulated proteins (Fig. 6B). Additionally, several proteins involved in starch and Suc metabolism were differentially regulated by N deficiency, specifically, β-glucosidase26 and galactinol-sucrose galactosyltransferase5. The latter is involved in the synthesis of raffinose, and both interacted with β-fructofuranosidase (BFRUCT4; Fig. 6B), with possible roles in the continued mobilization of Suc to sink organs contributing to root elongation.
Interestingly, DEPs involved in phenylpropanoid biosynthesis interacted tightly together and clustered into two large units. Similar to Figure 5B, most of the DEPs that participated in the phenylpropanoid pathway (lignin precursor biosynthesis) were up-regulated, whereas O-methyltransferase1 (OMT1; i.e. an enzyme catalyzing the methylation of lignin precursors, monolignols) seemed an important link between these two units (Fig. 6B). Furthermore, interaction networks also found that PODs were linked with each other, and most of them were down-regulated by N deficiency (Fig. 6B).
Changes in POD Activity of Rapeseed under N Deficiency
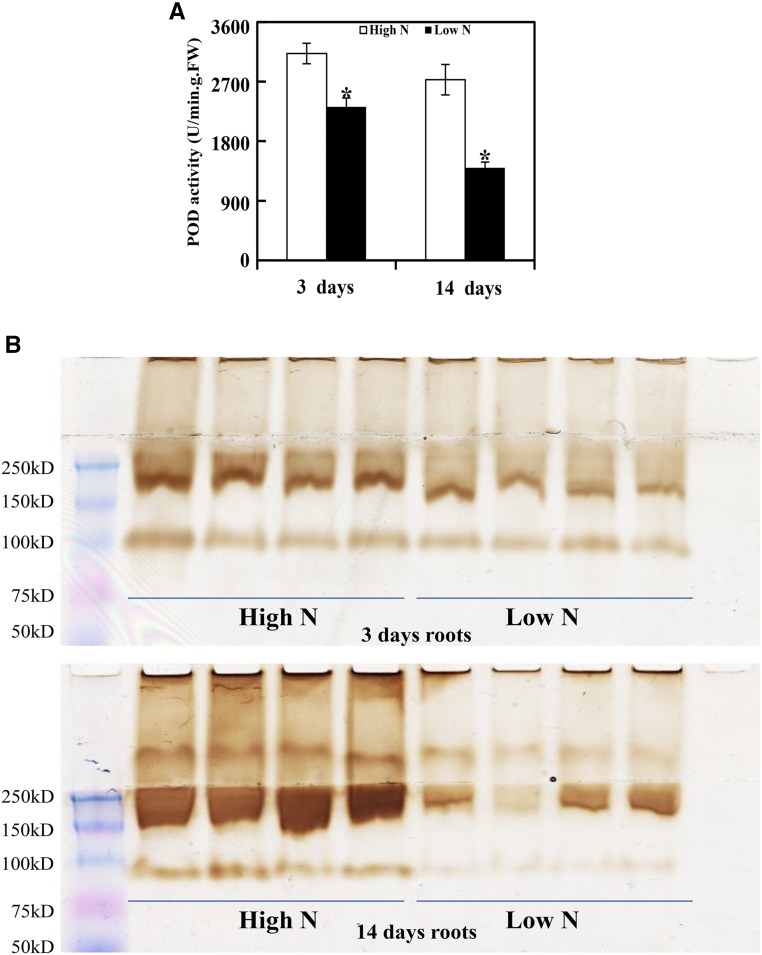
PODs are widely reported to catalyze the last enzymatic step in the biosynthesis of lignin and also have regulatory roles in regulating cell wall loosing and stiffening. In order to further validate the results from the proteomic analysis in this study, POD activity in N-deficient rapeseed roots was determined through colorimetric and in gel assays. As shown in Figure 7A, the POD activity in N-deficient roots determined via a colorimetric spectrophotometry assay was 25% and 48% less than in N-sufficient roots after 3 and 14 d, respectively. Furthermore, an in gel assay also displayed reduced POD activity in N-deficient roots (Fig. 7B). Overall, these results indicate that POD activity in rapeseed roots is reduced with the onset of N deficiency, which is consistent with the decreased abundance of PODs identified via proteomic analysis in rapeseed roots under N deficiency conditions.
Figure 7.
Assay of POD activity in N-deficient rapeseed roots. POD activity assays in N-deficient rapeseed roots were determined by a colorimetric enzyme assay (A) and staining in gel (B). For the colorimetric enzyme assay, POD activities were determined by monitoring A470 using the guaiacol method. Data represent means ± sd (n = 4). Asterisks indicate significant differences between N treatments at the same sampling day (Student’s t test, *, P < 0.05). For POD activity staining in gel, extracted proteins were equally loaded and separated on a native PAGE gel at 4°C and then incubated with staining substrates (0.03% [v/v] H2O2 and 1 mm diaminobenzidine). Four biological replicates for each treatment also were used for in gel assays. FW, Fresh weight.
Cell Wall Composition Is Altered in N-Deficient Rapeseed Roots
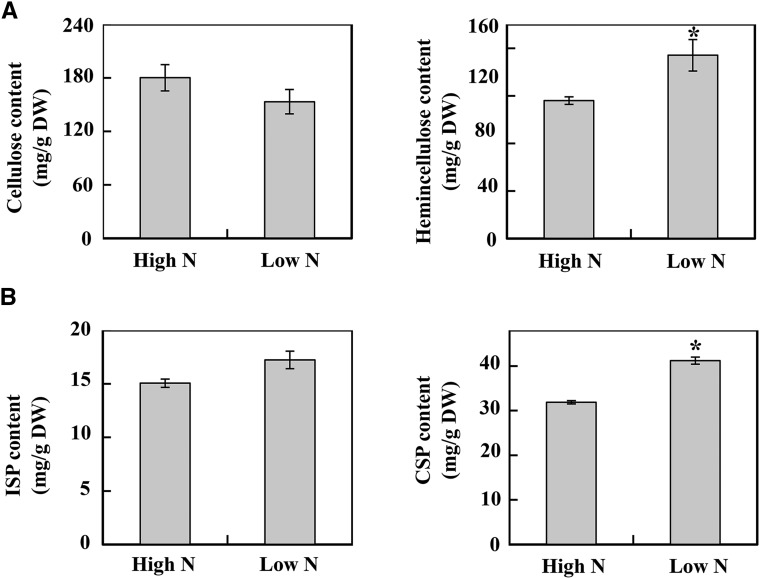
The cell walls of plants determine cell size and shape, which determine tissue morphology. To further find the correlation between DEPs involved in cell wall metabolism and the components of cell walls in N-deficient rapeseed roots, the four major components, cellulose, hemicellulose, pectin, and lignin, were measured in roots of rapeseed after 14 d in the different N treatments. As shown in Figure 8A, the hemicellulose content was enhanced significantly in N-deficient roots, while the cellulose content did not seem to be influenced by N deficiency. As for pectin, two kinds of pectin were detected in this study, as seen in Figure 8B, and the content of ISP was increased under N deficiency conditions, while the CSP also was affected significantly by N supply, whose content was increased 29.2% in the N-deficient roots of rapeseed.
Figure 8.
Changes of cell wall components in rapeseed roots in response to N deficiency. A, Cellulose and hemicellulose contents. B, Ionic bound pectin (ISP) and covalently bound pectin (CSP) contents. Rapeseed roots were collected at 14 d after high-N (7,500 μm N) and low-N (190 μm N) treatments. Data represent means ± sd (n = 4). Asterisks indicate significant differences between the two N treatments (Student’s t test, *, P < 0.05). DW, Dry weight.
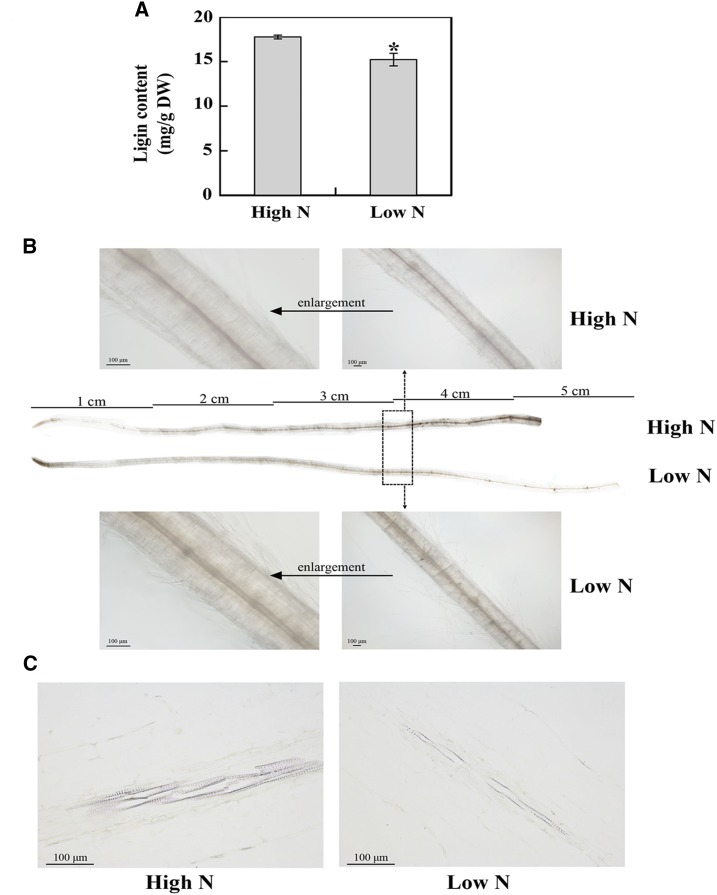
However, the lignin content in N-deficient roots was 11.2% less than that in N-sufficient roots (Fig. 9A). Moreover, as shown in Figure 9B, the stele of the N-sufficient rapeseed roots was pink after phloroglucinol staining, whereas the color was relatively weak under N deficiency conditions, especially for the region 3 cm away from the root tip. Root segments (the root region between 3 and 4 cm away from the root tip, as shown in Fig. 9B) were further stained with phloroglucinol after paraffin sectioning; lignified vessel elements in the xylem of N-sufficient rapeseed roots showed deeper color than those of N-deficient roots (Fig. 9C), suggesting that N deficiency might influence the lignin synthesis of rapeseed roots. Thus, it was consistent with the abundant changes of DEPs involved in lignin synthesis and the morphological changes of N-deficient rapeseed roots.
Figure 9.
Changes of lignin content in rapeseed roots in response to N deficiency. A, Lignin content. Rapeseed roots were collected 14 d after the N treatments and analyzed by the acetyl bromide method for measuring lignin content. Data represent means ± sd (n = 4). The asterisk indicates a significant difference between the two N treatments (Student’s t test, *, P < 0.05). DW, Dry weight. B, Lignin histochemistry staining in rapeseed roots. Three days after the N treatments, roots (eight individual rapeseed seedlings for each treatment) were stained and imaged using an optical microscope (Olympus BX53F). Lignin was visualized by pink staining. The area within the black dotted rectangle (root segments between 3 and 4 cm away from the root tip) was magnified as shown above and below. C, Lignin deposition in the root xylem tissues. Three days after the N treatments, root segments (between 3 and 4 cm away from the root tip) that were visibly different between the two N treatments based on staining were selected for longitudinal slicing and then were observed with an optical microscope (Nikon Eclipse Ci). High N, 7,500 μm N; low N, 190 μm N.
Functional Analysis of Two Candidate DEPs in Transgenic Arabidopsis
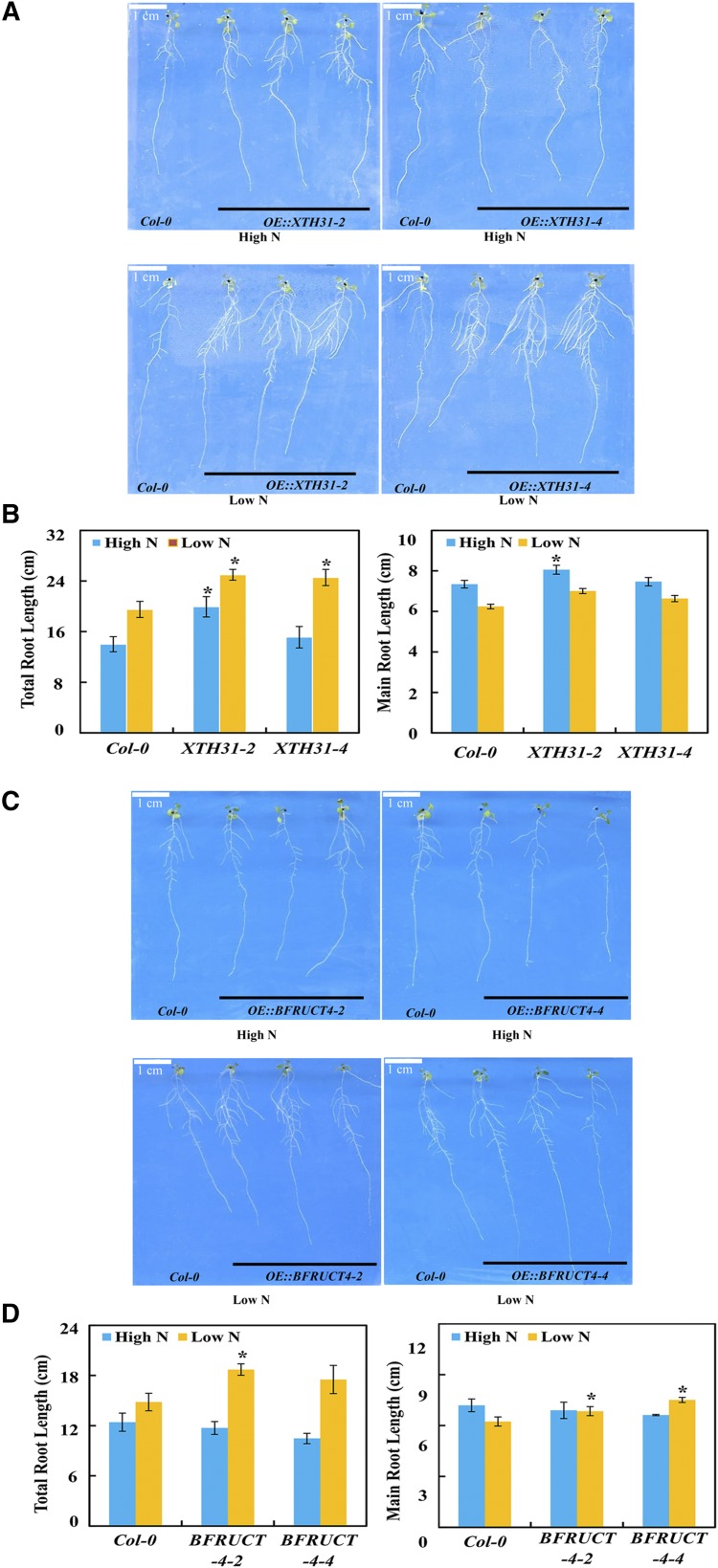
To further verify the functions of DEPs from proteomic analysis in root growth, two putative root-related DEPs were selected, XTH31 and BFRUCT4, which had been reported previously to be associated with cell wall organization (Zhu et al., 2012) and glycan metabolism (Sergeeva et al., 2006) in Arabidopsis, respectively. Transgenic Arabidopsis plants harboring the coding region of BnaXTH31 or BnaBFRUCT4 under the control of the 35S promoter were generated and evaluated for changes in root growth under both high-N (60 mm) and low-N (2 mm) conditions. The results showed that the overexpression of BnaXTH31 promoted total root growth of Arabidopsis after 7 d, especially under low-N conditions (Fig. 10, A and B), while overexpressing BnaBFRUCT4 increased main root growth only at the low-N level (Fig. 10, C and D). These results indicated that these two root-related proteins contribute to stimulate plant root growth under N deficiency stress.
Figure 10.
Root growth performance of BnaXTH31 and BnaBFRUCT4 ectopically expressed in Arabidopsis. A, Phenotypes of wild-type Columbia-0 (Col-0) and transgenic lines expressing BnaXTH31. B, Root length. C, Phenotypes of wild-type (Col-0) and transgenic lines expressing BnaBFRUCT4. D, Root length. Arabidopsis plants were grown in high-N (60 mm N) or low-N (2 mm N) Murashige and Skoog medium for 7 d. Data represent means ± sd (n = 12). Asterisks indicate significant differences between wild-type and transgenic plants at the same N level (Student’s t test, *, P < 0.05).
DISCUSSION
N deficiency is a major constraint for rapeseed production. Yet, most published reports on N deficiency responses in rapeseed have focused on leaves, especially after long-term N starvation with obvious N-deficient symptoms (Kim et al., 2009). Still, root systems are important for acquiring nutrients from soils, and root system alterations in response to N availability have been noted in many crops, including rice (Oryza sativa) and wheat (Triticum spp.; Wang et al., 2002; Rasmussen et al., 2015). In these studies, root morphological traits, such as main root length and total root length, were determined by using 2D root observation systems (Wang et al., 2002; Rasmussen et al., 2015). However, the spatial distribution of roots in soils is three-dimensional. In recent years, several research efforts have shown that root architecture can be influenced significantly by nutrient availability (Walch-Liu et al., 2006; Giehl et al., 2014; Saengwilai et al., 2014). In maize, root architectures with deeper roots were more effective at N acquisition from low-N soils (Saengwilai et al., 2014). In this study, a 3D in situ quantification system was applied to investigate the root architectural responses of rapeseed to N deficiency. Then, the mechanisms underlying rapeseed root responses to N deficiency were investigated using TMT-based proteomic techniques.
As reported previously, low N stimulates the elongation of main and lateral roots, but not the density of lateral roots, in Arabidopsis (López-Bucio et al., 2003; Gruber et al., 2013). In this study, we observed that N deficiency not only stimulated the elongation of main and lateral roots of rapeseed but also increased root tip count (Fig. 1B). It is well known that root morphogenesis is controlled by the regulation of cell division and expansion (Strader et al., 2010). As expected, not only the cell number in the meristematic zone of rapeseed roots was enhanced but also the cell size in the elongation zone was enlarged by N starvation (Fig. 2). This suggested that the increased root growth of N-deficient rapeseed seedlings might be caused by the stimulation of both cell division and expansion.
Moreover, the cell wall influences cell size and shape, which determine the tissue morphology of plants. A previous review summarized that cell wall architecture and cell wall metabolism often are affected by various abiotic stresses, such as drought, flooding, heat, cold, salt, heavy metals, light, and air pollutants (Le Gall et al., 2015). In our study, the root growth of rapeseed seedlings under N deficiency was accompanied by the increased expression of several cell wall-related proteins (e.g. XTH and EXP; Table 1). Several studies have reported that the abundance of XTH increased in osmotically stressed plants, resulting in striking cell wall remodeling, thus promoting cell growth and strengthening of the cell wall (Tenhaken, 2015). For example, Arabidopsis XTH31 showed predominant xyloglucan endohydrolase activity in vitro, and XTH31 has been found to be involved in cell wall modification and cell elongation through modulating xyloglucan endotransglucosylase action under aluminum stress (Zhu et al., 2012). In this study, functional analysis of BnaXTH31 in transgenic Arabidopsis demonstrated that this gene contributes to the enhanced root growth upon N deprivation stress (Fig. 10, A and B). EXPs are a group of proteins involved in the irreversible extension of cell walls and associated cell enlargement in plants (Cosgrove, 1996). They also have been reported to be involved in root architectural modifications in soybean (Glycine max; Guo et al., 2011). In this study, three EXP proteins were identified only in long-term N-deficient roots displaying dramatic root morphological changes (Table 1). And the enhanced expression of these cell wall-related proteins was in line with the increased content of hemicellulose in N-deficient rapeseed roots (Fig. 8A). Moreover, several other proteins involved in cell wall formation also were identified in this study. For instance, rapeseed homologs of the Arabidopsis proteins cellulose synthase, CESA1 and CEV1, which function in cellulose and primary cell wall formation (Pysh et al., 2012), as well as the chitinase family protein POM1, which participates in radical cell expansion in Arabidopsis (Scheres et al., 2002), were all up-regulated in day-14 N-deficient rapeseed roots.
Here, several cell wall-related DEPs involved in cytoskeleton organization also were identified with increased abundance in the 14-d N-deficient rapeseed roots (Table 1), such as actin11 (ACT11), which is an essential component of the cell cytoskeleton and plays important roles in cell shape determination, cell division, organelle movement, and extension growth (Hussey et al., 2006). Microtubules are major structural components of eukaryotic cells and play a key role in regulating cell division, cell proliferation, and cell morphology (Wasteneys and Yang, 2004). In this study, two tubulin proteins, β-6 tubulin (TUB6) and TUB8, were identified in N-deficient rapeseed roots. Interestingly, ACT11 and two TUB proteins were predicted to interact tightly with each other by STRING analysis (Fig. 6A). Additionally, TUB6 and TUB8 potentially interacted with several Rab GTPases (Fig. 6A; Supplemental Table S1), which belong to a family of monomeric small GTPases of the Ras superfamily. It has been reported that several Rab GTPases are crucial for cytokinesis and function in regulating cell division (Qi and Zheng, 2013; Gibieža and Prekeris, 2018). Therefore, the response of root elongation in rapeseed to N deficiency might be achieved through a coordination of these relevant proteins involved in both cell division and cell expansion. This might ultimately trigger root morphological changes under low-N conditions.
According to 3D in situ quantification, roots became wider but softer under N deficiency conditions, and the solidity of roots decreased significantly within 3 d under the low-N treatment (Fig. 1B). PODs are ubiquitous to all organisms and are encoded by large multigene families (Francoz et al., 2015). Several studies suggest that PODs function in cell wall lignification, auxin catabolism, defensive responses, and salt tolerance (Hiraga et al., 2001; Shigeto and Tsutsumi, 2016). It is well known that cell expansion is linked to processes controlling cell wall loosening and stiffening, and the balance between these two processes can be regulated by reactive oxygen species homeostasis (Tsukagoshi et al., 2010), which is controlled by PODs; thus, the balance between cell wall loosening and stiffening can be influenced by the activities of class III PODs (Tenhaken, 2015). In this study, a large number of PODs were characterized as down-regulated DEPs in response to N deficiency (Table 2), which is consistent with previous results in maize (Trevisan et al., 2015). In addition, POD enzyme activity in rapeseed roots was repressed significantly by N deficiency in this work (Fig. 7). According to a previous study, oxygen radicals and H2O2 are two major reactive oxygen species that are differentially distributed within the root tissues of Arabidopsis (Dunand et al., 2007). In this study, we speculated that the down-regulation of PODs and the reduction of POD enzyme activity may facilitate root growth by the decrease of apoplastic H2O2 and the production of oxygen radicals to cleave the cell wall polymers, then promote cell wall loosing and root growth. Additionally, the STRING analysis of protein-protein interaction networks predicts that down-regulated PODs interact with proteins in lignin synthesis (e.g. OMT1) and catalytic enzymes in the biosynthesis of lignin precursors (e.g. 4CL1, 4CL2, 4CL5, PAL1, PAL4, and CCoAMOT1; Fig. 6B). Although all the identified enzymes involved in the biosynthesis of lignin precursors were up-regulated by N deficiency, most of the enzymes that participate in the last step of lignin synthesis were suppressed by N deficiency, which might lead to the decrease of the lignin content observed in N-deficient rapeseed roots in this study (Figs. 5B and 9). Furthermore, one of the down-regulated PODs is a homolog of AtPRX71, which has been demonstrated to restrict cell expansion in Arabidopsis (Raggi et al., 2015). Therefore, the down-regulation of PODs herein might promote root elongation in concert with the stimulation of EXPs and XTHs while also decreasing root solidity through the inhibition of lignin biosynthesis under low-N conditions. Our results suggest that N deficiency may make plant roots softer, which might be attributable to the down-regulation of PODs.
In conclusion, these results provide a global view of architectural changes as well as the underlying molecular mechanisms involved in the adaption of rapeseed roots to N deficiency. Changes in the abundance of cell wall-related proteins and PODs quite likely alter the rigidity of the cell wall to promote root cell division and expansion, leading to increased root elongation, which could contribute to the longer and thinner rapeseed lateral roots induced by N deficiency.
MATERIALS AND METHODS
Dynamic Changes of Root Architecture in N-Deficient Rapeseed Plants
For 3D in situ quantification of N deficient roots, a hydroponic experiment was conducted using a Chinese-grown rapeseed (Brassica napus) cultivar, Zhongshuang 11. Seeds were sterilized and germinated in one-quarter-strength modified Hoagland nutrient solution for 3 d before transplanting into one-half-strength nutrient solution with different N treatments (high N, 7,500 μm N; or low N, 190 μm N; pH 5.8, refreshed weekly). Plants were grown in a 24°C/20°C day/night temperature regime with a 14-h/10-h light/dark photoperiod. Root images were acquired daily from days 1 to 7 after implementing the N treatments. The imaging system consisted of a Nikon D800E Digital SLR camera with a Nikon 105 mm f/2.8D AF ED-IF lens. Prior to 3D reconstruction, each series of 100 2D images was identically cropped, adjusted to a resolution of 200 mm per pixel, and converted to grayscale using Adobe Photoshop. The images then were thresholded and reconstructed. Finally, root traits such as total root length, total root volume, maximum depth, maximum width, minimum width, solidity, and tip count were computed in RootReader3D (Clark et al., 2011; Piñeros et al., 2016), and lateral root length was the total length of all roots minus the maximum depth (namely main root length).
After 3 d of N treatment, cell number and cell size were investigated in 0.5-cm root segments starting from the root tip. The root segments (eight individual rapeseed seedlings for each treatment) were stained with 10 μg mL−1 PI for 3 min as described by Bai et al. (2009) with a modification for staining time and then were observed with a confocal laser scanning microscope (Zeiss LSM 710). To quantify cell number in the meristematic zone, root samples (eight individual rapeseed seedlings for each treatment) were fixed overnight in 50% (v/v) formaldehyde-acetic acid, dehydrated in a graded ethanol series, embedded in paraffin wax, and longitudinally sectioned (5 µm thick) with a microtome (Leica RM2016). Sections were dried onto slides at 37°C and stained with Fast Green/safranin prior to acquiring images with an optical microscope (Nikon Eclipse Ci). Then, photographs taken by the microscope were used to count the number of cortex cells in the meristematic zone of root tips at high- and low-N levels.
Experimental Design, Protein Preparation, and TMT10-plex Labeling
For proteomics analysis, a hydroponics experiment was conducted using the same rapeseed cultivar and N treatments as mentioned above. Roots from three independent biological replicates were harvested separately 3 d (short term) and 14 d (long term) after initiating the N treatments for subsequent TMT-based proteomic analysis.
An overview of the proteomics experimental design and workflow is shown in Supplemental Figure S1. Specifically, samples were collected from rapeseed roots after 3 and 14 d of growth under N-deficient and nutrient-sufficient conditions. Three biological replicates for each treatment led to nine samples being collected at each time point. Proteins were extracted according to the previous method with some modifications (Yang et al., 2007). Each sample was powdered in liquid N2 followed by suspension in extraction buffer (0.5 m Tris-HCl, pH 7.5) containing 50 mm EDTA, 0.1 m KCl, 0.7 m Suc, 10 mm DTT, 1 mm phenylmethylsulfonyl fluoride, and 1% (w/v) polyvinylpyrrolidone. An equal volume of Tris-EDTA-saturated phenol (pH 8) was added followed by shaking for 30 min at 4°C. The homogenate was centrifuged at 4,000 rpm for 30 min at 4°C. The phenolic phase was collected, and the extraction was repeated. Five volumes of 0.1 m ammonium acetate in methanol was added to the collected phenolic phase to precipitate proteins at −20°C overnight. After centrifugation for 40 min, the supernatant was discarded and the resulting pellet was washed once with chilled methanol and twice with chilled acetone. The pellet was air dried and stored at −80°C.
Protein pellets were resuspended in lysis buffer, then protein quantitation, digestion, and TMT labeling were performed as reported previously (Chen et al., 2013), except that TMT10-plex (Thermo Fisher Scientific) tagging was used in this work. Specifically, 126-tag, 127N-tag, and 127C-tag were used for low-N samples and 129C-tag, 130N-tag, and 130C-tag were used for N-sufficient samples. The remaining 131-tag in each set of TMT10-plex was used for a mixture of equal amounts of protein from each of the 12 samples to bridge the results collected between days 3 and 14. Then, the 10 labeled samples were pooled and evaporated to dryness before being subjected to cation-exchange chromatography using an SCX cartridge (PolyLC) and desalted as described previously (Chen et al., 2013) for subsequent high-pH reverse-phase chromatography.
High-pH Reverse-Phase Fractionation and Nano-Scale Reverse-Phase Chromatography-Tandem Mass Spectrometry
All high-pH reverse-phase chromatography was performed with a Dionex UltiMate 3000 HPLC system as reported previously (Yang et al., 2011). The TMT10-plex-tagged tryptic peptides described above were reconstituted and loaded onto an XTerra MS C18 column from Waters. Forty-eight fractions were collected at 1-min intervals and pooled into a total of 18 fractions based on the UV A214 and with the multiple fraction concatenation strategy as published (Wang et al., 2011). All fractions were dried and reconstituted for nano-scale reverse-phase chromatography-tandem mass spectrometry (MS/MS) analysis.
The nano-scale reverse-phase chromatography-MS/MS analysis and Orbitrap Elite settings were the same as published recently (Wang et al., 2014), with slight modifications incorporated specifically for running TMT10-plex-labeled peptides. The Orbitrap Elite device was operated using an fourier transform mass analyzer for a one-survey MS scan, followed by higher energy collisional dissociation-MS/MS scans on the top 15 precursor peptides with multiple charged ions above a threshold ion count of 8,000 at a normalized collision energy of 37.5%. MS survey scans were set at a resolving power of 60,000 (full width at half maximum at mass-to-charge ratio [m/z] 400) for the mass range of m/z 375 to 1,800 with automatic gain control = 1e6 and maximum injection time = 100 ms. MS/MS scans were set at 30,000 resolution with 1.5 atomic mass units of isolation width at automatic gain control = 1e5 and maximum injection time = 250 ms for the mass range m/z 100 to 2,000. Dynamic exclusion parameters were set at repeat count 1 with a 30-s repeat duration, an exclusion list size of 500, and a 70-s exclusion duration with ±10 ppm exclusion mass width. The activation time was 0.1 ms for higher energy collisional dissociation analysis. All data were acquired using the Xcalibur 2.2 operation software (Thermo Fisher Scientific).
Data Processing and Protein Identification
All MS and MS/MS raw spectra from each set of TMT10-plex experiments were processed using Sequest HT software within Proteome Discoverer 1.4. The database of rapeseed peptide sequences (http://www.genoscope.cns.fr/brassicanapus/data/) was used for searches. The default search settings were as follows: two miscleavages for full trypsin with fixed carbamidomethyl modification of Cys, fixed TMT10-plex modifications on Lys and N-terminal amines, variable modifications of Met oxidation, and deamidation of Asn and Gln residues. The peptide mass tolerance and fragment mass tolerance values were 10 ppm and 50 mD, respectively. Only high-confidence peptides defined by Sequest HT with a 1% FDR by Percolator were considered for peptide identification. The TMT10-plex quantification method within Proteome Discoverer 1.4 was used to calculate the reporter ratios with a mass tolerance ±10 ppm without applying isotopic correction factors. Only peptide spectra containing all reporter ions were designated as quantifiable spectra and used for peptide/protein quantitation. Values were expressed as the ratio in each sample relative to the value of the pooled bridge sample. A protein ratio was expressed as the median value of the ratios for all quantifiable spectra of unique peptides pertaining to that protein. A precursor coisolation filter of 50% also was applied to minimize ratio compression caused by the coisolation of precursor ions. For each relative ratio group, normalization on protein median was applied. Matching of at least two peptides was required for each protein to continue analysis. All statistical analyses were performed with R scripts and in Microsoft Excel. Differential expression between low N and high N was determined for each peptide using a combination of Student’s t tests and empirically derived ratio thresholds to control the false discovery rate based on previous reports (Janvilisri et al., 2012; Chen et al., 2013). Briefly, proteins were considered differentially expressed if (1) Student’s t test P values were less than 0.05 and (2) the ratio of expression between low N and high N was beyond the range of ratios observed for 95% of the ratios observed between replicates within N treatments. For the day-3 treatment, proteins with a fold change larger than 1.6 and P < 0.05 were considered to be significantly differentially expressed. For the day-14 treatment, the fold change ratio threshold for DEPs was 1.51.
Functional Annotation and Bioinformatics Analysis
To determine the functional properties of identified DEPs, all rapeseed peptides and GO annotations were downloaded from the rapeseed genome database and BLAST searched against the Swiss-Prot database release-2015_07 using BLASTP, returning the top 10 hits with an e value cutoff of 10−10. The GO terms were categorized into third-level parent terms through is a relations within the molecular function and biological process ontologies. All peptides also were used to query the KEGG database through BlastKOALA (http://www.kegg.jp/blastkoala/). Representation analysis for GO and KEGG was performed using hypergeometric tests with FDR-corrected q values (Benjamini and Hochberg, 1995). Categories were considered significantly overrepresented if the q value was less than 0.05 and the proportion in the category was greater in DEPs than in the set of all proteomic proteins. Protein-protein interaction networks were constructed using the STRING program (http://string-db.org/) with the Arabidopsis (Arabidopsis thaliana) database and confidence scores greater than 0.7. Network files were downloaded from STRING, and network images were made in Cytoscape 3.3.0.
POD Activity Assays in N-Deficient Rapeseed Roots
To quantitatively assay POD activity, roots of day-3 and day-14 N-sufficient and N-deficient rapeseed plants were homogenized in ice-cold phosphate-buffered saline solution (50 mm, pH 7) and centrifuged at 12,000 rpm for 30 min at 4°C. The supernatant was used immediately to determine POD activity by monitoring A470 using the guaiacol method (Fang and Kao, 2000).
To measure POD activity in gel, proteins were extracted by grinding 100 mg of roots in an equal volume of extraction buffer (25 mm Tris-HCl, pH 8.5, 5 mm EDTA, 85 mm NaCl, 15 mm MgCl2, and protease inhibitor). The method for in gel assays of POD activity was described previously (Den Herder et al., 2007). The protein concentration of tissue extracts was determined according to the method of Bradford (Bradford, 1976) using BSA as a protein standard. Protein samples were diluted with ultrapure water to achieve equal protein concentrations. Proteins were equally loaded per lane, then separated on a native PAGE gel at 4°C before incubating in staining solution containing substrates (0.03% [v/v] H2O2 and 1 mm diaminobenzidine). Four biological replicates for each treatment were used for quantitative and in gel assays.
Measurement of Root Lignocellulosic Components
The hemicellulose and cellulose contents were measured using a previously reported method (Foster et al., 2010b). Briefly, rapeseed roots were collected at 14 d after initiating N treatments, then 0.5-g dry root samples were powdered and analyzed. First, the cell wall materials of the roots were isolated using 0.01% (w/v) sodium azide, and the cell wall starch was digested. After that, the cell wall material was hydrolyzed by 2 m trifluoroacetic acid, and after centrifugation, the precipitate and the supernatant were separately incubated and pretreated for the measurement of cellulose and hemicellulose. The released Glc was measured by an anthrone assay at 625 nm.
The ISP and CSP contents of the collected root samples were assayed according to the previous method (Szatanik-Kloc et al., 2017). Cell wall materials were prepared as described above. The ISP and CSP pectin fractions were extracted from dried cell wall samples with two different extractants. For the ISP fraction, a 5-mg sample was added with 1 mL of 50 mm cyclohexane diamine tetraacetic acid (pH 6.5). For the CSP fraction, a 5-mg sample was added with 1 mL of 50 mm Na2CO3 (containing 2 mm cyclohexane diamine tetraacetic acid). The supernatants from different extractions were determined using the sulfuric acid-carbazole colorimetry method (Szatanik-Kloc et al., 2017).
The lignin content of the root samples was determined by the acetyl bromide method as described previously (Hatfield et al., 1999; Foster et al., 2010a). A sample of 0.5 g of dry roots from the different N treatments also was powdered for lignin content measurement. First, the cell wall of the roots was isolated using 0.01% (w/v) sodium azide, and the cell wall starch was digested. Then, lignin was quantified with the acetyl bromide method with absorbance values at 280 nm. The histochemistry staining for lignin was modified slightly from Chinwang et al. (2011). Briefly, roots (eight individual rapeseed seedlings for each treatment) were stained in 1% (w/v) phloroglucinol in 95% (v/v) ethanol for 3 min, and the phloroglucinol was removed and then immediately incubated with 6 mm HCl. The roots were mounted on microscope slides and imaged using an optical microscope (Olympus BX53F). Lignin was visualized by pink or red staining. To further examine lignin deposition, root segments were prepared and fixed with paraffin and then longitudinally sectioned (5 µm thick) with a microtome (Leica RM2016). After removal of the paraffin, the root samples (six individual rapeseed seedlings for each treatment) were stained with 1% (w/v) phloroglucinol in 95% (v/v) ethanol for 3 min, and the phloroglucinol was removed and incubated with 6 mm HCl. The root sections were stained and then observed with an optical microscope (Nikon Eclipse Ci).
Vector Construction and Plant Transformation
In order to investigate the possible roles of the detected DEPs in root growth in response to N deficiency, BnaXTH31-overexpression and BnaBFRUCT4-overexpression constructs were generated. In short, an 879-bp full-length coding sequence of BnaXTH31 was amplified from cDNA of cv Zhongshuang 11 and inserted into the SacI/BamHI cloning sites of the pBI121S vector driven by the 35S promoter. And a 1,977-bp full-length coding sequence of BnaBFRUCT4 also was amplified and inserted into the XhoI cloning sites of the pBI121S vector. The sequences of primers used for plasmid construction are listed in Supplemental Table S2. To generate transgenic Arabidopsis plants overexpressing BnaXTH31 or BnaBFRUCT4, Agrobacterium tumefaciens strain GV3101 harboring the plasmid pBI121S was used to transform Col-0 plants through floral dip-mediated infiltration (Clough and Bent, 1998). Independent T3 homozygous transgenic lines were acquired through kanamycin-resistant screening and molecular verification and were used in the experiment of root growth analysis.
Root Growth of Transgenic Arabidopsis Plants
For root growth analysis in response to N deficiency stress, Arabidopsis wild-type Col-0 and transgenic plants were grown under high-N (60 mm) and low-N (2 mm) conditions for 1 week. Seeds were surface sterilized and sown on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 0.8% (w/v) agar and 1% (w/v) Suc. The sown seeds were vernalized for 3 d in the dark at 4°C and then grown in a 24°C/20°C day/night temperature regime with a 16-h/8-h light/dark photoperiod. Root images were acquired after implementing N treatments for 1 week by an EPSON scanner (EPSON V700 J221A), and the root length was calculated by image-analysis software (ImageJ).
Statistical Analyses
All means and se values of data in this study except proteomics were calculated in Microsoft Excel 2010. All comparisons between high-N and low-N treatments in this study were performed using Student’s t test in Microsoft Excel 2010 with P < 0.05 considered statistically significant, and the comparison of root growth between the wild type and transgenic lines at the same N level in Arabidopsis also was analyzed as above.
Accession Numbers
Sequence data of the major proteins discussed in this work can be found in the rapeseed genome database (http://www.genoscope.cns.fr/brassicanapus/) with the protein identifiers listed in Table 1, and the sequence data of corresponding homologous proteins in Arabidopsis can be found in the National Center for Biotechnology Information database with the accession numbers also listed in Table 1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Experimental design and workflow diagram for the TMT10-plex labeling-based methods used in this study.
Supplemental Table S1. DEPs involved in signal transduction and transcription identified in this study.
Supplemental Table S2. Primer sequences used for the overexpression transgenic Arabidopsis study.
Supplemental Table S3. Full names of DEPs shown in the protein-protein interaction networks (Fig. 6).
Acknowledgments
We thank Eric Craft (Robert Holley Center for Agriculture and Health, U.S. Department of Agriculture-Agricultural Research Service) for technical support regarding imaging and analysis of root system morphology and architecture. We also thank Xiaomin Jia (Robert Holley Center for Agriculture and Health, U.S. Department of Agriculture-Agricultural Research Service) for technical support in the proteomic experiment as well as Xiaonan Ma (Henan University) and Xinjie Shen (Oil Crops Research Institute) for technical support in the PI staining experiment.
Footnotes
This work was supported by grants from the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2013-OCRI) and the Excellent Young Scientist Fund (1610172015004) of the Chinese Academy of Agricultural Sciences and by the National Key Research and Development Program (2016YFD0100202-5).
Articles can be viewed without a subscription.
References
- Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, Wu Z, Song CP (2009) Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J 60: 314–327 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discover rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chen JW, Scaria J, Mao C, Sobral B, Zhang S, Lawley T, Chang YF (2013) Proteomic comparison of historic and recently emerged hypervirulent Clostridium difficile strains. J Proteome Res 12: 1151–1161 [DOI] [PubMed] [Google Scholar]
- Chen S, Ding G, Wang Z, Cai H, Xu F (2015) Proteomic and comparative genomic analysis reveals adaptability of Brassica napus to phosphorus-deficient stress. J Proteomics 117: 106–119 [DOI] [PubMed] [Google Scholar]
- Chen Z, Cui Q, Liang C, Sun L, Tian J, Liao H (2011) Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics 11: 4648–4659 [DOI] [PubMed] [Google Scholar]
- Chinwang U, Siriphanich J, Chairat R (2011) Enzymatic browning of fresh-cut galangal (Alpinia siamense K. Schum) and its relationship to oxidative enzymes. J Jpn Soc Hortic Sci 80: 103–112 [Google Scholar]
- Clark RT, MacCurdy RB, Jung JK, Shaff JE, McCouch SR, Aneshansley DJ, Kochian LV (2011) Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol 156: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (1996) Plant cell enlargement and the action of expansins. BioEssays 18: 533–540 [DOI] [PubMed] [Google Scholar]
- Den Herder J, Lievens S, Rombauts S, Holsters M, Goormachtig S (2007) A symbiotic plant peroxidase involved in bacterial invasion of the tropical legume Sesbania rostrata. Plant Physiol 144: 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: Possible interaction with peroxidases. New Phytol 174: 332–341 [DOI] [PubMed] [Google Scholar]
- Fan SC, Lin CS, Hsu PK, Lin SH, Tsay YF (2009) The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Kao CH (2000) Enhanced peroxidase activity in rice leaves in response to excess iron, copper and zinc. Plant Sci 158: 71–76 [DOI] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010a) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: Lignin. J Vis Exp 37: e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010b) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: Carbohydrates. J Vis Exp 37: e1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz E, Ranocha P, Nguyen-Kim H, Jamet E, Burlat V, Dunand C (2015) Roles of cell wall peroxidases in plant development. Phytochemistry 112: 15–21 [DOI] [PubMed] [Google Scholar]
- Gao K, Chen F, Yuan L, Zhang F, Mi G (2015) A comprehensive analysis of root morphological changes and nitrogen allocation in maize in response to low nitrogen stress. Plant Cell Environ 38: 740–750 [DOI] [PubMed] [Google Scholar]
- Gibieža P, Prekeris R (2018) Rab GTPases and cell division. Small GTPases 9: 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, Gruber BD, von Wirén N (2014) It’s time to make changes: Modulation of root system architecture by nutrient signals. J Exp Bot 65: 769–778 [DOI] [PubMed] [Google Scholar]
- Gruber N, Galloway JN (2008) An Earth-system perspective of the global nitrogen cycle. Nature 451: 293–296 [DOI] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhao J, Li X, Qin L, Yan X, Liao H (2011) A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J 66: 541–552 [DOI] [PubMed] [Google Scholar]
- Hao J, Guo H, Shi X, Wang Y, Wan Q, Song YB, Zhang L, Dong M, Shen C (2017) Comparative proteomic analyses of two Taxus species (Taxus × media and Taxus mairei) reveals variations in the metabolisms associated with paclitaxel and other metabolites. Plant Cell Physiol 58: 1878–1890 [DOI] [PubMed] [Google Scholar]
- Hatfield RD, Grabber J, Ralph J, Brei K (1999) Using the acetyl bromide assay to determine lignin concentrations in herbaceous plants: Some cautionary notes. J Agric Food Chem 47: 628–632 [DOI] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Hinsinger P, Betencourt E, Bernard L, Brauman A, Plassard C, Shen J, Tang X, Zhang F (2011) P for two, sharing a scarce resource: Soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiol 156: 1078–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of class III plant peroxidases. Plant Cell Physiol 42: 462–468 [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ (2006) Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Janvilisri T, Scaria J, Teng CH, McDonough SP, Gleed RD, Fubini SL, Zhang S, Akey B, Chang YF (2012) Temporal differential proteomes of Clostridium difficile in the pig ileal-ligated loop model. PLoS ONE 7: e45608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Shibato J, Kim DW, Oh MK, Kim MK, Shim IeS, Iwahashi H, Masuo Y, Rakwal R (2009) Gel-based proteomics approach for detecting low nitrogen-responsive proteins in cultivated rice species. Physiol Mol Biol Plants 15: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Fukushima A, Redestig H, Saito K (2011) Metabolomic approaches toward understanding nitrogen metabolism in plants. J Exp Bot 62: 1439–1453 [DOI] [PubMed] [Google Scholar]
- Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C (2015) Cell wall metabolism in response to abiotic stress. Plants (Basel) 4: 112–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zeng R, Liao H (2016) Improving crop nutrient efficiency through root architecture modifications. J Integr Plant Biol 58: 193–202 [DOI] [PubMed] [Google Scholar]
- Liu JC, Li JS, Chen FJ, Zhang FS, Ren TH, Zhuang ZJ, Mi GH (2008) Mapping QTLs for root traits under different nitrate levels at the seedling stage in maize (Zea mays L.). Plant Soil 305: 253–265 [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Møller AL, Pedas P, Andersen B, Svensson B, Schjoerring JK, Finnie C (2011) Responses of barley root and shoot proteomes to long-term nitrogen deficiency, short-term nitrogen starvation and ammonium. Plant Cell Environ 34: 2024–2037 [DOI] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P (2014) Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 37: 162–174 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 495–497 [Google Scholar]
- Paul MJ, Driscoll SP (1997) Sugar repression of photosynthesis: The role of carbohydrates in signalling nitrogen deficiency through source:sink imbalance. Plant Cell Environ 20: 110–116 [Google Scholar]
- Piñeros MA, Larson BG, Shaff JE, Schneider DJ, Falcão AX, Yuan L, Clark RT, Craft EJ, Davis TW, Pradier PL, et al. (2016) Evolving technologies for growing, imaging and analyzing 3D root system architecture of crop plants. J Integr Plant Biol 58: 230–241 [DOI] [PubMed] [Google Scholar]
- Pysh L, Alexander N, Swatzyna L, Harbert R (2012) Four alleles of AtCESA3 form an allelic series with respect to root phenotype in Arabidopsis thaliana. Physiol Plant 144: 369–381 [DOI] [PubMed] [Google Scholar]
- Qi X, Zheng H (2013) Functional analysis of small Rab GTPases in cytokinesis in Arabidopsis thaliana. Methods Mol Biol 1043: 103–112 [DOI] [PubMed] [Google Scholar]
- Raggi S, Ferrarini A, Delledonne M, Dunand C, Ranocha P, De Lorenzo G, Cervone F, Ferrari S (2015) The Arabidopsis class III peroxidase AtPRX71 negatively regulates growth under physiological conditions and in response to cell wall damage. Plant Physiol 169: 2513–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen IS, Dresboll DB, Thorup-Kristensen K (2015) Winter wheat cultivars and nitrogen (N) fertilization: Effects on root growth, N uptake efficiency and N use efficiency. Eur J Agron 68: 38–49 [Google Scholar]
- Rellán-Alvarez R, Andaluz S, Rodríguez-Celma J, Wohlgemuth G, Zocchi G, Alvarez-Fernández A, Fiehn O, López-Millán AF, Abadía J (2010) Changes in the proteomic and metabolic profiles of Beta vulgaris root tips in response to iron deficiency and resupply. BMC Plant Biol 10: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengwilai P, Tian X, Lynch JP (2014) Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiol 166: 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan L (2002) Root development. The Arabidopsis Book 1: e0101, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJB, Bentsink L, Vonk J, van der Plas LHW, Koornneef M, Vreugdenhil D (2006) Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA 103: 2994–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeto J, Tsutsumi Y (2016) Diverse functions and reactions of class III peroxidases. New Phytol 209: 1395–1402 [DOI] [PubMed] [Google Scholar]
- Strader LC, Chen GL, Bartel B (2010) Ethylene directs auxin to control root cell expansion. Plant J 64: 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatanik-Kloc A, Szerement J, Cybulska J, Jozefaciuk G (2017) Input of different kinds of soluble pectin to cation binding properties of roots cell walls. Plant Physiol Biochem 120: 194–201 [DOI] [PubMed] [Google Scholar]
- Tenhaken R. (2015) Cell wall remodeling under abiotic stress. Front Plant Sci 5: 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S, Manoli A, Ravazzolo L, Botton A, Pivato M, Masi A, Quaggiotti S (2015) Nitrate sensing by the maize root apex transition zone: A merged transcriptomic and proteomic survey. J Exp Bot 66: 3699–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Turek I, Wheeler JI, Gehring C, Irving HR, Marondedze C (2015) Quantitative proteome changes in Arabidopsis thaliana suspension-cultured cells in response to plant natriuretic peptides. Data Brief 4: 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Behrens I, Komatsu M, Zhang Y, Berendzen KW, Niu X, Sakai H, Taramino G, Hochholdinger F (2011) Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J 66: 341–353 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG (2006) Nitrogen regulation of root branching. Ann Bot 97: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sun X, Xie Y, Li M, Chen W, Zhang S, Liang D, Ma F (2014) Melatonin regulates proteomic changes during leaf senescence in Malus hupehensis. J Pineal Res 57: 291–307 [DOI] [PubMed] [Google Scholar]
- Wang XB, Wu P, Hu B, Chen QS (2002) Effects of nitrate on the growth of lateral root and nitrogen absorption in rice. Acta Bot Sin 44: 678–683 [Google Scholar]
- Wang X, Bian Y, Cheng K, Zou H, Sun SS, He JX (2012) A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. J Proteome Res 11: 2301–2315 [DOI] [PubMed] [Google Scholar]
- Wang X, Chang L, Tong Z, Wang D, Yin Q, Wang D, Jin X, Yang Q, Wang L, Sun Y, et al. (2016) Proteomics profiling reveals carbohydrate metabolic enzymes and 14-3-3 proteins play important roles for starch accumulation during cassava root tuberization. Sci Rep 6: 19643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, et al. (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11: 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO, Yang Z (2004) New views on the plant cytoskeleton. Plant Physiol 136: 3884–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Thannhauser TW, Li L, Zhang S (2007) Development of an integrated approach for evaluation of 2-D gel image analysis: Impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28: 2080–2094 [DOI] [PubMed] [Google Scholar]
- Yang Y, Qiang X, Owsiany K, Zhang S, Thannhauser TW, Li L (2011) Evaluation of different multidimensional LC-MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. J Proteome Res 10: 4647–4660 [DOI] [PubMed] [Google Scholar]
- Zheng M, Zheng H, Wu Y, Xiao Y, Du Y, Xu W, Lu F, Wang X, Ouyang Z (2015) Changes in nitrogen budget and potential risk to the environment over 20 years (1990-2010) in the agroecosystems of the Haihe Basin, China. J Environ Sci (China) 28: 195–202 [DOI] [PubMed] [Google Scholar]
- Zhu XF, Shi YZ, Lei GJ, Fry SC, Zhang BC, Zhou YH, Braam J, Jiang T, Xu XY, Mao CZ, et al. (2012) XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 24: 4731–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieske LR. (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot 57: 1501–1508 [DOI] [PubMed] [Google Scholar]