Abstract
Multiple abi4 alleles fail to show a deficiency in chloroplast-to-nucleus retrograde signalling indicating that, contrary to contemporary models, ABI4 is not a component of this signalling pathway
Chloroplast-to-nucleus retrograde signaling pathways function during chloroplast development to enable coordination of the nuclear and chloroplast genomes for the assembly of the photosynthetic apparatus (Chan et al., 2016). This coordination is extremely important for seedling survival, as misregulation of photosynthetic development can lead to severe photooxidative damage and seedling lethality. The pathways mediating chloroplast-to-nucleus retrograde signaling during chloroplast development, termed biogenic signaling, are still poorly understood, but the transcription factor ABSCISIC ACID-INSENSITIVE4 (ABI4) has been proposed as an important downstream component (Koussevitzky et al., 2007) and features prominently in all published models (Chan et al., 2016; de Souza et al., 2017; Brunkard and Burch-Smith, 2018; Hernández-Verdeja and Strand, 2018). However, we had observed that chloroplast-to-nucleus retrograde signaling was not affected in abi4 mutants. Given the prevalence of ABI4 in retrograde signaling models, we have now systematically assessed the phenotype of abi4 mutants in an attempt to clarify the role of ABI4 in this signaling pathway. Here, we have analyzed the expression of eight retrograde-regulated nuclear genes following treatments with norflurazon (NF) and lincomycin (Lin), which block chloroplast development, in multiple abi4 alleles and in four different laboratories. Our analyses show no consistent effect of abi4 mutations on the retrograde response and do not support a role for ABI4 in this pathway. Therefore, we propose that ABI4 be omitted from future models of biogenic chloroplast-to-nucleus retrograde signaling.
Biogenic chloroplast-to-nucleus retrograde signaling pathways have been demonstrated using mutants that lack normal chloroplast development or the application of treatments such as the carotenoid synthesis inhibitor NF or the plastid translation inhibitor Lin. Both chemical treatments lead to chloroplast damage and a photobleached phenotype and result in a severe reduction in the expression of most photosynthesis-related nuclear genes (Koussevitzky et al., 2007; Woodson et al., 2013). The signaling pathway mediating this response remains unknown, but clues have come from the isolation of mutants that show less inhibition of nuclear gene expression after chloroplast damage. These mutants, termed genomes uncoupled or gun mutants, were identified originally as having elevated expression of the nuclear gene LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN1.2 (LHCB1.2) after NF treatment, a response that has become known as a gun phenotype. The original screens resulted in six loci that are important in retrograde signaling: five of these encode components of the tetrapyrrole biosynthesis pathway and rescue expression on NF (Mochizuki et al., 2001; Larkin et al., 2003; Woodson et al., 2011), while the sixth, gun1, lacks a pentatricopeptide repeat protein and can rescue expression on both NF and Lin (Koussevitzky et al., 2007). Based on these discoveries, the current model for chloroplast-to-nucleus retrograde signaling during chloroplast biogenesis is that signals from different sources, including tetrapyrrole biosynthesis, are integrated by GUN1 and relayed to the nucleus (Chan et al., 2016).
ABI4 was first identified in a screen for mutants that could germinate in the presence of abscisic acid (Finkelstein, 1994) and subsequently was shown to be related to a family of transcription factors containing an APETALA2 (AP2) domain, one of 147 AP2/ethylene response element-binding proteins in the Arabidopsis (Arabidopsis thaliana) genome (Nakano et al., 2006). ABI4 has been implicated in many growth and developmental responses in plants, with abi4 mutants also being identified independently in screens for sugar signaling mutants (León et al., 2013). These roles include signaling from the mitochondria to regulate ALTERNATIVE OXIDASE1a (Giraud et al., 2009) and chloroplast-to-nucleus retrograde signaling during chloroplast development (Koussevitzky et al., 2007). A role for ABI4 in chloroplast-to-nucleus signaling was first proposed by Nott et al. (2006) based on the reduced inhibition of a heterologous RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN (RBCS)-GUS reporter in an abi4 mutant background after NF treatment (Acevedo-Hernández et al., 2005; although no effect of abi4 was seen for an NF-responsive minimal CMA5 promoter construct) and their own data, later published as Koussevitzky et al. (2007), showing that abi4 also rescued LHCB expression after Lin treatment. From that point, ABI4 became established as a signaling intermediate in biogenic retrograde signaling and is included routinely in all published models. Despite this, the evidence for a role for ABI4 in chloroplast-to-nucleus signaling is not undisputed. Although some recent studies support a role for ABI4 (Sun et al., 2011; Zhang et al., 2013; Guo et al., 2016), others have not observed a gun phenotype on NF or Lin when looking at the expression of CARBONIC ANHYDRASE1 (CA1; Cottage and Gray, 2011), LHCB1.1 (Kerchev et al., 2011), or GOLDEN2-LIKE1 (GLK1; Martín et al., 2016). An abi4 mutant also was unable to rescue the loss of nuclear gene expression in the ppi2 mutant, in contrast to gun1 (Kakizaki et al., 2009). We also independently observed that abi4 mutants did not show a gun phenotype in our assays. Therefore, to try and resolve the question of whether ABI4 is required for biogenic retrograde signaling, we systematically assessed the phenotypes of four different abi4 alleles across four different research laboratories in three locations (Southampton, United Kingdom; Kyoto, Japan; and Munich, Germany).
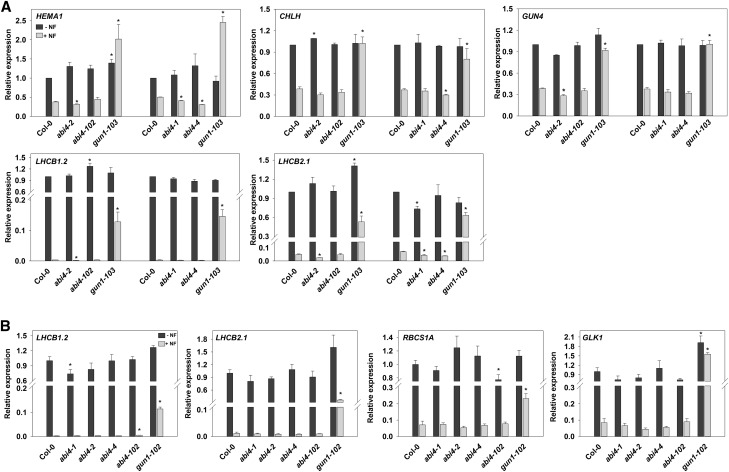
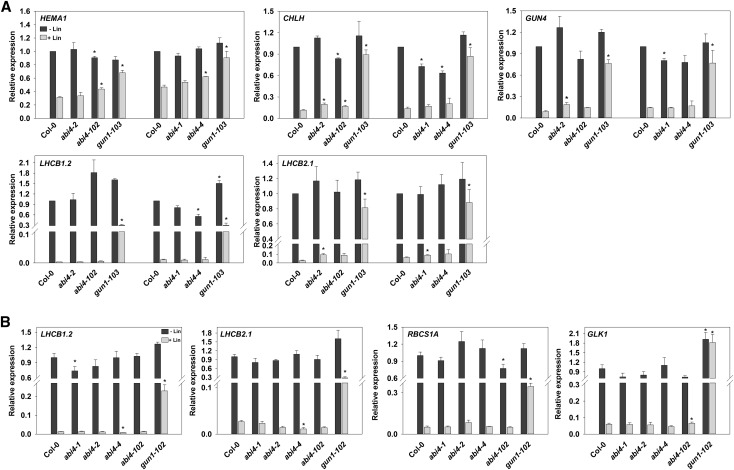
The four different alleles of abi4 used in this study were the abi4-102 allele used by Koussevitzky et al. (2007), the abi4-1 allele used by Sun et al. (2011), and two alleles that have not been characterized previously in terms of retrograde signaling, abi4-2 and abi4-4 (Supplemental Fig. S1; Supplemental Table S1; Supplemental Methods). Previous studies supporting a role for ABI4 based their conclusions on changes in LHCB expression measured by RNA gel blotting (Koussevitzky et al., 2007) or reverse transcription quantitative PCR (RT-qPCR) experiments with LHCB2.1 (Sun et al. [2011] used a primer pair that most closely matched this gene) or LHCB1.2 (Guo et al., 2016) in the presence of Suc. Therefore, we included both of these genes in our analysis, which also was performed in the presence of Suc (for a summary of the conditions used in this study, see Supplemental Table S2). As shown in Figure 1A, the expression of LHCB1.2, LHCB2.1, and three additional chlorophyll synthesis genes, HEMA1, CHLH, and GUN4, which show a strong dependence on GUN-mediated retrograde signaling (Moulin et al., 2008; Page et al., 2017), was strongly down-regulated in the presence of NF in wild-type seedlings with no increase in expression observed in any of the four abi4 alleles tested. In contrast, the gun1-103 mutant showed a strong rescue of nuclear gene expression in all cases. In parallel experiments performed in Kyoto, which included two additional NF down-regulated genes, RBCS1A and GLK1, and gun1-102 as a control, identical results were observed, although a small but statistically significant increase was seen for LHCB1.2 in the abi4-102 mutant only (Fig. 1B). Similar experiments using Lin to inhibit nuclear gene expression also showed essentially the same results, except that a very small, but significant, gun phenotype was observed in abi4-2 for LHCB2.1, CHLH, and GUN4 and in abi4-102 for HEMA1 and CHLH in the experiments performed in Southampton (Fig. 2A). This was under conditions in which gun1-103 rescued expression almost completely (Fig. 2A). However, no gun phenotype was observed in the experiments performed in Kyoto, including for LHCB2.1 in abi4-2 (Fig. 2B). To confirm that the lack of a gun phenotype was not due to the choice of reference gene, we replotted the data in Figure 2A using ACTIN2 (Sun et al., 2011) instead of YLS8. This made no difference to the conclusion, with a small, but significant, response seen only for the LHCB2.1 gene in abi4-2 (Supplemental Fig. S2A).
Figure 1.
abi4 mutants do not show a gun phenotype on NF. A, Seedlings were grown on one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc and 1% (w/v) agar (pH 5.8) with (light gray bars) or without (dark gray bars) 1 µm NF for 2 d of dark followed by 3 d of continuous white light (WLc; 100 µmol m−2 s−1). B, Seedlings were grown on Murashige and Skoog medium supplemented with 2% (w/v) Suc and 0.8% (w/v) agar (pH 5.8) with (light gray bars) or without (dark gray bars) 2.5 µm NF and grown for 4 d in WLc (100 µmol m−2 s−1). Expression was determined by RT-qPCR and is relative to wild-type Columbia-0 (Col-0) −NF and normalized to YELLOW LEAF SPECIFIC GENE8 (YLS8, At5g08290; A) or to TUBULIN BETA-CHAIN2 (TUB2, At5g62690; B). Data shown are means + se of three independent biological replicates. Asterisks denote significant differences versus the wild type for the same treatment (−NF or +NF) by Student’s t test (P < 0.05).
Figure 2.
abi4 mutants do not show a gun phenotype on Lin. A, Seedlings were grown on one-half-strength Linsmaier and Skoog medium supplemented with 2% (w/v) Suc and 0.8% (w/v) agar (pH 5.8) with (light gray bars) or without (dark gray bars) 0.5 mm Lin for 5 d of dark after an initial 2 h of WLc treatment (120 µmol m−2 s−1). B, Seedlings were grown on Murashige and Skoog medium supplemented with 2% (w/v) Suc and 0.8% (w/v) agar (pH 5.8) with or without 450 µm Lin for 4 d in WLc (100 µmol m−2 s−1). Expression was determined by RT-qPCR and is relative to wild-type Columbia-0 (Col-0) −Lin and normalized to YLS8 (At5g08290; A) or to TUB2 (At5g62690; B). The expression values for the control condition −Lin in B are the same as those shown in Figure 1B (−NF; dark gray bars). Data shown are means + se of three independent biological replicates. Asterisks denote significant differences versus the wild type for the same treatment (−Lin or +Lin) by Student’s t test (P < 0.05).
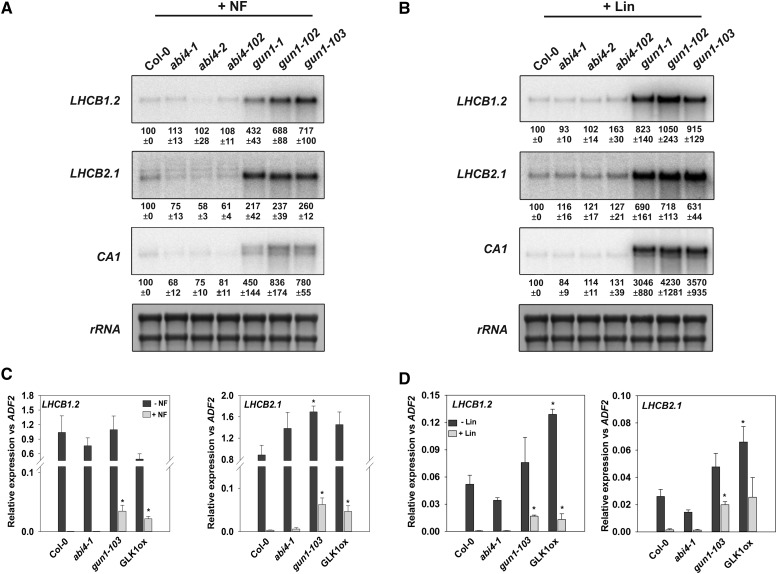
In the final set of experiments to test for a gun phenotype in abi4, which were performed in Munich, analysis was conducted using both RNA gel-blot analysis, as used in the original Koussevitzky et al. (2007) study, and RT-qPCR (Fig. 3). RNA gel-blot analyses of LHCB1.2, LHCB2.1, and CA1 showed no evidence for elevated gene expression after NF treatment in three abi4 alleles, while three gun1 alleles all showed strong responses (Fig. 3A). Similar results were observed after a shorter 6-d treatment with NF and WLc (Supplemental Fig. S2B). After Lin treatment, a very small increase in expression was observed for LHCB1.2 and LHCB2.1, but not for CA1, and only in abi4-102, not in abi4-1 or abi4-2 (Fig. 3B). Since Koussevitzky et al. (2007) used the abi4-102 allele, this result may account for their observations, but with the absence of a phenotype in the other abi4 alleles tested, it cannot be interpreted as supporting a role for ABI4. Finally, analysis of abi4-1 (used by Sun et al., 2011) did not show a gun phenotype for either LHCB1.2 or LHCB2.1 after NF or Lin treatment under conditions in which the positive controls gun1-103 and a GLK1-overexpressing line (Leister and Kleine, 2016; Martín et al., 2016) both resulted in a strong rescue of gene expression (Fig. 3, C and D). This result was not dependent on the reference gene used (Supplemental Fig. S2, C and D). Interestingly, simultaneous analysis of expression in the ptm1 mutant (Supplemental Fig. S2, E and F) confirmed that a third laboratory failed to see a gun phenotype for this mutant, consistent with our previous study (Page et al., 2017).
Figure 3.
abi4 mutants do not show a gun phenotype after NF or Lin treatment. A and B, Expression of photosynthetic genes after NF and Lin treatments determined by RNA gel-blot analysis. Seedlings were grown on one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc and 1% (w/v) agar (pH 5.8) with or without 5 µm NF (A) or 0.5 mm Lin (B). For NF treatments (A), seedlings were grown for 4 d of dark and 3 d in WLc (100 µmol m−2 s−1), and for Lin treatments (B), seedlings were grown for 6 d in WLc (100 µmol m−2 s−1). Five micrograms of total RNA was loaded per sample with Methylene Blue staining of rRNA as a loading and RNA-transfer control. Results from one of three independent experiments are shown, with values indicating means ± se of densitometric scans from all three experiments. C and D, Expression of photosynthetic genes after NF and Lin treatments determined by RT-qPCR. Seedlings were grown on one-half-strength Murashige and Skoog medium supplemented with 2% (w/v) Suc and 0.8% (w/v) agar (pH 5.8) with or without 5 µm NF (C) or 0.5 mm Lin (D) under the same growth conditions as for A and B. Expression is relative to ACTIN DEPOLYMERIZING FACTOR2 (ADF2, At3g46000), and data shown are means + se of three independent biological replicates. Asterisks denote significant differences versus wild-type Columbia-0 (Col-0) for the same treatment (NF or Lin) by Student’s t test (P < 0.05).
In the original study by Koussevitzky et al. (2007), it was reported that there was significant overlap of gun1- and abi4-regulated genes (approximately 50% of derepressed or repressed genes) following transcriptome analysis, and this finding was used to support the hypothesis that they act in the same retrograde pathway. Here, we reanalyzed this data set and compared the response to Lin in abi4-102 and gun1-1. As shown in Supplemental Figure S3, the response in abi4-102 clustered with the wild type after Lin treatment in contrast to gun1-1, but it did show some difference from the wild type in control conditions (Supplemental Fig. S3, A and B). Expression analysis after Lin treatment correlated well between the wild type and abi4-102 but not between the wild type and gun1-1 overall (Supplemental Fig. S3C), and this could be seen clearly when changes in the expression of individual photosynthesis (Supplemental Fig. S3D) and tetrapyrrole biosynthesis (Supplemental Fig. S3E) genes were analyzed. Similar conclusions were drawn from this data set by Martín et al. (2016) in the context of PHYTOCHROME INTERACTING FACTOR-regulated genes. Therefore, these results do not support a role for ABI4 in the same retrograde pathway as GUN1.
One of the observations that supported a prominent role for ABI4 in chloroplast-to-nucleus retrograde signaling was that ABI4 gene expression was strongly up-regulated on NF and Lin and that this response was completely absent in the gun1 mutant (Sun et al., 2011). This followed on from initial observations that ABI4 expression was reduced in gun1 in the presence and absence of Lin (Koussevitzky et al., 2007). We tested this response in our assays and observed very different results. In this case, treatment with NF or Lin resulted in a 34- or 6-fold increase in ABI4 expression in wild-type seedlings, respectively, and expression was even more strongly up-regulated in the three different gun1 alleles tested (Supplemental Fig. S4A). An increase in ABI4 expression also was observed in gun1 mutants when analyzed by RNA gel-blot analysis (Supplemental Fig. S4B). Thus, although the induction of ABI4 expression under these stress conditions was confirmed in this study, the response in gun1 was opposite to that reported previously and not consistent with the regulation of ABI4 via a GUN1-mediated retrograde signaling pathway.
Recent models for biogenic chloroplast-to-nucleus retrograde signaling have ABI4 acting downstream of GUN1 in a PTM-dependent pathway. While the strong gene expression phenotype of different gun1 mutant alleles has been verified in many studies, including this one, further analysis of the role of PTM in retrograde signaling has not supported such a model (Page et al., 2017; this study). Here, we have reevaluated the role of ABI4 in biogenic retrograde signaling using the same basic experimental conditions, such as the presence of Suc and developmental age of the seedlings, and by testing the same genes. If ABI4 has a major role in this signaling pathway (and previous studies have shown the response to be almost as strong as that of gun1; Sun et al., 2011), then we would expect to see some response under the conditions tested across the three different locations in which our experiments were conducted. The results presented here show no consistent or strong gun phenotype for multiple abi4 alleles across multiple laboratories and, therefore, do not support a role for ABI4 in biogenic retrograde signaling. As noted earlier, other studies also have reported a lack of a gun phenotype for abi4 mutants (Cottage and Gray, 2011; Kerchev et al., 2011; Martín et al., 2016), and our results can be considered to be in agreement with these. Therefore, we recommend that ABI4 should be omitted from future models of chloroplast-to-nucleus retrograde signaling. There have been some significant recent developments in our understanding of the importance of tetrapyrroles and chloroplast protein homeostasis in plastid retrograde signaling (Woodson et al., 2011; Tadini et al., 2016; Paieri et al., 2018; Wu et al., 2018), and attention can now focus on these areas of research.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of the four abi4 mutant alleles used in this study.
Supplemental Figure S2. Additional analyses of retrograde regulation of photosynthetic gene expression after NF and Lin treatments.
Supplemental Figure S3. Reanalysis of microarray data from Koussevitzky et al. (2007).
Supplemental Figure S4. Regulation of ABI4 gene expression by retrograde signaling.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Summary of treatment conditions used in this study.
Supplemental Methods. Supplemental materials and methods.
Acknowledgments
We thank Eiji Nambara (RIKEN) for the abi4-4 (E14-7) mutant allele. D.L. thanks Lixin Zhang (Chinese Academy of Sciences) for the abi4-1 and ptm mutant alleles used in Munich and Jane Langdale (University of Oxford) for the GLK1ox line. N.M. also thanks Lixin Zhang for the abi4-1 mutant allele.
Footnotes
The work was supported by JSPS KAKENHI Grant JP 17K07444 to N.M. S.M.K. was supported by the Gatsby Charitable Foundation. Work on retrograde signalling by M.J.T. is supported by the UK Biotechnology and Biological Sciences Research Council. T.K., B.N., and D.L. are supported by the Deutsche Forschungsgemeinschaft (KL 2362/1-1 and TRR175, projects C01 and C05).
Articles can be viewed without a subscription.
Senior author.
References
- Acevedo-Hernández GJ, León P, Herrera-Estrella LR (2005) Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J 43: 506–519 [DOI] [PubMed] [Google Scholar]
- Brunkard JO, Burch-Smith TM (2018) Ties that bind: The integration of plastid signalling pathways in plant cell metabolism. Essays Biochem 62: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Cottage A, Gray JC (2011) Timing the switch to phototrophic growth: A possible role of GUN1. Plant Signal Behav 6: 578–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A, Wang JZ, Dehesh K (2017) Retrograde signals: Integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol 68: 85–108 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Giraud E, Van Aken O, Ho LH, Whelan J (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Feng P, Chi W, Sun X, Xu X, Li Y, Ren D, Lu C, Rochaix JD, Leister D, et al. (2016) Plastid-nucleus communication involves calcium-modulated MAPK signaling. Nat Commun 7: 121–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Verdeja T, Strand Å (2018) Retrograde signals navigate the path to chloroplast development. Plant Physiol 176: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che FS, Terauchi R, Inaba T (2009) Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol 151: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev PI, Pellny TK, Vivancos PD, Kiddle G, Hedden P, Driscoll S, Vanacker H, Verrier P, Hancock RD, Foyer CH (2011) The transcription factor ABI4 is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in Arabidopsis. Plant Cell 23: 3319–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Leister D, Kleine T (2016) Definition of a core module for the nuclear retrograde response to altered organellar gene expression identifies GLK overexpressors as gun mutants. Physiol Plant 157: 297–309 [DOI] [PubMed] [Google Scholar]
- León P, Gregorio J, Cordoba E (2013) ABI4 and its role in chloroplast retrograde communication. Front Plant Sci 3: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG (2008) Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ (2017) Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol 174: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paieri F, Tadini L, Manavski N, Kleine T, Ferrari R, Morandini P, Pesaresi P, Meurer J, Leister D (2018) The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol 176: 634–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 477. [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, et al. (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R (2018) Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol 176: 2472–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZW, Feng LY, Cheng J, Tang H, Xu F, Zhu F, Zhao ZY, Yuan M, Chen YE, Wang JH, et al. (2013) The roles of two transcription factors, ABI4 and CBFA, in ABA and plastid signalling and stress responses. Plant Mol Biol 83: 445–458 [DOI] [PubMed] [Google Scholar]