Image-based phenotyping facilitates the genetic dissection of uni- and multi-dimensional traits of sorghum panicles.
Abstract
Because structural variation in the inflorescence architecture of cereal crops can influence yield, it is of interest to identify the genes responsible for this variation. However, the manual collection of inflorescence phenotypes can be time consuming for the large populations needed to conduct genome-wide association studies (GWAS) and is difficult for multidimensional traits such as volume. A semiautomated phenotyping pipeline, TIM (Toolkit for Inflorescence Measurement), was developed and used to extract unidimensional and multidimensional features from images of 1,064 sorghum (Sorghum bicolor) panicles from 272 genotypes comprising a subset of the Sorghum Association Panel. GWAS detected 35 unique single-nucleotide polymorphisms associated with variation in inflorescence architecture. The accuracy of the TIM pipeline is supported by the fact that several of these trait-associated single-nucleotide polymorphisms (TASs) are located within chromosomal regions associated with similar traits in previously published quantitative trait locus and GWAS analyses of sorghum. Additionally, sorghum homologs of maize (Zea mays) and rice (Oryza sativa) genes known to affect inflorescence architecture are enriched in the vicinities of TASs. Finally, our TASs are enriched within genomic regions that exhibit high levels of divergence between converted tropical lines and cultivars, consistent with the hypothesis that these chromosomal intervals were targets of selection during modern breeding.
The grass family (Poaceae) includes maize (Zea mays), wheat (Triticum aestivum), rice (Oryza sativa), sorghum (Sorghum bicolor), and other cereal crops, which collectively provide 56% of the calories consumed by humans in developing countries and over 30% in developed countries (Amine et al., 2003; Bruinsma, 2003). The development of the grain-bearing inflorescences of cereals begins with the transition of the vegetative shoot apical meristem (SAM) into an inflorescence meristem, which later forms into branch meristems and further generates spikelet meristems (Zhang and Yuan, 2014). Variation in these developmental processes accounts for the substantial interspecific and intraspecific variability in inflorescence architecture observed among the cereals (Vollbrecht et al., 2005; Huang et al., 2009; Youssef et al., 2017). The transition of the SAM into an inflorescence meristem is regulated by genes that affect both the identity and maintenance of meristems (Pautler et al., 2013; Zhang and Yuan, 2014). For example, in Arabidopsis (Arabidopsis thaliana), meristem identity is regulated primarily by a negative feedback loop between CLAVATA (CLV) and the homeobox gene WUSCHEL (WUS), which prevents the misspecification of meristem cells and the premature termination of floral and shoot meristems (Laux et al., 1996; Mayer et al., 1998; Pautler et al., 2013; Tanaka et al., 2013). Mutations of CLV genes often result in larger inflorescence meristems (Clark et al., 1997; Fletcher et al., 1999; Jeong et al., 1999). Similarly, mutations of the CLV1 homolog in maize, thick tassel dwarf1 (TD1), and the CLV2 homolog, fasciated ear2, produce tassels with more spikelets and fasciated ears with extra rows of kernels (Taguchi-Shiobara et al., 2001; Bommert et al., 2005). The KNOTTED1-like homeobox genes also affect inflorescence development by altering the establishment and maintenance of SAM tissues (Vollbrecht et al., 2000; Bolduc and Hake, 2009; Bolduc et al., 2012). Mutations of the maize knotted1 (Kn1) and the rice Oryza sativa homeobox1 genes both exhibit a sparse-inflorescence phenotype caused by reduced meristem maintenance (Kerstetter et al., 1997; Tsuda et al., 2011). The null allele kn1-E1 is epistatic to the null allele td1-glf in maize ear development and suggests the importance of Kn1 in regulating both meristem identity and lateral organ initiation (Lunde and Hake, 2009).
These functional studies of large-effect or qualitative mutants have greatly enhanced our understanding of the developmental processes underlying inflorescence development and architecture. Even so, because many quantitative traits also are affected by large numbers of small-effect genes (Buckler et al., 2009; Danilevskaya et al., 2010; Brown et al., 2011; Li et al., 2012), there remains the opportunity to expand our understanding of inflorescence development via the application of genome-wide association studies (GWAS) to identify associations between specific loci and quantitative phenotypic variation. GWAS has been used to identify genes associated with inflorescence architecture in multiple crops (Brown et al., 2011; Morris et al., 2013; Crowell et al., 2016; Wu et al., 2016; Zhao et al., 2016; Xu et al., 2017). Given that thousands or millions of markers can now be readily discovered and genotyped, phenotyping is typically the bottleneck for conducting GWAS. Traditionally, crop scientists have collected unidimensional traits, such as spike length, spike width, and branch length and number, manually. This is time consuming for large populations. Therefore, to fully utilize the advantages of GWAS, there is a need for accurate, high-throughput phenotyping platforms.
Computer vision has been shown to be efficient in isolating inflorescences (Aquino et al., 2015; Zhao et al., 2015; Millan et al., 2017), and several studies have attempted to extract inflorescence features from images of rice and maize (AL-Tam et al., 2013; Crowell et al., 2014; Zhao et al., 2015; Gage et al., 2017). The complexity of inflorescence architecture complicates the accurate extraction of phenotypes from images. To date, two studies have applied image-based phenotyping to the genetic analyses of crop inflorescences, and they focused either only on artificially flattened rice inflorescences (Crowell et al., 2016) or on unidimensional traits of maize such as tassel length and central spike length (Gage et al., 2018). Considering that inflorescences are 3D structures, phenotyping strategies that flatten an inflorescence or focus on only a single plane will inevitably fail to capture a considerable amount of phenotypic variation and, therefore, will reduce the probability of discovering genes involved in inflorescence architecture. This limitation highlights the need for new automated phenotyping platforms that accurately collect inflorescence traits, especially multidimensional traits, which have not been collected by previous automated phenotyping projects.
Sorghum is the world’s fifth most important cereal crop and is a major food crop in some developing countries (Hariprasanna and Rakshit, 2016). It is evolutionarily closely related to the other well-studied cereals such as maize, wheat, and rice (Paterson et al., 2009; Schnable et al., 2012; Choulet et al., 2014; Schnable, 2015). Studies conducted to date on the genetic architecture of sorghum panicles have focused on unidimensional traits such as panicle length, panicle width, and branch length (Hart et al., 2001; Brown et al., 2006; Srinivas et al., 2009; Morris et al., 2013; Nagaraja Reddy et al., 2013; Zhang et al., 2015; Zhao et al., 2016). Due to the challenges associated with collecting multidimensional traits such as panicle area, volume, and compactness, our understanding of the genetic architectures of these traits is limited.
Sorghum exhibits extensive population structure associated with both morphological type and geographic origin (Bouchet et al., 2012; Morris et al., 2013), which has the potential to introduce false-positive signals into GWAS analyses unless properly controlled (Yu et al., 2006; Zhang et al., 2010). In addition, the average extent of linkage disequilibrium (LD) decay in sorghum is substantially greater than in maize, due at least in part to its mode of reproduction (Chia et al., 2012; Morris et al., 2013). Therefore, each trait-associated single-nucleotide polymorphism (TAS) identified via GWAS is likely to be linked to a large genomic region, making it challenging to identify candidate genes. However, quantitative trait locus (QTL) studies have identified syntenic chromosomal regions that control inflorescence traits in both maize and sorghum (Brown et al., 2006). Furthermore, it is possible to identify conserved sorghum homologs of genes that regulate inflorescence architecture in maize and rice (Paterson et al., 2009; Schnable et al., 2012; Zhang et al., 2015). These findings suggest that scanning chromosomal regions surrounding sorghum TASs for homologs of maize and rice genes with known functions in inflorescence architecture could overcome the challenges in identifying candidate genes caused by sorghum’s high LD.
In this study, we developed and deployed a high-resolution imaging pipeline to collect panicle phenotypes from a subset of the Sorghum Association Panel (SAP; Casa et al., 2008) consisting of 272 accessions. We used a semiautomated procedure to extract both unidimensional and multidimensional traits from these images. The resulting phenotypic data were used to perform GWAS, which identified TASs, some of which are located within chromosomal regions identified via previously published QTL and GWAS analyses of sorghum for similar traits. In addition, a statistically enriched fraction of these TASs are located near sorghum homologs of maize or rice genes known to influence inflorescence architecture. A genome-wide analysis of population differentiation suggests that the genomic regions that contain the TASs have undergone artificial selection during sorghum breeding.
RESULTS
Phenotyping
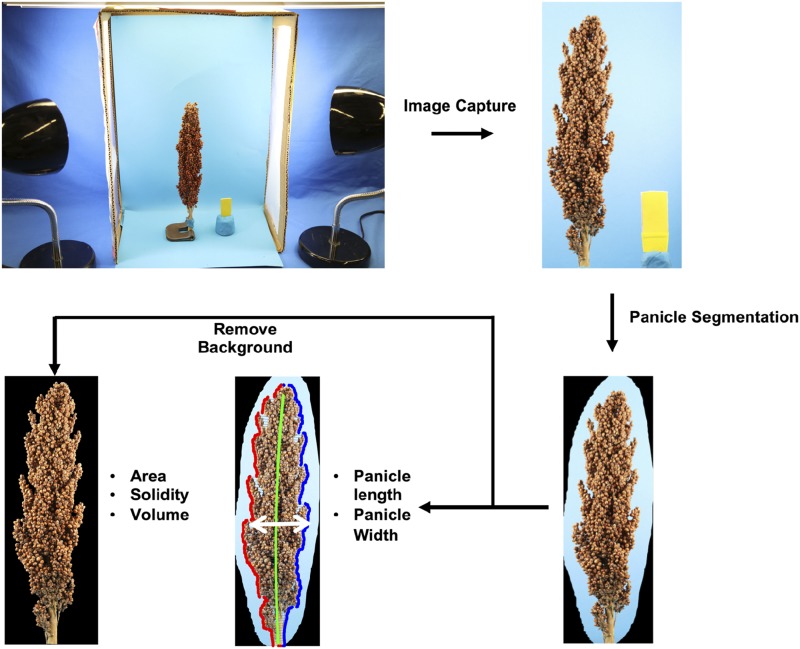
The front and side planes of 1,064 panicles from 272 genotypes (designated SAP-FI; Supplemental Fig. S1) were imaged (Fig. 1; see “Materials and Methods”). A MATLAB app from TIM (Toolkit for Inflorescence Measurement) was used to mark the approximate boundary of each panicle that was subsequently cropped from the original image. A fully automated trait extraction protocol (see “Materials and Methods”) then was used to extract length and width directly from cropped front plane images.
Figure 1.
Phenotyping pipeline used to extract traits from 1,064 panicles of 272 genotypes. Each panicle was photographed from both the front and side planes. Segmentation was conducted manually for subsequent automatic trait extraction.
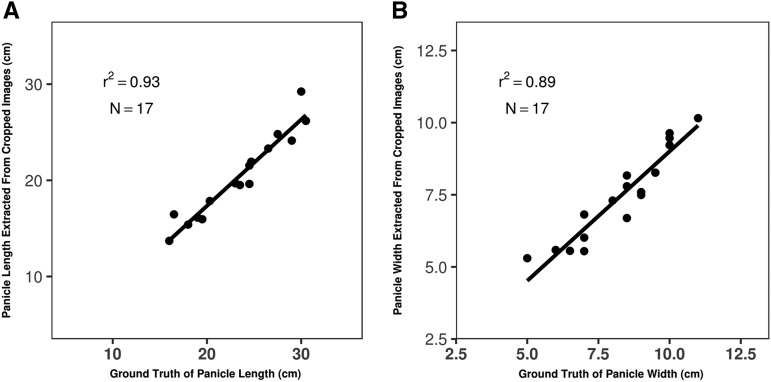
To evaluate the accuracy of our semiautomated image-processing method, we manually measured the length and width of single panicles from 17 genotypes randomly selected from the SAP-FI. The resulting trait values were compared with the corresponding values automatically extracted from images of the same panicles. The coefficient of determination (r2) between the values of the autoextracted length and width.front versus ground truth were 0.93 and 0.89, respectively (Fig. 2), indicating that our pipeline can accurately extract panicle length and width.
Figure 2.
Comparison of ground-truth measurements and trait values extracted from images of 17 randomly chosen panicles. A, Panicle length. B, Panicle width of front plane.
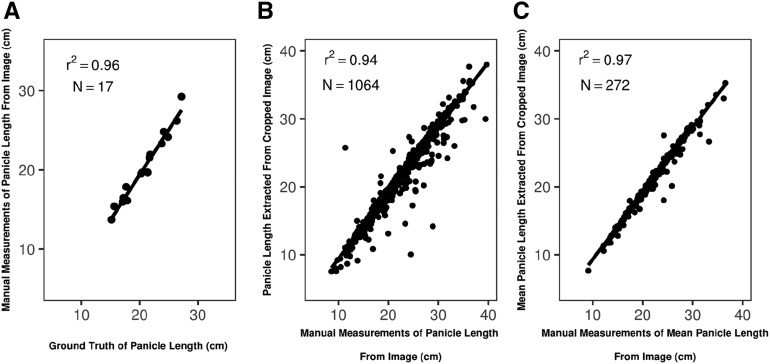
Differences between the values for autoextracted traits and ground truth as shown in Figure 2 consist of two components: (1) variation between trait measurements of a 3D panicle and the representation of these trait measurements in a 2D image, and (2) errors in autoextracting trait values from a 2D image. To evaluate the first source of variation, we manually measured lengths from 2D panicle images of the above 17 genotypes using our MATLAB app and compared the resulting trait values with panicle lengths of ground truth. The 0.96 value for r2 (Fig. 3) indicated that little variation was introduced via 2D imaging.
Figure 3.
Comparison of panicle lengths measured manually from images using a custom MATLAB app and extracted from the same images using our automated image-processing pipeline. A, Comparison of ground-truth measurements and panicle length measured manually from images of 17 randomly chosen panicles (front plane). B, Panicle lengths from 1,064 individual images (front plane). C, Panicle lengths using mean values of 272 genotypes.
To quantify the second source of variation, we also manually measured lengths from 2D images of panicles using our MATLAB app and compared the resulting trait values with lengths autoextracted from cropped images. Considering all 1,064 panicles, the r2 was 0.94 (Fig. 3). The r2 value could be increased further to 0.97 by comparing extracted trait values of a genotype averaged across replications and locations (Fig. 3). These results demonstrate that the second source of error was minimal and could be well controlled under our experimental design. Mean trait values were used for all subsequent phenotypic analyses, including GWAS.
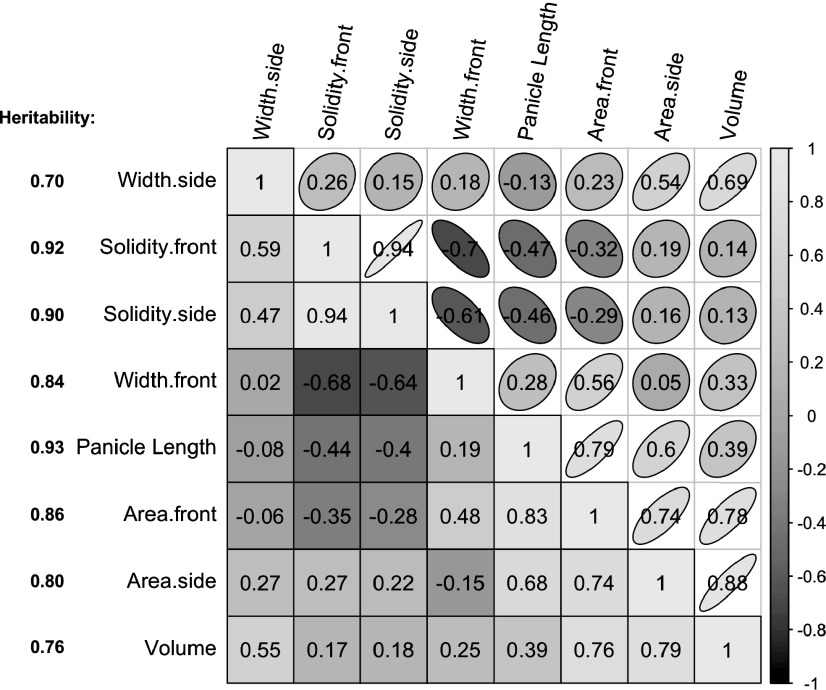
In addition to length and width.front, six other panicle traits were extracted from cropped panicle images. Width.side was extracted using the same criteria as width.front but using a panicle’s side plane image. Because the same pipeline was used to extract width.front and width.side, we did not collect ground-truth data for the latter trait. Area and solidity are features of a 3D object projected onto a 2D plane, so we chose these two traits to reflect structural variation in panicle size and compactness projected on the front and side planes, yielding the traits area.front, area.side, solidity.front, and solidity.side. Panicle volume was estimated using information extracted from both planes (see “Materials and Methods”). No ground-truth data were collected for the five multidimensional traits due to the challenge of measuring them by hand. All eight traits exhibited high heritabilities (Fig. 4), with the highest for panicle length (H2 = 0.93) and lowest for width.side (H2 = 0.7).
Figure 4.
Phenotypic and genetic correlations among panicle traits. Traits are ordered using the hclust function of R, with phenotypic and genetic correlations indicated by ellipses and squares, respectively. Traits extracted from the front or side plane are distinguished by the extensions .front and .side, respectively. The heritability of each trait is shown on the left.
Phenotypic and Genetic Correlations of Panicle Traits
The phenotypic correlation between two traits is determined by their genetic and nongenetic correlations and heritabilities (Bernardo, 2010). The genetic correlation between two traits is a function of pleiotropy (i.e. the effect of a gene on multiple traits; Mackay et al., 2009). Correlations among the eight traits were determined by analyzing all pairwise comparisons. The phenotypic correlations among panicle traits were strongly associated with their corresponding genotypic correlations (Fig. 4). For example, the 10 pairs of traits with the highest phenotypic correlations also exhibited the highest genetic correlations.
For solidity and area, values of the front plane were highly correlated with those of the side plane (r = 0.94 and 0.74, respectively; Fig. 4). In contrast, as expected based on the definition of front and side planes (see “Materials and Methods”), there was only a low correlation (r = 0.18) between the panicle widths of each plane. Although width.front was positively correlated with area.front (r = 0.56), it was poorly correlated with area.side (r = 0.05), despite the reasonably high phenotypic correlation between the two area traits (r = 0.74). The genotypes within the SAP-FI panel exhibit a great deal of variation in panicle architecture (Supplemental Fig. S1). Some of these genotypes exhibit a flat shape, in which the width.side is much smaller than width.front (see, for example, the three genotypes displayed on the right of Supplemental Fig. S1). Inspection of individual panicles confirmed that flat genotypes contribute to the poor correlation between width.front and area.side (Supplemental Fig. S2). Similarly, both solidity traits were negatively correlated with traits from the front plane (panicle length, width.front, and area.front) while being positively correlated with width.side and area.side. Because these phenotypic correlations are consistent with their genetic correlations (Fig. 4), we hypothesize that some genes have functions in both planes.
GWAS
To identify loci that affect panicle architecture, GWAS was performed on the SAP-FI panel for the eight panicle traits using an improved version of FarmCPU (Liu et al., 2016) termed FarmCPUpp (Kusmec and Schnable, 2018). These analyses identified 49 associations (Supplemental Fig. S3) representing 46 nonredundant TASs. A bootstrapping process was used to filter false-positive signals (see “Materials and Methods”). Of the 46 single-nucleotide polymorphisms (SNPs) detected in the GWAS of the SAP-FI panel, 38 associations, representing 35 high-confidence, nonredundant TASs, also exhibited a resample model inclusion probability (RMIP) ≥ 0.05 in the bootstrap experiment (see “Materials and Methods”; Table 1).
Table 1. List of 35 significant TASs affecting 38 trait associations.
| Trait | SNP | −log10(P value) | Estimate Effect | Standardized Effect Size | RMIP |
|---|---|---|---|---|---|
| Area.front | S1_3176715 | 9.80 | −5.75 | 0.27 | 0.47 |
| Area.front | S4_12278584a | 6.56 | −4.16 | 0.20 | 0.13 |
| Area.front | S4_60591319 | 6.88 | −5.38 | 0.25 | 0.19 |
| Area.front | S10_1793055 | 6.58 | 4.96 | 0.23 | 0.22 |
| Area.side | S3_3201555b | 9.47 | 6.55 | 0.51 | 0.16 |
| Area.side | S3_58994806a | 8.07 | 3.53 | 0.28 | 0.12 |
| Area.side | S5_17518239 | 6.74 | 4.07 | 0.32 | 0.44 |
| Area.side | S6_49755954 | 7.91 | −3.28 | 0.26 | 0.30 |
| Panicle length | S1_8065027a,c | 6.69 | 0.98 | 0.22 | 0.07 |
| Panicle length | S2_63553713a,c | 15.21 | 3.36 | 0.74 | 0.75 |
| Panicle length | S3_58920501a | 6.92 | 0.90 | 0.20 | 0.05 |
| Panicle length | S8_14556347c | 7.28 | −1.67 | 0.37 | 0.40 |
| Panicle length | S10_50809289 | 10.68 | 1.31 | 0.29 | 0.19 |
| Panicle length | S10_54184063 | 6.88 | −0.84 | 0.18 | 0.17 |
| Solidity.front | S2_10933862b | 15.07 | −0.07 | 0.71 | 0.82 |
| Solidity.front | S2_70599334 | 10.56 | −0.03 | 0.32 | 0.09 |
| Solidity.front | S2_76284762a | 9.48 | 0.04 | 0.41 | 0.05 |
| Solidity.front | S3_56132328 | 6.59 | 0.02 | 0.16 | 0.27 |
| Solidity.front | S4_56171463 | 15.34 | −0.03 | 0.36 | 0.45 |
| Solidity.front | S6_60219786a | 7.39 | 0.02 | 0.26 | 0.07 |
| Solidity.front | S8_6056282b | 7.40 | −0.03 | 0.36 | 0.13 |
| Solidity.front | S10_1149373 | 6.62 | −0.02 | 0.24 | 0.05 |
| Solidity.side | S1_19664643a | 7.20 | −0.04 | 0.59 | 0.31 |
| Solidity.side | S2_10933862b | 10.13 | −0.05 | 0.66 | 0.81 |
| Solidity.side | S7_54381792 | 9.13 | 0.04 | 0.56 | 0.31 |
| Solidity.side | S8_6056282b | 7.57 | −0.03 | 0.41 | 0.38 |
| Volume | S3_3201555b | 9.34 | 33.33 | 0.53 | 0.45 |
| Width.front | S2_68297155 | 14.17 | 0.53 | 0.38 | 0.72 |
| Width.front | S3_3724913 | 8.87 | 0.48 | 0.34 | 0.48 |
| Width.front | S3_57037006 | 10.99 | 0.71 | 0.50 | 0.69 |
| Width.front | S4_65197133 | 7.61 | 0.53 | 0.38 | 0.10 |
| Width.front | S5_11264509 | 7.98 | 0.58 | 0.41 | 0.15 |
| Width.front | S6_60559938 | 6.62 | 0.42 | 0.30 | 0.17 |
| Width.front | S8_26248649 | 8.04 | 0.58 | 0.42 | 0.18 |
| Width.front | S8_45726712 | 9.35 | 0.44 | 0.31 | 0.12 |
| Width.front | S9_3027904 | 7.81 | 0.62 | 0.44 | 0.24 |
| Width.side | S3_2724080a | 9.57 | 0.27 | 0.43 | 0.19 |
| Width.side | S10_60236042a | 6.82 | 0.15 | 0.23 | 0.08 |
Have the candidate gene located within 350 kb on either side of the TAS (Table 2).
Pleiotropic TASs in GWAS of the full set.
TASs that overlap with previously reported QTLs or GWAS intervals (Supplemental Table S1).
Sixty-six percent (n = 23) of these 35 high-confidence, nonredundant TASs were associated with three of the eight traits: length (n = 6), solidity.front (n = 8), and width.front (n = 9). For the three traits measured in both planes, approximately twice as many TASs were detected for front plane traits (n = 21) as compared with the corresponding side plane (n = 10; Table 1). This probably reflects the lower heritabilities of the side plane traits relative to the front plane (Fig. 4).
We compared the TASs associated with panicle length and width.front with published QTL and GWAS results (Hart et al., 2001; Brown et al., 2006; Srinivas et al., 2009; Morris et al., 2013; Nagaraja Reddy et al., 2013; Zhang et al., 2015; Zhao et al., 2016). Encouragingly, two of our six TASs for panicle length are located within previously reported QTL intervals (Supplemental Table S1; Hart et al., 2001; Srinivas et al., 2009), and another (at position 8,065,027 bp on chromosome 1) is located within the interval reported by a previous GWAS for panicle length (Zhang et al., 2015). One of the nine TASs identified for width.front (at position 3,724,913 bp on chromosome 3) was located within regions identified via previous QTL and GWAS on panicle width and branch length (Hart et al., 2001; Brown et al., 2006; Morris et al., 2013; Zhang et al., 2015). Given the differences in genetic materials used in these studies and the fact that SAP-FI is only a subset of the original SAP used for GWAS by others, it is not surprising that the results of these studies do not fully overlap.
Only three TASs were associated with multiple traits: two were associated with both solidity.front and solidity.side (Table 1) and one with both area.side and volume. The limited number of overlapping SNPs was unexpected given the substantial correlations among traits (Fig. 4). We hypothesized that this low overlap is due to false-negative associations that arose as a consequence of the modest effect sizes of many of the detected TASs (standardized effect sizes < 0.5; Supplemental Fig. S4) in combination with the relatively small size of our panel, which limited the power of statistical tests. To evaluate this hypothesis, we compared the high-confidence TASs for one trait with all significant TASs identified via bootstrapping that exhibited an RMIP ≥ 0.05 for a second trait highly correlated with the first one. This process identified nine additional TASs (Supplemental Table S2) affecting highly correlated traits, supporting our hypothesis that our stringent control of false-positive signals reduced the ability to detect TASs that associated with multiple traits. Similar to previous studies on the effect sizes of pleiotropic TASs for maize inflorescences (Brown et al., 2011), the standardized effect sizes of the 12 sorghum TASs that affect multiple traits were similar for both members of pairs of affected traits (Supplemental Fig. S5).
Identification of Candidate Genes
The sorghum genome exhibits extensive LD (Morris et al., 2013). Although estimates of LD vary across the genome (Hamblin et al., 2005; Bouchet et al., 2012; Morris et al., 2013), we elected to use a genome-wide average estimate of LD in our search for candidate genes because our local estimates of LD surrounding TASs were noisy, perhaps due to the relatively small number of SNPs per Mb (200 = 146,865/730-Mb genome) and missing data in this genotyping by sequencing-derived SNP set. Hence, we screened 700-kb windows centered on each TAS for candidate genes (see “Materials and Methods”).
The limited number of sorghum genes that have been subjected to functional analyses complicates the identification of candidate genes. We overcame this challenge by looking for sorghum homologs of maize and rice genes (Supplemental Table S3) associated previously with inflorescence architecture (Vollbrecht et al., 2005; Tanaka et al., 2013; Zhang and Yuan, 2014) located within the 700-kb windows surrounding TASs (see “Materials and Methods”).
Using this procedure, nine candidate genes were identified (Table 2). A permutation test (see “Materials and Methods”; P = 0.026) indicated that sorghum homologs of maize and rice genes known to affect inflorescence architecture are enriched in chromosomal regions surrounding TASs. In addition, the maize and rice homologs of each of the nine sorghum candidate genes have functions consistent with the traits used to identify associations (Table 2), and five of these maize and/or rice genes were associated with relevant traits based on GWAS conducted in those species (Table 2).
Table 2. List of nine candidate genes for panicle architecture.
| Trait | SNP | Sorghum Candidate Gene Identifier (V1.4) | Annotation of Sorghum Candidate Gene | Distance from SNP to Candidate Gene (kb) | Homolog of Candidate Gene in Maize (Zm) or Rice (Os) | Function of Homolog in Maize/Rice Inflorescence Development | Traits Associated with Homolog Gene via GWAS in Maize/Rice | References |
|---|---|---|---|---|---|---|---|---|
| Panicle length | S1_8065027 | Sb01g009480 | Homeobox domain-containing protein | 157.6 | Kn1 (Zm) | Meristem maintenance and regulate the transition from SAM into inflorescence meristem | Central spike length | Vollbrecht et al. (2000); Brown et al. (2011); Xu et al. (2017) |
| Solidity.side | S1_19664643 | Sb01g018890 | Dicer-like3 | 144.3 | dicer-like3 (DCL3; Os) | Regulate GA and BR homeostasis; mutation causes reduced secondary branches | (Wei et al., 2014) | |
| Panicle length | S2_63553713 | Sb02g028420 | SBP box gene family member | 43.0 | tsh4 (Zm) | Lateral meristem initiation and stem elongation | Tassel length | (Chuck et al., 2010); Brown et al. (2011); Xu et al. (2017) |
| Solidity.front | S2_76284762 | Sb02g042400 | AP2 domain-containing protein | 146.1 | BD1 (Zm) | Specification of spikelet meristem identity and inhibit the formation of axillary meristems | Secondary branching/branch length | (Chuck et al., 2002); Brown et al. (2011); Crowell et al. (2016); Wu et al. (2016); Xu et al. (2017) |
| Width.side | S3_2724080 | Sb03g002525 | MADS box family gene with MIKCc-type box | 307.8 | MADS3 (Os), zmm2 (Zm) | Specification of spikelet meristem identity | (Yamaguchi et al., 2006) | |
| Area.side | S3_58994806 | Sb03g030635 | Homeobox3 | 142.8 | DWT1 (Os) | Stem cell elongation and cell proliferation | Wang et al. (2014) | |
| Panicle length | S3_58920501 | Sb03g030635 | Homeobox3 | 68.5 | DWT1 (Os) | Stem cell elongation and cell proliferation | Wang et al. (2014) | |
| Area.front | S4_12278584 | Sb04g009700 | Gal oxidase/Kelch repeat superfamily protein | 3.3 | Larger panicle (LP; Os) | Modulate cytokinin level in inflorescence and branch meristems | Panicle size/panicle length | (Li et al., 2011; Crowell et al., 2016; Wu et al., 2016 |
| Solidity.front | S6_60219786 | Sb06g031880 | Homeobox domain-containing protein | 40.2 | WUS (Os) | Inhibit differentiation of the stem cells | (Nardmann and Werr, 2006) | |
| Width.side | S10_60236042 | Sb10g030270 | Receptor protein kinase CLAVATA1 precursor | 273.1 | Td1 (Zm) | Regulate shoot and floral meristem size | Cob diameter/kernel row number | Bommert et al. (2005); Brown et al. (2011) |
Population Differentiation
Because inflorescence traits can influence per plant yield, these traits have likely been targets of selection. According to Casa et al. (2008), the SAP-FI (n = 272) used in this study contains 55 elite inbred lines and landraces that were developed in the United States, 40 cultivars collected worldwide, and 177 converted tropical lines. The latter group was generated by the Sorghum Conversion Program (SCP), which aimed to introduce novel genetic variation from exotic, tropical germplasm collected from around the world into modern U.S. breeding lines (Stephens et al., 1967). During this process, tropical lines were backcrossed to a single adapted line (BTx406) and selection was performed only for flowering time and plant height. Specifically, there was no direct selection for panicle architecture; thus, much of the natural genetic variation present in the tropical lines associated with panicle traits presumably would have been retained in the converted tropical lines (Casa et al., 2008; Thurber et al., 2013). As a group, the 177 converted tropical lines have shorter and narrower panicles as well as smaller cross-sectional areas compared with the 40 cultivars (Supplemental Fig. S6). The origins of the 40 cultivars and exotic donors of the 177 converted tropical lines have similar geographic distributions (Casa et al., 2008). Hence, by identifying chromosomal regions that exhibit statistically significant differences in allele frequencies between the two groups (see “Materials and Methods”), we can identify regions that have putatively been under selection. We could then ask whether such chromosomal regions are enriched for our TASs.
A prior comparison of the parental and converted tropical lines generated in the SCP identified chromosomal regions on the long arms of chromosomes 6, 7, and 9 that exhibit signatures of selection. Consistent with the nature of the SCP, these regions contain clusters of genes (Ma1, Ma6, Dw1, Dw2, and Dw3) that regulate plant height and flowering time (Thurber et al., 2013). Our comparison between the 177 converted tropical lines and 40 cultivars also identified the regions (Fig. 5) associated with flowering time and plant height described by Thurber et al. (2013). At first, this result was surprising, but subsequent analyses on previously reported plant height loci (Brown et al., 2008; Li et al., 2015) revealed that the 40 cultivars have lower frequencies of the favorable alleles of three height genes located on chromosomes 6, 7, and 9 that were contributed by BTx406 than the converted tropical lines.
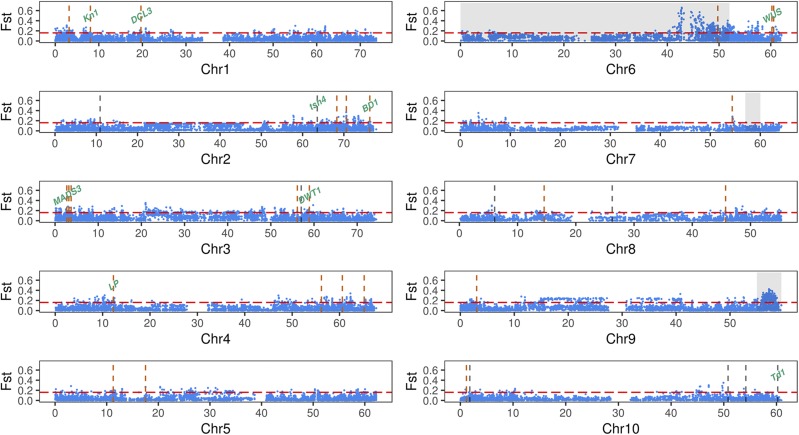
Figure 5.
Identification of chromosomal regions that exhibit evidence of selection. Allele frequencies were compared between 177 converted tropical lines and 40 cultivars. A 99.5% fixation index cutoff is indicated by horizontal dashed lines. Vertical dashed lines designate the positions of 35 TASs. TASs located within divergent regions are indicated in red. Two TASs on chromosome 3 at 58,920,501 and 58,994,806 bp are represented by a single red dashed line. Candidate genes are labeled. Gray shading on chromosomes 6, 7, and 9 indicates genomic regions identified by Thurber et al. (2013) carrying maturity and plant height genes introduced in the SCP.
In addition to the adaptation genes located on chromosomes 6, 7, and 9, we also identified genomic regions that exhibit signatures of selection that were not identified by Thurber et al. (2013); Fig. 5). These regions probably reflect selection during modern breeding for agronomic traits, potentially including panicle architecture. Consistent with the hypothesis that selection has occurred for panicle traits, 26 of our 35 TASs colocalized within chromosomal regions that exhibit evidence of selection. Eight of these 26 TASs are linked to candidate genes (Fig. 5). Based on permutation tests, more overlap was observed than would be expected if the TASs had not been under selection (P < 0.001).
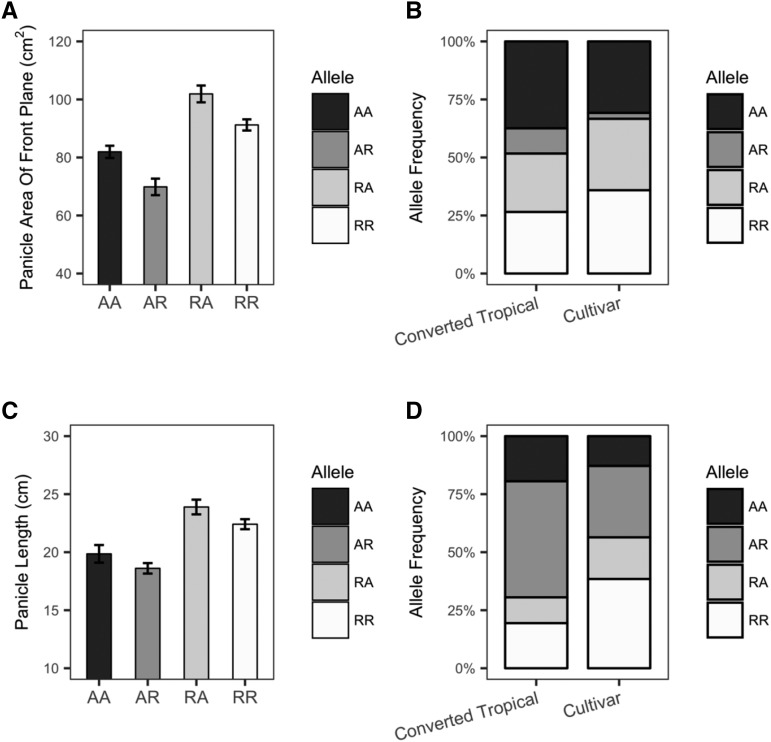
To further test this hypothesis, we determined trait values for area.front for each of the four potential genotypes associated with the two TASs for this trait, both of which exhibited signatures of selection. For the first TAS (S1_3176715), the reference (R) allele is associated with larger area.front. In contrast, the alternative (A) allele of the second TAS (S4_12278584) is associated with larger area.front. Considering all members of the SAP-FI, the RA and AR genotypes have the largest and smallest area.front values, respectively (Fig. 6). Consistent with our hypothesis that differences in allele frequencies of these two TASs in the two groups are due to selection for panicle architecture, the favorable R allele of SNP S1_3176715 exhibits a higher (29%) frequency in the cultivars (26 of 40) than in the converted tropical lines (89 of 177). The frequency of the favorable A allele of SNP S4_12278584 is not higher in the cultivars than in the converted tropical lines; only one of the 40 cultivars carries the least favorable genotype of both SNPs (AR). Although not statistically significant (Fisher’s exact test, P = 0.069), this frequency (one of 40) is 5 times lower than that observed in the converted tropical lines (20 of 177; Fig. 6). Similarly, the frequency of the least favorable genotypic class determined by the two SNPs associated with panicle length was reduced in cultivars as compared with the converted tropical lines (Fig. 6). These results at least suggest the possibility that chromosomal regions surrounding some of the TASs identified in this study may have been targets of selection during modern breeding.
Figure 6.
Evidence of selection against alleles for small panicle areas and short panicles. The allele from the reference genome of BTx623 is designated as the reference (R) allele, and other alleles are designated as alternative (A) alleles. A and B, Panicle area. C and D, Panicle length. A and C, Phenotypes associated with all four genotypes of TASs S1_3176715 and S4_12278584 (A) and TASs S1_8065027 and S3_58920501 (C) in the complete panel. B and D, Genotype frequencies of TASs in the converted tropical lines and cultivars.
DISCUSSION
Given the substantial reductions in the cost of genotyping large panels, phenotyping has become the bottleneck for large-scale genetic analyses of crops. Two groups have reported pipelines (P-TRAP and PANorama) to capture phenotypes of rice panicles using photographs of flattened panicles (AL-Tam et al., 2013; Crowell et al., 2014). In both cases, only a single image was analyzed per flattened panicle. Hence, these pipelines are not suitable for extracting multidimensional traits associated with the 3D structures of panicles. Others have used complex imaging chambers to measure 3D structures of limited numbers of Arabidopsis inflorescences (Hall and Ellis, 2012), but it would be difficult to scale this approach to phenotype large diversity panels.
Here, we present a low-cost, easy-to-replicate pipeline that was used to phenotype panicles in a sorghum diversity panel. To explore variation in panicle architecture among the members of this diversity panel, we captured images from the two planes of a panicle. This enabled us to extract not only unidimensional traits, such as panicle length and width, but also multidimensional traits, such as area.front, area.side, solidity.front, solidity.side, and volume. Extracted trait values for panicle length and width exhibited high accuracies (0.93 and 0.89, respectively) as compared with ground-truth measurements of intact panicles. Because it is not possible to extract ground-truth data for multidimensional traits, we could not determine the accuracy of our pipeline for these traits. Even so, since the heritabilities were high (0.7–0.93), we conducted a GWAS for these traits and identified candidate genes associated with some of the TASs.
Given the extensive LD present in sorghum, it is not possible to conclude that any particular candidate gene affects the associated trait. However, the enrichment of sorghum homologs for maize and rice genes known to affect inflorescence architecture near these TASs, and the correspondence of their functions in maize and rice with the associated phenotypes in sorghum (Table 2), support the accuracy of our phenotyping pipeline and the hypothesis that at least some of the candidate genes are causative.
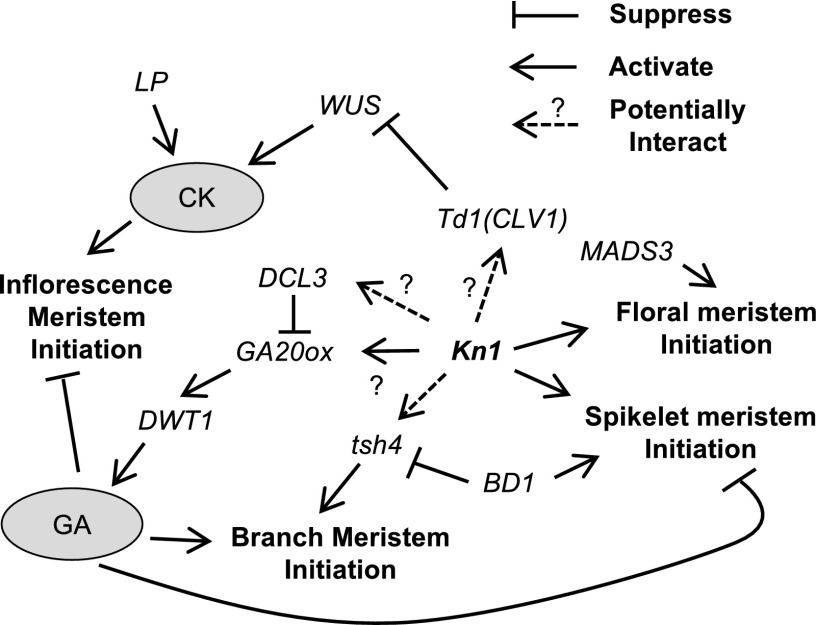
Functional studies (Tanaka et al., 2013; Zhang and Yuan, 2014) have demonstrated that many genes that regulate inflorescence development are functionally conserved among grass species. More specifically, GWAS suggest the conservation of genetic architecture between rice panicles and maize tassels (Brown et al., 2011; Crowell et al., 2016; Xu et al., 2017). Hence, we prioritized our selection of candidates by identifying sorghum homologs of inflorescence genes discovered previously in maize and rice. In this manner, we identified nine candidate genes, most of which are related to Kn1 (Fig. 7).
Figure 7.
Potential relationships among nine candidate genes (plus ga20ox) and their functions in inflorescence development. Kn1 is a key regulator of floral transition (Vollbrecht et al., 2000; Zhang and Yuan, 2014) that is essential for meristem maintenance and is involved in the initiation of spikelet and floral meristems in rice and maize (Kerstetter et al., 1997; Tsuda et al., 2011). Analyses of maize mutants demonstrate that Kn1 regulates the cytokinin (CK) pathway, perhaps via interactions with Td1, a suppressor of the WUS gene (Bommert et al., 2005; Lunde and Hake, 2009). Kn1 also participates in regulating GA levels in the SAM through ga20ox and various KNOTTED1-like homeobox proteins (Bolduc and Hake, 2009). In sorghum, the expression of Kn1 is strongly correlated with DCL-3, which is involved in the production of 24-nucleotide siRNAs that target genes (e.g. ga20ox) involved in homeostasis of the GA and brassinosteroid pathways to alter branch meristems in rice (Wei et al., 2014). In addition, GWAS on panicle area identified one WUS-related homeobox transcription factor, DWT1, which in rice is associated with GA signaling, participates in panicle meristem elongation, and is potentially downstream of ga20ox (Wang et al., 2014). In maize, tsh4 regulates the establishment of lateral meristem boundaries followed by meristem initiation and is regulated via the expression of Kn1 and Bd1 (Chuck et al., 2010). In rice and maize, Kn1 participates in spikelet meristem initiation and floral meristem initiation, which are positively regulated by Bd1 and MADS3 (Kerstetter et al., 1997; Yamaguchi et al., 2006; Bolduc and Hake, 2009).
In maize, both Kn1- and Ra1-related genes affect tassel development, and many auxin-related genes are targeted by both the RA1 and KN1 proteins (Vollbrecht et al., 2005; McSteen, 2006; Eveland et al., 2014). Although our GWAS associated many Kn1-related sorghum genes with panicle architecture, no TASs were found in LD with members of the ramosa pathway. This was surprising because, in maize, this pathway contributes to the formation of spikelet pair meristems and has been associated with tassel architecture via GWAS (Brown et al., 2011; Wu et al., 2016; Xu et al., 2017). Given the level of functional conservation between these two species, it is unlikely that the ramosa pathway does not contribute to the development of sorghum panicles. Instead, the failure of our GWAS to detect associations with genes in the ramosa pathway suggests that differentially functional alleles of these genes are not segregating at high frequencies in the SAP-FI. This finding highlights the value of combining high-throughput phenotyping with GWAS to identify standing variation that can be used to select targets for marker-assisted breeding.
On average, cultivars have larger panicles than converted tropical lines, and our results suggest that, by employing phenotypic selection, breeders have selected for favorable alleles of genes that regulate panicle architecture. Even so, our results also indicate that cultivars have not been purged of unfavorable alleles of these genes. This is likely because most TASs identified in this study exhibit only moderate effect sizes and, consequently and consistent with expectation (Bernardo, 2008, 2016; Heffner et al., 2009), traditional phenotypic selection has not yet fixed favorable alleles at all relevant loci. This presents an opportunity to employ the TASs identified in our GWAS to design a marker-assisted selection program to improve panicle architecture traits with impact on final grain yield.
The general imaging and data analysis approach reported here could be used to phenotype other grass species with panicle structures similar to those of sorghum. Often, academic software is not supported on the latest versions of operating systems. Our code is based on built-in MATLAB functions, which does not put the burden of long-term maintenance on the developers or users. Hence, anyone in possession of our scripts can run them on MATLAB, which is commercially maintained and routinely updated to cater to all operating systems. In addition, our semiautomated trait extraction pipeline requires no advanced hardware; any commercially available laptop is capable of completing trait extractions from 1,000 images within 24 h. As such, it should be possible to deploy this method even at remote breeding stations in developing countries.
MATERIALS AND METHODS
Imaging Panicles
A total of 302 sorghum (Sorghum bicolor) genotypes from the SAP (Casa et al., 2008) were grown at two Iowa State University experimental farms (Kelley Farm in Ames, Iowa, and Burkey Farm in Boone, Iowa) in 2015. This panel was planted in a randomized complete block design with two replications at each location, as described previously by Salas Fernandez et al. (2017). One panicle per replicate per location was harvested during the third week of October and imaged using a light box. A few genotypes had some panicles with drooping primary branches. To avoid artifacts due to these drooping panicles, we selected for phenotyping within such genotypes only those specific panicles that did not droop. The light box was constructed with a Pacific Blue background to ensure easy segmentation of panicles in subsequent image-processing steps. This box was designed to accommodate the diversity of panicles in the SAP (Fig. 1), with dimensions of about 45 cm tall, 36 cm wide, and 24 cm deep. All images were captured using a single Canon 5DS DSLR camera with EF100 mm f/2.8L Macro IS USM lens. The camera was mounted at a fixed height and angle to ensure consistent imaging.
Each panicle was mounted upright in the center of the light box with a reference scale of known dimensions placed next to it. The reference scale allows the automated conversion of size-dependent traits from the number of pixels occupied in the image to real lengths (i.e. inches or centimeters). The reference scale was initially a 6.5-cm-wide white scale but was later changed to a 2.54-cm-wide (1 inch) yellow scale, for easier extraction from the blue background. Because the images were 2D projections of the panicles, the panicles were imaged along two perpendicular planes, based on a front view and a side view. The plane that exhibited the largest cross-sectional area, as determined via visual inspection, was designated the front plane and the plane perpendicular to it as the side plane. The front plane of a panicle was always imaged first.
Extraction of Traits from Images
We developed a pipeline, TIM (available at the Schnable Lab’s GitHub page: https://github.com/schnablelab), to semiautomatically extract traits from panicle images. The trait extraction via TIM involved the following steps: (1) segmentation of the panicle and the reference scale from the background using a custom-built MATLAB app (Supplemental Methods); (2) measurement of traits on the segmented panicle; and (3) conversion of trait values from pixels to metric measurements using a reference scale (Fig. 1; Supplemental Methods). Trait measurement was performed in a fully automated manner using standard image-processing algorithms (described in Supplemental Methods). Image processing is particularly suited to traits that can be cumbersome to measure by hand, such as the exact cross-sectional area of the panicle, solidity, etc. However, some traits, such as length and width, can be challenging to extract via automated image processing from strongly curved panicles whose tips touched the bottom of the light box, making it difficult to distinguish the first internode of a panicle’s rachis. Therefore, we removed 30 genotypes that consistently exhibited U-shaped panicles across locations and repetitions; thus, 272 genotypes and 1,064 panicles were used subsequently for image processing and data analyses. Due to missing data, the total number of imaged panicles was 1,064, rather than 1,188 (= 272 × 2 repetitions/location × 2 locations). In this report, this subset of the SAP (n = 272 genotypes) will be referred to as SAP-FI.
Trait Extraction
Eight panicle traits were extracted from front and side plane images of the 1,064 panicles as described below. These are length, width, area, volume, and solidity. Of these, width, area, and solidity were extracted from both the front and side planes. Length is defined as the length of the main panicle axis from the region of the lowest branch point to the tip of the panicle. Because length is expected to be the same in the two planes, it was extracted from only the front plane. Volume is a derived quantity that is estimated jointly from the front and side plane images. The front plane and side plane images were first matched to each other height-wise. Subsequently, the widths from the front plane and side plane were extracted along the length of the panicle in slices from the top to the bottom (Supplemental Methods). Assuming that the maximum and minimum widths represent the major axis and the minor axis of an ellipse, it is possible to calculate the elliptical cross-sectional area (an ellipse being an approximation of the shape of the panicle). Integrating the cross-sectional area over the height of the panicle provides an approximate volume of the panicle.
The remaining traits, width, area, and solidity, were extracted from both the front and side plane images (width.front, width.side, area.front, area.side, solidity.front, and solidity.side). Width represents the maximum value of the width of the panicle obtained from the longest line that can be drawn between the two boundaries of the panicle and that is perpendicular to the panicle’s length (Fig. 1). Area refers to the projected area of the panicle on a 2D plane, because the panicle is a 3D structure. Separate area measurements were obtained from the number of pixels contained within the projections of the panicle on the front and side planes. Solidity is a measure of whether the panicle is loosely or tightly packed. It is derived from the convex hull, which is the smallest polygonal bounding curve that encompasses the shape of the panicle. Solidity is the ratio of the projected area of the panicle to the area of its convex hull.
With the exception of solidity, which is a dimensionless quantity, all trait values were measured in units of pixels. The pixel-to-cm conversion was obtained from the pixel width of the 1-inch yellow scale in each image (pixel/cm = pixel width of yellow scale/2.54) to obtain the actual lengths of these panicles. Phenotype values of every trait were then transformed into standard units (cm for length and width, cm2 for area, and cm3 for volume) based on the pixel/cm ratio calculated for each image.
Analysis of Phenotypic Data
For each phenotype, variance components were estimated using the lme4 package (Bates et al., 2015) in R (version 3.4.2) using the following function: lmer(phenotype ∼ (1|genotype) +(1|location) + (1|Location/Rep) + (1| genotype: location)). Entry mean-based heritability values (H2) were calculated from variance components estimates (Bernardo, 2010) as follows: H2 = VG/[VG + (VGE/n) + (Ve/(nr)], where VG is the genotypic variance, VGE is the variance of genotype by location interaction, Ve is the residual variance, n is the number of locations, and r is the number of replications. Phenotypic correlations among eight traits were estimated using Pearson correlations between two traits using the mean value of each genotype via the cor function of R.
Genotyping Data
We used SNP data generated and imputed by Morris et al. (2013). These SNPs were filtered to retain only those with a minor allele frequency > 0.05 and a missing rate of less than 60% in the SAP-FI. The remaining 146,865 SNPs were used to calculate genetic correlations among the eight panicle traits using bivariate genomic relatedness-based restricted maximum likelihood analysis from GCTA (Yang et al., 2010, 2011).
GWAS
GWAS was conducted using the 146,865 SNPs in R using a version of FarmCPU (Liu et al., 2016) termed FarmCPUpp, which was modified to increase computational efficiency and reduce run times (Kusmec et al., 2017). FarmCPU combines SNP-based covariate selection (MLMM; Segura et al., 2012) and restricted kinship matrices (SUPER; Wang et al., 2014) to reduce false positives and negatives. Simulations demonstrate that FarmCPU achieves both of these goals in addition to increasing computational efficiency (Liu et al., 2016). Principal component analysis was conducted on the SNP data using the R prcomp function. The first three principal components were used as covariates to control for population structure. FarmCPUpp’s optimum bin selection procedure was conducted using bin sizes of 50, 100, and 500 kb and pseudoquantitative trait nucleotide values of 3, 6, 9, 12, and 15. Statistical significance was determined after Bonferroni correction: α = (0.05/total number of SNPs).
Initially, GWAS was conducted using all 272 accessions of the SAP-FI (Supplemental Table S4). To identify high-confidence SNPs, bootstraps were conducted for each phenotype following previously described methods (Brown et al., 2011; Wallace et al., 2014). The full panel was first divided into four subpanels determined via fastSTRUCTURE (Raj et al., 2014). Then, we performed 100 bootstraps with each iteration randomly assigned 10% of the phenotypic values within each subpanel as missing. Then, for each iteration, GWAS was repeated using the same parameters as for the full panel. The RMIP was calculated based on the fraction of bootstraps in which an SNP was associated significantly with the phenotype. Only SNPs with RMIP ≥ 5 (five out of 100 iterations) were considered for further analysis. We also made this process more stringent by requiring these hits to be significant in the GWAS conducted on the full panel. These SNPs were defined as high-confidence TASs and used to screen for candidate genes.
Colocalization of TASs and Candidate Genes
Genome-wide LD was assessed using PLINK (version 1.9; www.cog-genomics.org/plink/1.9/; Chang et al., 2015) by calculating the pairwise r2 of every SNP within 1 Mb using 25-kb steps. The average length of the interval within which r2 fell below 0.1 was 350 kb. Hence, we scanned 350 kb upstream and downstream of each TAS for candidate genes.
Potential candidate genes were first identified by screening the literature for cloned maize (Zea mays) and rice (Oryza sativa) genes with known functions in inflorescence architecture (Supplemental Table S3). The corresponding sorghum homologs, based on sorghum genome V1.4 from PlantGDB (http://www.plantgdb.org/SbGDB/), of the resulting 67 maize and 17 rice genes were identified using MaizeGDB (Schnable et al., 2012) and the rice genome annotation project (Kawahara et al., 2013), respectively. To test whether the distribution of candidate genes was enriched in regions surrounding the TASs, we performed 1,000 permutations during which 35 SNPs (equal to the number of TASs detected via GWAS) were selected randomly from the 146,865 SNPs for each permutation. We then recorded the number of permutations in which nine or more candidate genes were present within 700-kb windows surrounding the randomly sampled SNPs. This process was repeated for 10 iterations, and the median P value from these iterations was used to evaluate the significance.
Population Differentiation
A total of 177 converted tropical lines and 40 cultivars from the SAP-FI were used to analyze genome-wide population differentiation. Genotypic data of the two subpanels were first filtered to have a missing rate of less than 5% within each subpanel. The Hudson estimator of fixation index (Hudson et al., 1992) was calculated using the 74,142 filtered SNPs shared by both subpanels to estimate the genome-wide divergence between the two, following previously described methods (Bhatia et al., 2013).
Accession Numbers
Sequence data used in this article can be found in the Sequence Read Archive database (www.ncbi.nlm.nih.gov/sra) under accession number SRA062716.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Example showing the structural diversity of SAP-FI.
Supplemental Figure S2. Phenotypic correlation between width.front and area.side, with examples showing genotype panicle shapes that are circled in the scatterplot.
Supplemental Figure S3. Manhattan and QQ plots for eight panicle traits.
Supplemental Figure S4. Standardized effect sizes of 38 TASs.
Supplemental Figure S5. Standardized effect sizes of SNPs associated with multiple traits.
Supplemental Figure S6. Box plot of values of eight panicle traits in 177 converted tropical lines and 40 cultivars.
Supplemental Table S1. TASs that overlapped with previously reported QTL and GWAS intervals on panicle length, panicle width, and branch length.
Supplemental Table S2. List of TASs with pleotropic effect in 100 bootstraps.
Supplemental Table S3. List of sorghum homologs of maize or rice genes with known functions on inflorescence architecture.
Supplemental Table S4. List of mean phenotypic values of eight panicle traits of 272 genotypes used in GWAS.
Supplemental Methods. The instructions to ToolKit for Inflorescence Measurements (TIM).
Acknowledgments
We thank Lisa Coffey (Schnable laboratory) and Nicole Lindsey (Salas Fernandez laboratory) for assistance with designing and conducting the sorghum field experiments; graduate student Zihao Zheng (Schnable laboratory) for assistance harvesting panicles; and undergraduate students Stephanie Shuler, Jodie Johnson, and Katherine Lenson (Schnable laboratory) for assistance collecting image data.
Footnotes
Senior author.
This work was supported in part by the National Institute of Food and Agriculture, U.S. Department of Agriculture (award no. 2012-67009-19713 to P.S.S. and M.G.S.F. and colleagues) and additional funding from Iowa State University’s Plant Sciences Institute to P.S.S., M.G.S.F., and B.G.
Articles can be viewed without a subscription.
References
- AL-Tam F, Adam H, dos Anjos A, Lorieux M, Larmande P, Ghesquière A, Jouannic S, Shahbazkia HR (2013) P-TRAP: A panicle trait phenotyping tool. BMC Plant Biol 13: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amine EK, Baba NH, Belhadj M, Deurenberg-Yap M, Djazayery A, Forrestre T, Galuska DA, Herman S, James WPT, M’Buyamba Kabangu JR (2003), World Health Organization Technical Report Series Diet, nutrition and the prevention of chronic diseases. World Health Organization, Geneva [Google Scholar]
- Aquino A, Millan B, Gaston D, Diago MP, Tardaguila J (2015) vitisFlower®: Development and testing of a novel Android-smartphone application for assessing the number of grapevine flowers per inflorescence using artificial vision techniques. Sensors (Basel) 15: 21204–21218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67: [Google Scholar]
- Bernardo R. (2008) Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci 48: 1649–1664 [Google Scholar]
- Bernardo R. (2010) Breeding for Quantitative Traits in Plants, Ed 2 Stemma Press, Woodbury, MN [Google Scholar]
- Bernardo R. (2016) Bandwagons I, too, have known. Theor Appl Genet 129: 2323–2332 [DOI] [PubMed] [Google Scholar]
- Bhatia G, Patterson N, Sankararaman S, Price AL (2013) Estimating and interpreting FST: The impact of rare variants. Genome Res 23: 1514–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Hake S (2009) The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W (2005) thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bouchet S, Pot D, Deu M, Rami JF, Billot C, Perrier X, Rivallan R, Gardes L, Xia L, Wenzl P, et al. (2012) Genetic structure, linkage disequilibrium and signature of selection in sorghum: Lessons from physically anchored DArT markers. PLoS ONE 7: e33470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Klein PE, Bortiri E, Acharya CB, Rooney WL, Kresovich S (2006) Inheritance of inflorescence architecture in sorghum. Theor Appl Genet 113: 931–942 [DOI] [PubMed] [Google Scholar]
- Brown PJ, Rooney WL, Franks C, Kresovich S (2008) Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics 180: 629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, Holland JB, Flint-Garcia S, McMullen MD, Buckler ES, et al. (2011) Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet 7: e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma J. (2003) World Agriculture: Towards 2015/2030. Earthscan Publications Ltd, London [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48: 30–40 [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia JM, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, Elshire RJ, Gaut B, Geller L, Glaubitz JC, et al. (2012) Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet 44: 803–807 [DOI] [PubMed] [Google Scholar]
- Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, Pingault L, Sourdille P, Couloux A, Paux E, et al. (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science 345: 1249721. [DOI] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Chuck G, Whipple C, Jackson D, Hake S (2010) The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Crowell S, Falcão AX, Shah A, Wilson Z, Greenberg AJ, McCouch SR (2014) High-resolution inflorescence phenotyping using a novel image-analysis pipeline, PANorama. Plant Physiol 165: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell S, Korniliev P, Falcão A, Ismail A, Gregorio G, Mezey J, McCouch S (2016) Genome-wide association and high-resolution phenotyping link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nat Commun 7: 10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Ananiev EV (2010) Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol 153: 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Goldshmidt A, Pautler M, Morohashi K, Liseron-Monfils C, Lewis MW, Kumari S, Hiraga S, Yang F, Unger-Wallace E, et al. (2014) Regulatory modules controlling maize inflorescence architecture. Genome Res 24: 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gage JL, Miller ND, Spalding EP, Kaeppler SM, de Leon N (2017) TIPS: A system for automated image-based phenotyping of maize tassels. Plant Methods 13: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JL, White MR, Edwards JW, Kaeppler S, de Leon N (2018) Selection Signatures Underlying Dramatic Male Inflorescence Transformation During Modern Hybrid Maize Breeding. Genetics 210: 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall H, Ellis B (2012) Developmentally equivalent tissue sampling based on growth kinematic profiling of Arabidopsis inflorescence stems. New Phytol 194: 287–296 [DOI] [PubMed] [Google Scholar]
- Hamblin MT, Salas Fernandez MG, Casa AM, Mitchell SE, Paterson AH, Kresovich S (2005) Equilibrium processes cannot explain high levels of short- and medium-range linkage disequilibrium in the domesticated grass Sorghum bicolor. Genetics 171: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariprasanna K, Rakshit S (2016) Economic importance of sorghum. In Rakshit S, Wang YH, eds, The Sorghum Genome. Springer, Cham, Switzerland, pp 1–25 [Google Scholar]
- Hart GE, Schertz KF, Peng Y, Syed NH (2001) Genetic mapping of Sorghum bicolor (L.) Moench QTLs that control variation in tillering and other morphological characters. Theor Appl Genet 103: 1232–1242 [Google Scholar]
- Heffner EL, Sorrells M, Jannink JL (2009) Genomic selection for crop improvement. Crop Sci 49: 1–12 [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S (1997) Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kusmec A, Schnable PS (2018) FarmCPUpp: Efficient large-scale genomewide association studies. Plant Direct 2: e00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmec A, Srinivasan S, Nettleton D, Schnable PS (2017) Distinct genetic architectures for phenotype means and plasticities in Zea mays. Nat Plants 3: 715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Li M, Tang D, Wang K, Wu X, Lu L, Yu H, Gu M, Yan C, Cheng Z (2011) Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol J 9: 1002–1013 [DOI] [PubMed] [Google Scholar]
- Li X, Zhu C, Yeh CT, Wu W, Takacs EM, Petsch KA, Tian F, Bai G, Buckler ES, Muehlbauer GJ, et al. (2012) Genic and nongenic contributions to natural variation of quantitative traits in maize. Genome Res 22: 2436–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li X, Fridman E, Tesso TT, Yu J (2015) Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proc Natl Acad Sci USA 112: 11823–11828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12: e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde C, Hake S (2009) The interaction of knotted1 and thick tassel dwarf1 in vegetative and reproductive meristems of maize. Genetics 181: 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: Challenges and prospects. Nat Rev Genet 10: 565–577 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2006) Branching out: The ramosa pathway and the evolution of grass inflorescence morphology. Plant Cell 18: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan B, Aquino A, Diago MP, Tardaguila J (2017) Image analysis-based modelling for flower number estimation in grapevine. J Sci Food Agric 97: 784–792 [DOI] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, et al. (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 110: 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja Reddy R, Madhusudhana R, Murali Mohan S, Chakravarthi DVN, Mehtre SP, Seetharama N, Patil JV (2013) Mapping QTL for grain yield and other agronomic traits in post-rainy sorghum [Sorghum bicolor (L.) Moench]. Theor Appl Genet 126: 1921–1939 [DOI] [PubMed] [Google Scholar]
- Nardmann J, Werr W (2006) The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol Biol Evol 23: 2492–2504 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pautler M, Tanaka W, Hirano HY, Jackson D (2013) Grass meristems I: Shoot apical meristem maintenance, axillary meristem determinacy and the floral transition. Plant Cell Physiol 54: 302–312 [DOI] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK (2014) fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 197: 573–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas Fernandez MG, Bao Y, Tang L, Schnable PS (2017) A high-throughput, field-based phenotyping technology for tall biomass crops. Plant Physiol 174: 2008–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC. (2015) Genome evolution in maize: From genomes back to genes. Annu Rev Plant Biol 66: 329–343 [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M, Lyons E (2012) Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol Evol 4: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M (2012) An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet 44: 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas G, Satish K, Madhusudhana R, Reddy RN, Mohan SM, Seetharama N (2009) Identification of quantitative trait loci for agronomically important traits and their association with genic-microsatellite markers in sorghum. Theor Appl Genet 118: 1439–1454 [DOI] [PubMed] [Google Scholar]
- Stephens JC, Miller FR, Rosenow DT (1967) Conversion of alien sorghums to early combine genotypes. Crop Sci 7: 396 [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Pautler M, Jackson D, Hirano HY (2013) Grass meristems II: Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol 54: 313–324 [DOI] [PubMed] [Google Scholar]
- Thurber CS, Ma JM, Higgins RH, Brown PJ (2013) Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol 14: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Ito Y, Sato Y, Kurata N (2011) Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S (2000) Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES IV, Martienssen R (2005) Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126 [DOI] [PubMed] [Google Scholar]
- Wallace JG, Bradbury PJ, Zhang N, Gibon Y, Stitt M, Buckler ES (2014) Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet 10: e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Tian F, Pan Y, Buckler ES, Zhang Z (2014) A SUPER powerful method for genome wide association study. PLoS ONE 9: e107684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Gu L, Song X, Cui X, Lu Z, Zhou M, Wang L, Hu F, Zhai J, Meyers BC, Cao X (2014) Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc Natl Acad Sci USA 111: 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Shi Y, Song Y, Zhang D, Li C, Buckler ES, Li Y, Zhang Z, Wang T (2016) Joint-linkage mapping and GWAS reveal extensive genetic loci that regulate male inflorescence size in maize. Plant Biotechnol J 14: 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Wang X, Huang C, Xu D, Li D, Tian J, Chen Q, Wang C, Liang Y, Wu Y, et al. (2017) Complex genetic architecture underlies maize tassel domestication. New Phytol 214: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY (2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef HM, Eggert K, Koppolu R, Alqudah AM, Poursarebani N, Fazeli A, Sakuma S, Tagiri A, Rutten T, Govind G, et al. (2017) VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat Genet 49: 157–161 [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zhang D, Yuan Z (2014) Molecular control of grass inflorescence development. Annu Rev Plant Biol 65: 553–578 [DOI] [PubMed] [Google Scholar]
- Zhang D, Kong W, Robertson J, Goff VH, Epps E, Kerr A, Mills G, Cromwell J, Lugin Y, Phillips C, et al. (2015) Genetic analysis of inflorescence and plant height components in sorghum (Panicoidae) and comparative genetics with rice (Oryzoidae). BMC Plant Biol 15: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Mantilla Perez MB, Hu J, Salas Fernandez MG (2016) Genome-wide association study for nine plant architecture traits in sorghum. Plant Genome 9: 1–14 [DOI] [PubMed] [Google Scholar]
- Zhao S, Gu J, Zhao Y, Hassan M, Li Y, Ding W (2015) A method for estimating spikelet number per panicle: Integrating image analysis and a 5-point calibration model. Sci Rep 5: 16241. [DOI] [PMC free article] [PubMed] [Google Scholar]