Genetic and molecular analysis of the maize REL2 transcriptional corepressor defines a function for the WD40 domain of TOPLESS proteins and reveals a major role for REL2 in meristem development.
Abstract
Transcriptional repression in multicellular organisms orchestrates dynamic and precise gene expression changes that enable complex developmental patterns. Here, we present phenotypic and molecular characterization of the maize (Zea mays) transcriptional corepressor RAMOSA1 ENHANCER LOCUS2 (REL2), a unique member of the highly conserved TOPLESS (TPL) family. Analysis of single recessive mutations in rel2 revealed an array of vegetative and reproductive phenotypes, many related to defects in meristem initiation and maintenance. To better understand how REL2-mediated transcriptional complexes relate to rel2 phenotypes, we performed protein interaction assays and transcriptional profiling of mutant inflorescences, leading to the identification of different maize transcription factors and regulatory pathways that employ REL2 repression to control traits directly impacting maize yields. In addition, we used our REL2 interaction data to catalog conserved repression motifs present on REL2 interactors and showed that two of these, RLFGV- and DLN-type motifs, interact with the C-terminal WD40 domain of REL2 rather than the N terminus, which is known to bind LxLxL EAR motifs. These findings establish that the WD40 domain of TPL family proteins is an independent protein interaction surface that may work together with the N-terminal domain to allow the formation of large macromolecular complexes of functionally related transcription factors.
Transcriptional repression is an essential and widespread mechanism regulating many eukaryotic developmental pathways (Krogan and Long, 2009; Payankaulam et al., 2010). Many of the eukaryotic corepressors identified to date, including Tup1 from yeast, Groucho/TLE from Drosophila and humans, and the mammalian TBL/SMRT/NCOR complex, contain conserved structural features despite showing limited sequence homology (Buscarlet and Stifani, 2007; Martin-Arevalillo et al., 2017). In plants, several different types of corepressors have also been reported, the best studied of which are those belonging to the highly conserved TOPLESS (TPL) family (Long et al., 2002, 2006; Liu and Karmarkar, 2008; Lee and Golz, 2012). Like their fungal and animal counterparts, TPL proteins do not bind DNA directly but instead are recruited by diverse DNA-binding transcription factors or other transcriptional regulators (hereafter collectively referred to as transcription factors, TFs; Causier et al., 2012a). Processes that use TPL corepressors to repress transcriptional outputs include TFs from nearly all of the major plant hormone-signaling pathways, as well as stem cell-related and organ-specific developmental pathway proteins. Previous data has shown that these TFs generally contain at least one of four small repression motifs that are responsible for mediating the interaction with TPL proteins. The most common of the repression motifs is the EAR (ERF-associated amphiphilic repression) motif, comprising the consensus amino acid sequence LxLxL, which is predicted to occur on ∼25% of all TFs (Ohta et al., 2001; Szemenyei et al., 2008; Causier et al., 2012a). Additional repression motifs include a DLNxxP-type motif, an RLFGV-type motif, and the WUS-box, containing the sequence TLxLFP (Kieffer et al., 2006; Ikeda and Ohme-Takagi, 2009; Causier et al., 2012a). Appending either the LxLxL or RLFGV motif, or TPL itself, to certain TFs is sufficient to confer transcriptional repression to specific pathways and has been used as a tool for engineering transcriptional outcomes (Ikeda and Ohme-Takagi, 2009; Mahfouz et al., 2012; Khakhar et al., 2018).
For many TFs, these repression motifs are highly conserved across species, such as in the case of Aux/IAA repressors, which perform an essential function in the auxin signal transduction pathway (Szemenyei et al., 2008). In other cases, the presence of one or more motifs appears to be species specific, found for example among monocots but not dicots (Gallavotti et al., 2010; Ma et al., 2017). In general, the short, degenerate nature of the motifs often makes them difficult to predict, highlighting the need for empirical studies to be carried out in diverse species other than Arabidopsis (Arabidopsis thaliana). The fact that large numbers of diverse TFs contain one or more of these repression motifs has raised many intriguing questions about how this family of corepressors operate.
All TPL and TPL-RELATED (TPR) family proteins contain several highly conserved domains: an N-terminal region comprised of LisH, CTLH, and CRA domains, and a C-terminal region defined by two blocks of WD40 repeats (Ke et al., 2015; Martin-Arevalillo et al., 2017). The two halves of the protein are separated by a flexible Prorich middle region. Several crystallization studies have established that the N-terminal region is sufficient to bind LxLxL-type EAR repression motifs, while also mediating tetramerization (Ke et al., 2015; Ma et al., 2017; Martin-Arevalillo et al., 2017). The N terminus has also been shown to bind nucleosomes and histone tails (Ma et al., 2017). These data, together with genetic and molecular evidence indicating that TPL-repressive function is carried out in conjunction with histone deacetylase HDA19 (Long et al., 2006; Krogan et al., 2012), suggest TPL proteins are involved in regulating chromatin structure. However, the C-terminal WD40 domain, while speculated to play a role in this process as well, has not been characterized.
The TPL family was originally identified based on the unique tpl-1 mutant allele of the Arabidopsis TPL, which acted as a dominant negative toward its related family members. Single recessive tpl mutants did not show a phenotype, suggesting genetic redundancy in Arabidopsis (Long et al., 2002, 2006). In contrast, a maize homolog of TPL, RAMOSA1 ENHANCER LOCUS2 (REL2), was first isolated as an enhancer of the classic maize ramosa1 (ra1) mutant, with genetic analysis also revealing that rel2 mutants had an inflorescence phenotype independent of ra1 (Vollbrecht et al., 2005; Gallavotti et al., 2010). Furthermore, the finding that the rice (Oryza sativa) ortholog of REL2 also shows a phenotype (Yoshida et al., 2012) suggests certain TPL family proteins may harbor unique functions that are present in many other plant species but absent in Arabidopsis.
In this study, we report the characterization of the vegetative and reproductive phenotypes present in single rel2 recessive mutants. We show that many rel2 phenotypes result from meristem initiation and maintenance defects and involve pathways regulating tassel and ear architecture, traits known to directly impact maize yield. Protein interaction screening, together with transcriptional profiling of rel2 inflorescences, identified many important players and pathways that use REL2-mediated transcriptional repression for the formation of fully fertile maize inflorescences. Furthermore, by defining both the TF motifs and the REL2 domains required for their interaction, we show that the C-terminal WD40 domain of REL2 binds TFs containing RLFGV- and DLN-type motifs in a manner analogous to Gro/TLE corepressors (Pickles et al., 2002; Jennings et al., 2006). The two separate interaction domains of REL2 (an N terminus that binds to LxLxL motifs and a C terminus that binds to RLFGV and DLN-type motifs) suggest that REL2 may be a large scaffold that simultaneously hosts numerous functionally related TFs. These findings resolve several questions regarding REL2/TPL-mediated transcriptional repression in plants and point to a fundamental mechanism underlying eukaryotic transcriptional repression.
RESULTS
rel2 Mutants Show Background-Dependent Pleiotropic Phenotypes in Vegetative and Reproductive Development
Three mutant alleles of rel2 were originally reported in a genetic screen for enhancers of ra1 (Gallavotti et al., 2010) and subsequently found to harbor an independent inflorescence phenotype. Further genetic screens for inflorescence mutants in ethyl methanesulfonate (EMS) mutagenized populations identified three additional alleles of rel2, rel2-341, rel2-203, and rel2-114. Each of the six recovered alleles contained single point mutations within the REL2 coding region, with all but the rel2-203 allele producing a premature stop codon (Fig. 1A). To better characterize rel2 mutants, we carried out detailed phenotypic analysis using three of the six alleles: rel2-SLO73, a predicted null (Q14 > STOP); rel2-ref, a C-terminal truncation missing most of the WD40 domains (Y527 > STOP); and rel2-203, which contains a missense mutation (G368D) located in the first WD40 repeat domain of the REL2 protein (Fig. 1A). The three rel2 alleles were introgressed into two morphologically distinct inbred backgrounds, A619 and B73, and quantitatively scored for various vegetative and reproductive phenotypes (Fig. 1B, Supplemental Table S1).
Figure 1.
rel2 mutants show pleiotropic vegetative and reproductive phenotypes. A, Schematic representation of the REL2 domains and mutant alleles. In bold, alleles used in this study. B, The whole-plant phenotype of the rel2-ref allele in two different inbred backgrounds. Arrowheads point to mature ears. C, The tassel phenotype of rel2-ref in two different inbred backgrounds. D, rel2-ref mutants in B73 show earless stems with internode elongation defects. E, Seedling bases showing compressed internodes and vegetative axillary buds (arrowheads). Leaves have been removed. F, Only the rel2-203 allele shows consistent tassel feminization. wt, Wild type.
In general, all rel2 mutant alleles in both genetic backgrounds showed similar pleiotropic defects. These included an approximate 12% reduction in plant height relative to wild-type siblings, fewer internodes, and severely upright tassel branches borne with angles two to four times larger than wild-type siblings (Fig. 1C; Supplemental Table S1). rel2 mutant tassels were also shorter, producing fewer branches and about half as many spikelets (Fig. 1C; Supplemental Table S1). Occasionally, other defects were also observed in plant phyllotaxis, internode elongation (i.e. kneeling along the stem; Fig. 1D), and tassel sex determination. The latter phenotype was especially strong in the rel2-203 allele in the B73 background, which showed extreme feminization of tassel florets, frequently developing kernels and silks. The penetrance and strength of this phenotype in rel2-203 was enhanced when grown in our winter nursery in Molokai, HI, and was not observed with other alleles grown in the same environment (Fig. 1F). The rel2-203 allele also showed an increase in the number of tassel branches relative to wild-type, an opposite phenotype to that observed in the other rel2 alleles (Supplemental Table S1; Gallavotti et al., 2010).
One of the most striking phenotypes found in all rel2 mutants was a complete absence of ears in B73. This phenotype was not observed in A619 (Fig. 1B). Further investigation into this phenotype at early stages of development when the vegetative axillary meristems that will give rise to ears are first initiated (Supplemental Fig. S1A) revealed that while axillary buds were present in the lower nodes of the main shoot in both rel2 and wild-type B73, they were absent at the upper nodes in rel2 mutants (Fig. 1E). Specifically, axillary buds were never observed in positions after the first three basal nodes above the crown roots in rel2 mutants (Supplemental Fig. S1B; node 3 versus node 6 in wild-type B73). Given that in wild-type B73 plants, it is these upper axillary buds that will develop into ears (rather than the lower axillary buds that remain dormant), we conclude that the earless phenotype of rel2 in B73 is caused by a defect in upper vegetative axillary meristem initiation that cannot be compensated by basal axillary bud activity. However, these basal axillary buds could still be activated, as noticed when rel2 was crossed to the highly tillered tb1 mutant (Doebley et al., 1997). As shown in Supplemental Figure S1C, we found that rel2 tb1 double-mutant plants were still capable of forming tillers, although the number was reduced relative to tb1 single mutants.
In contrast to the situation in B73, rel2 mutants in the A619 background were able to produce ears, although they were consistently borne on lower nodes than their wild-type A619 siblings (Fig. 1B; Supplemental Fig. S1B; node 3 versus node 4 in wild-type A619). This suggested that rel2 A619 mutants also lacked upper axillary buds, but in contrast to the situation in B73, the mutants were able to initiate ears from more basal axillary buds. In addition to their unusually low position on plants in the A619 background, rel2 mutant ears were borne on extremely elongated shanks, approximately three to four times longer than wild-type A619 shanks (Fig. 1B; Supplemental Table S1). This finding suggests that REL2 is involved in repressing internode elongation in the shank.
Overall, the pleiotropic phenotypes observed in rel2 mutants suggest that REL2 is involved in numerous pathways regulating various aspects of vegetative and reproductive development. In particular, the strong tassel and ear phenotypes implicate REL2 as a major factor in regulating maize inflorescence architecture. These include many traits such as tassel branch number, branch angle, and spikelet number that have been targets of maize domestication and improvement due to their influence on maize yield (Xu et al., 2017). The background dependence of certain phenotypes such as the absence of ears in B73 indicates that genetic modifiers or additional homologous genes may be present in certain genetic backgrounds. Furthermore, since REL2 was transcribed normally in the rel2-SLO73, rel2-ref, and rel2-203 alleles (Supplemental Fig. S2), we assume their rel2 transcripts were translated, resulting in a severely truncated (null), C-terminally truncated, or C-terminal amino acid change-containing protein, respectively. Therefore, the largely similar phenotypes produced by these structurally different rel2 alleles suggest that the entire protein is needed for full functionality in many REL2-mediated processes.
rel2 Mutant Inflorescences Show Defects in Meristem Initiation and Maintenance
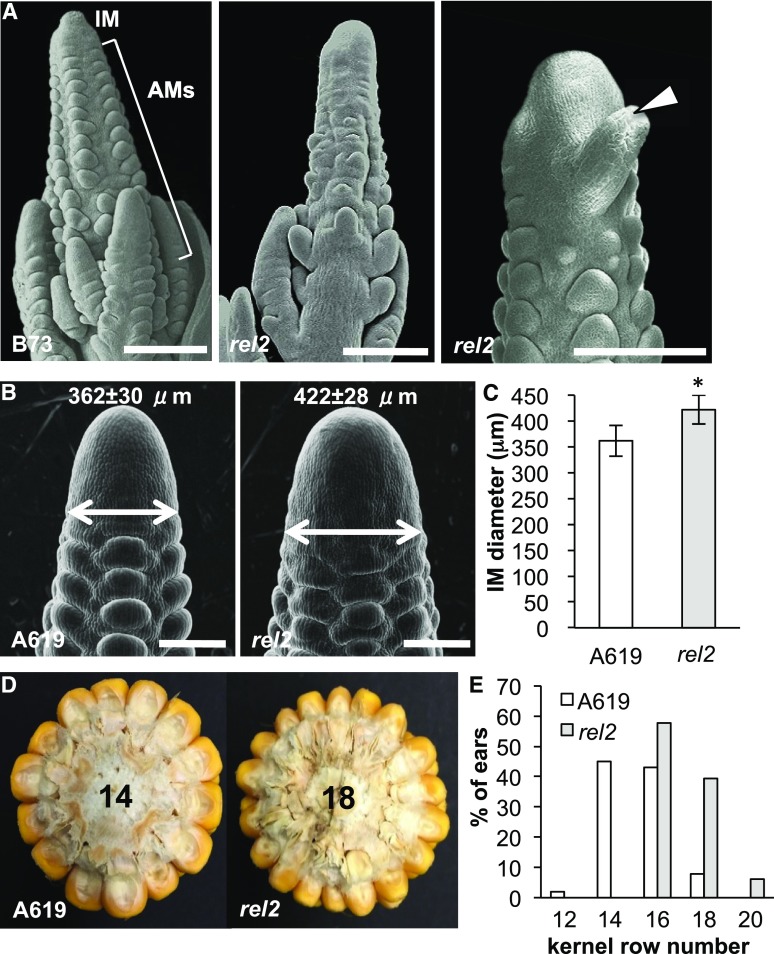
Many of the inflorescence phenotypes that were observed in rel2 mutants resembled those reported for meristem initiation and maintenance mutants. This prompted us to perform detailed morphological analysis on the early steps of inflorescence development to better understand the cause of these phenotypes. In wild-type tassels and ears, the apical inflorescence meristem (IM) gives rise to a series of reproductive axillary meristems (AMs) in its peripheral zone that eventually form a spikelet and its two florets (Fig. 2A). These AMs are regularly arranged along the main axis of growth in a basipetal developmental gradient. Scanning electron microscopy (SEM) of 2- to 3-mm rel2 tassels indicated that the regular initiation of AMs at the peripheral zone of the IM was disrupted and resulted in the formation of fewer AMs. At the same time, we also noticed defects in the IM itself, such as the occasional formation of abnormal meristems, suggesting that REL2 functions in both meristem initiation and maintenance in the tassel.
Figure 2.
rel2 mutants show defective axillary meristem initiation and inflorescence meristem maintenance defects. A, SEMs of immature tassels showing defective arrangement of axillary meristems (AMs) as well as abnormal primordia emerging at the peripheral zone of the inflorescence meristem (IM; arrowhead). Scale bars, 200 μm. B, SEMs of immature ears in wild-type and rel2-ref plants; note enlarged IM in rel2-ref. Scale bars, 200 μm. C, Quantification of ear IM size. n = 20; *P < 0.001 (Student’s t test). Error bars, sd. D, Cross section of mature ears showing additional rows of kernels in rel2 mutants. E, Quantification of the kernel row number phenotype. n ≥ 50 for A619 and rel2.
A similar role for REL2 in ear development was also apparent. In A619, where fertile ears did form, these ears were often slightly fasciated, a phenotype indicative of defects in meristem maintenance and size (Galli and Gallavotti, 2016). Quantification of this finding showed that the IM of rel2 mutant ears was ∼16% larger than in wild-type siblings (Fig. 2, B and C). As a consequence of the increased IM size, rel2 ears had an increased number of kernel rows (Fig. 2, D and E), an important agronomic trait that has undergone selection during maize domestication and improvement. Overall, these results suggest that REL2 functions in AM initiation as well as in the regulatory network controlling meristem size. It is interesting to note that in the A619 background where both ear and tassel structures were formed, we observed what appeared to be opposing IM-related phenotypes: an increase in kernel row number in the ear and a decrease in spikelet pair number in the tassel (Supplemental Table S1). This observation suggests differences in REL2-related transcriptional networks in the male and female inflorescences at early stages of reproductive development when many pathways are known to be highly conserved between the two structures.
The Maize Genome Contains Three Additional REL2-LIKE (RELK) Genes
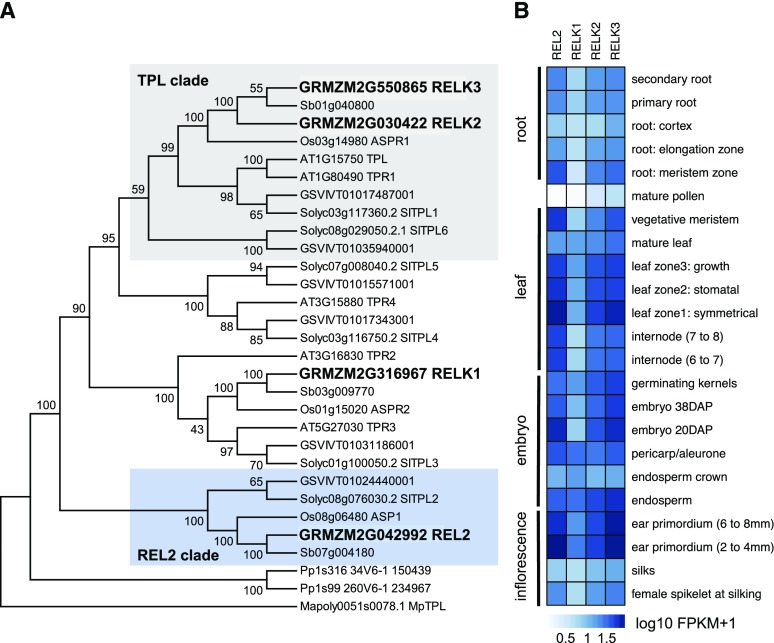
In most plant species, TPL family proteins exist as small gene families (Yoshida et al., 2012; Hao et al., 2014; Flores-Sandoval et al., 2015; Li et al., 2016). We identified three additional RELK genes in the maize reference B73v3 genome and named them RELK1 (GRMZM2G316967), RELK2 (GRMZM2G030422), and RELK3 (GRMZM2G550865). RELK1, RELK2, and RELK3 share 63%, 67%, and 68% identity with REL2, respectively. RELK2 and RELK3 are closely duplicated genes on chromosomes 1 and 9, encode proteins that share 97% identity, and are the most similar to Arabidopsis TPL (77% identity), whereas RELK1 on chromosome 3 is more similar to both TPR2 and TPR3 (Fig. 3A).
Figure 3.
REL2 is part of a small family of transcriptional corepressor proteins. A, Phylogenetic analysis of the TPL/REL2 family. In blue and gray are highlighted the REL2 and TPL clades, respectively. Pp, Physcomitrella; Mp, Marchantia; Sb, Sorghum bicolor; Os, rice; SI, tomato; GSVIV, grape; At, Arabidopsis thaliana; GRMZM maize. B, RNA-seq expression levels of REL2 and RELK genes in different tissues, from Walley et al. (2016).
Phylogenetic analysis of TPL family proteins from a diverse set of land plants showed that REL2 and ASP1, the rice ortholog of REL2 that also displays pleiotropic defects (Yoshida et al., 2012), formed a closely related but separate clade with no clear Arabidopsis ortholog (Fig. 3A). The RELKs on the other hand, grouped into clades with conserved TPL/TPR orthologs from Arabidopsis and other species. The REL2/ASP1 clade was not specific to grasses, as other eudicots, such as tomato (Solanum lycopersicum) and grape (Vitis vinifera), also appeared to have putative REL2 orthologs in their genome. It is therefore likely that the REL2/ASP1 ortholog was lost in the Arabidopsis lineage.
To investigate potential expression differences among different REL2/RELK family members, we used quantitative real-time reverse transcription PCR (RT-qPCR) in several maize tissues as well as published RNA-sequencing (RNA-seq) datasets (Walley et al., 2016). Both RT-qPCR and RNA-seq datasets showed that the RELK genes were broadly expressed similar to original reports for REL2 expression (Gallavotti et al., 2010; Fig. 3B; Supplemental Fig. S3A). Given the coexpression of the REL2/RELK family members and the finding that TPL proteins are able to form homotetrameric complexes (Ke et al., 2015; Martin-Arevalillo et al., 2017), we tested whether REL2 and RELK proteins could physically interact using a yeast 2-hybrid system (Y2H). Interactions were detected in all paired combinations but were somewhat weaker for RELK1 (Supplemental Fig. S3B).
Taken together, these results suggest that REL2 contains unique features that perform a specialized role in maize development that cannot be completely compensated by closely related family members, despite redundant expression patterns and potential in vivo oligomeric complex formation.
REL2 Interacts with TFs via Different Interaction Motifs
While TPL/REL2 proteins are well known to interact with TFs containing various repression motifs, no large-scale protein interaction screens have been carried out in species other than Arabidopsis, limiting our knowledge of functional repression motifs across species. Furthermore, because REL2 orthologs are absent from Arabidopsis, previously conducted screens with Arabidopsis TPL and TPRs may have overlooked possible REL2-specific interactors (Causier et al., 2012b). We therefore sought to empirically screen for maize interactors of REL2 reasoning that (1) experimentally supported cross species comparisons of interaction motifs would shed light on evolutionarily conserved aspects of TPL corepressor family interactions; and (2) such screens could help identify specific factors that contribute to the unique recessive phenotype of rel2 mutants.
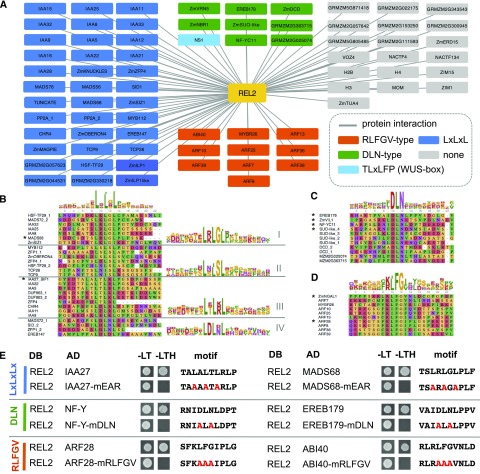
To this end, we performed Y2H screens using maize cDNA libraries from immature inflorescences and mixed tissue (Soderlund et al., 2009). In total, we identified 74 REL2-interacting proteins, the majority (68%) of which were TFs belonging to over 18 unique TF families (Fig. 4A; Supplemental Table S1). Many REL2 interactors corresponded to Arabidopsis homologs previously identified as interactors of TPL/TPRs, supporting an evolutionarily conserved repressive function (Causier et al., 2012a). These included members of the Aux/IAAs and clade B AUXIN RESPONSE FACTORs (ARFs) and known maize meristem-related regulators such as NARROWSHEATH1 and SISTER OF INDETERMINATE SPIKELET1 (SID1), homologs of WOX3 and AP2, respectively (Nardmann et al., 2004; Chuck et al., 2008). A maize homolog of the Arabidopsis C2H2 zinc finger KNUCKLES (GRMZM2G089448), known to act in meristem maintenance (Sun et al., 2009), was also among the REL2 interactors. Additional evolutionarily conserved interactors included the flowering time and floral development-related proteins ZmVERNALIZATION5, and MADS56 and MADS76 (co-orthologs of AtSOC1; Alter et al., 2016). Two additional floral regulators, TUNICATE/MADS72 and MADS68, maize co-orthologs of Arabidopsis SVP (not previously reported to interact with TPL/TPRs), were also identified in our REL2 screen. Other novel REL2 interactors included a DEVELOPMENTAL AND CELL DEATH DOMAIN protein (ZmDCD/GRMZM5G893913) and an EARLY RESPONSIVE TO DEHYDRATION15 (ZmERD15/GRMZM2G037189) TF, both shown to play a role in osmotic stress (Alves et al., 2011; Reis et al., 2016), and other uncharacterized proteins.
Figure 4.
REL2 interacts with many proteins containing different repressor motifs. A, Protein-protein interaction results from Y2H screens. In blue are highlighted proteins containing the canonical EAR motif (LxLxL), in green the DLN-type motif, in orange proteins containing RLFGV-type motifs, and in gray proteins with no obvious repression motif. B, C, and D, Alignments and position weight matrix of different type of repressor motifs. E, Mutation analysis of the repression motifs by direct Y2H.
Sequence alignment of REL2-interacting clones with closely related proteins from other species allowed us to identify highly conserved motifs within our recovered maize clones that generally matched at least one of the four known repression motifs (Fig. 4B; Supplemental Fig. S4, A–D; Supplemental Table S2). In several cases, the presence of the motif was not conserved in the Arabidopsis ortholog but was present in orthologs from other grasses or other species (Supplemental Fig. S4D). LxLxL-type repression motifs were most frequent, occurring on 44% of all interactors. RLFGV-type motifs were predominantly found on clade B ARFs as well as several other proteins. DLN-type motifs were found on over 13% of interactors and occasionally either overlapped or were found together with an LxLxL type motif (Supplemental Table S2; Supplemental Fig. S4E). The TLxLF WUS-box was found only on a single interactor in our screens and corresponded to NARROWSHEATH1, a WOX3 homolog. Given its infrequent occurrence in our dataset and previous characterization, we did not focus further on this motif (Kieffer et al., 2006). Alignment of the amino acid sequences for each of the three main motifs contained on REL2 interactors allowed us to derive consensus sequences for each motif type that differed slightly from those reported in Arabidopsis. EAR motifs were observed in four possible combinations, L R/K LGLx, LxLxSL, LxL R/K Lx, and LDL D/N L, while the DLN type was only observed as DLN and the RLFGV has a conserved R/K LFG core (Fig. 4, B–D).
To demonstrate that these motifs were responsible for mediating the interaction, we selected two representative TF clones each containing either the LxLxL, DLN-type and RLFGV motifs for further analysis. Mutational analysis of these motifs confirmed that they were necessary for the interaction with full-length REL2 (Fig. 4E), in agreement with previously published reports for TPL in Arabidopsis (Causier et al., 2012a). This analysis helps refine our understanding of the requirements for REL2/TPL recruitment and paves the way for discovering additional TFs in different plant species that contain these or similarly modified motifs.
Domain Dissection of REL2 Identifies Two Distinct TF Interaction Domains
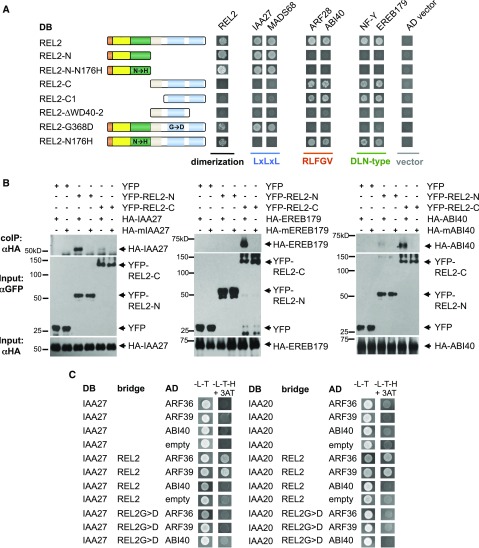
We next sought to understand whether each of the three main REL2-interacting motifs interacted with the same region of the REL2 protein. Like all TPL family proteins, REL2 contains an N-terminal domain comprised of a LisH, CTLH, and CRA domain, a Prorich middle region with relatively low conservation, and a C-terminal domain defined by two blocks of WD40 repeats that are predicted to form two seven-bladed β-propellers that fold into an open clamshell-shaped structure (Fig. 1A; Voegtli et al., 2003; Ke et al., 2015). Several crystallization studies have established that the N-terminal domain is responsible for interactions with the LxLxL repression motif (Ke et al., 2015; Ma et al., 2017; Martin-Arevalillo et al., 2017); however, no domain-mapping studies have been carried out for the DLN-type or RLFGV-type motifs. Given the structurally different properties of these two motifs relative to the LxLxL-type motif, we speculated that they might interact with a different region of REL2. To test this hypothesis, we created a series of REL2 constructs containing N-terminal and C-terminal truncations and performed Y2H experiments with our representative TFs containing the LxLxL, RLFGV, and DLN-type repressor motifs (Fig. 5A). As expected, LxLxL-containing TFs interacted with the REL2 N-terminal domain (REL2-N) but not the C terminus (REL2-C). In contrast, however, both DLN- and RLFGV-containing TFs interacted with REL2-C, but not REL2-N. In all cases, mutation of the repression motif abolished the domain-specific interaction (Supplemental Fig. S4F). Further domain dissection of the C terminus established that while the Prorich middle region was not necessary for the interaction, both WD40 repeat domains were required (Fig. 5A), in line with models of similar tandem WD40 domains in eukaryotes in which proper folding requires both β-propellers (Voegtli et al., 2003).
Figure 5.
Domain dissection of REL2 revealed different protein-protein interaction domains. A, Different TF repression motifs interact with different domains of the REL2 protein. B, Co-IP of different classes of TFs with REL2-N and -C terminus constructs. C, Y3H analysis of Aux/IAAs and clade B ARFs. REL2 proteins can bridge an interaction between Aux/IAAs and clade B ARFs.
To better understand the role of the REL2 C terminus in mediating protein interactions, we next tested the effect of the rel2-203 (G368D) mutation on the interaction with LxLxL, RLFGV, and DLN-type motif containing TFs (Fig. 5A). This single amino acid change is predicted to reside in the first blade of WD40 β-propeller1 and is highly conserved in TPL proteins (Supplemental Fig. S5A). Y2H assays with a full-length REL2 construct carrying the G368D mutation did not show interaction with RLFGV- or DLN-type repression motif TFs, supporting the finding that the C-terminal WD40 domain of REL2 mediates interactions with RLFGV- and DLN-type repression motifs (Fig. 5A). In contrast, LxLxL-type TFs did interact with REL2-G368D, confirming that the protein was stable and that the WD40 domain is not necessary for N-terminal TF interactions (Fig. 5A). The WD40 domain also did not appear to play a role in REL2 oligomerization as REL2-C constructs did not dimerize with full-length REL2 and the REL2-G368D construct showed a similar level of dimerization as with full-length REL2 (Fig. 5A).
Structural homology modeling showed that the loop region containing the G368 residue lies on the top face of β-propeller1, the region that most commonly mediates protein interactions in WD40 proteins (Supplemental Fig. S5, B and C). It is particularly interesting to note that the top face of the Groucho/TLE WD40 domain also binds two distinct short peptide motifs present on diverse animal TFs and that mutations on this exposed surface disrupt protein-protein interactions without causing gross disruption of the β-propeller structure (Pickles et al., 2002; Jennings et al., 2006). This highlights a conserved mechanistic feature among plant and animal corepressor proteins despite their lack of sequence homology.
We also tested whether the previously described tpl-1 mutation (REL2-N176H) affected the interaction with TFs containing the different repression motifs. As described previously for TPL/TPRs (Szemenyei et al., 2008; Ke et al., 2015; Martin-Arevalillo et al., 2017), N-terminal REL2 truncations containing the N176H mutation interacted with LxLxL motifs. In contrast, full length REL2 containing the N176H mutation did not interact with LxLxL motifs (Fig. 5A) but still interacted with TFs containing the RLFGV or DLN-type motifs. This finding suggests that in the context of the full-length REL2 protein, the N176 residue is not involved in RLFGV or DLN-type motif binding, but the large aggregates known to be formed by the N176H mutation (Ke et al., 2015; Ma et al., 2017) may render the LxLxL binding site inaccessible.
While these results showed that the REL2 C-terminal domain was sufficient for interaction with RLFGV and DLN-type motifs, yeast growth was weaker with the REL2-C construct relative to full-length REL2 (Fig. 5A). This suggested that REL2-C might be structurally unstable in yeast or that the N-terminal domain may help stabilize C-terminal interactions, possibly through its ability to mediate tetramerization (Ke et al., 2015; Martin-Arevalillo et al., 2017). We tested the latter hypothesis by creating a TraM-REL2-C fusion construct in which the REL2 N-terminal domain (position 1-229) was replaced with TraM, a small bacterial protein that drives tetramerization (Lu et al., 2006; Ke et al., 2015). A Y2H assay using the TraM-REL2-C construct and three clade B ARFs containing the RLFGV motif (ARF10,36,39) showed more robust interactions relative to the REL2-C construct although interactions were not as strong as with the full-length REL2 (Supplemental Fig. S6A). This finding suggests that REL2 tetramerization and other unidentified aspects of the N-terminal domain may stabilize C-terminal interactions.
To confirm our Y2H results showing that the WD40 domain of REL2 is responsible for mediating the interaction with RLFGV and DLN-type repression motifs, we performed coimmunoprecipitation experiments with transiently coexpressed proteins in tobacco (Nicotiana benthamiana) leaves (Fig. 5B). In agreement with our Y2H assays, an LxLxL-containing TF (IAA27/BIF1; Galli et al., 2015) interacted with REL2-N, while DLN-type (EREB179) and RLFGV-type (ABI40) TFs interacted only with REL2-C, and these interactions were disrupted when the DLN or RLFGV motifs were mutated (Fig. 5B). Overall, these data indicate that while the N terminus of REL2 interacts with the LxLxL motif, TFs containing the RLFGV and certain DLN-type motifs instead interact with the C terminus. The latter interaction was unexpected given that DLN-type motifs often overlap with LxLxL-type motifs and in such cases have been shown to interact with the N terminus of TPL or related homologs (Ke et al., 2015; Ma et al., 2017). We were unable to identify major sequence differences between the LDLNL- and the DLN-type motifs that dictate interaction with REL2-N or REL2-C (Supplemental Fig. S6B). We therefore hypothesize that the presence of critical amino acids surrounding the core DLN may influence binding domain specificity.
REL2 May Form Large Transcriptional Complexes with Related TFs
Overall our results show that REL2 has two independent interaction domains with which to engage TFs. This has important implications for several pathways known to involve TFs containing both LxLxL- and RLFGV-type motifs and raises the intriguing possibility that REL2 may be able to simultaneously bind two different TFs as a means to coordinate transcriptional responses of functionally related TFs. To test this possibility, we investigated the interaction of REL2 with two different gene families involved in auxin signaling that were well represented in our Y2H screening dataset: the Aux/IAA and clade B “repressor” ARF families. Aux/IAAs are well known to interact with TPL/REL2 via their LxLxL motif and to interact with clade A ARFs, two features which are important for their role as auxin coreceptors (Dharmasiri et al., 2005; Tan et al., 2007; Szemenyei et al., 2008; Calderón Villalobos et al., 2012). Clade B ARFs on the other hand are less well characterized. They do not generally interact directly with Aux/IAAs (Vernoux et al., 2011) but contain a highly conserved RLFGV-type motif that is not present on the clade A ARFs (Supplemental Fig. S6C; Ikeda and Ohme-Takagi, 2009). The role of this motif in auxin-related signaling has remained elusive despite being shown to interact with TPLs in previous studies (Causier et al., 2012a). Our finding that the clade B ARFs interact with the C-terminal WD40 domain of REL2 sheds light on their role as repressors and suggests that they could work together in the same complex with Aux/IAAs.
To explore this possibility, we used a yeast three-hybrid (Y3H) system and tested whether REL2 could bridge an interaction between Aux/IAAs and clade B ARFs (Fig. 5C). As expected, no interaction was detected between representative Aux/IAAs (BIF1/IAA27 and BIF4/IAA20; Galli et al., 2015) and clade B ARFs (ARF36 and ARF39). However, when REL2 was coexpressed together with the Aux/IAAs and ARFs, a positive interaction was observed, suggesting that Aux/IAAs and REL2 complexes can also contain clade B ARFs (Fig. 5C). These interactions were not detected with the rel2-203 (G368D) mutant version of REL2 or when a different RFLGV-containing TF was used (ABI40; Fig. 5C). These results show that REL2 binding by Aux/IAAs and clade B ARFs is not mutually exclusive and instead suggests that complexes containing clade A ARFs, Aux/IAAs, and REL2 may cooperate with clade B ARFs in vivo. It is important to point out, however, that REL2 was not able to bridge all of the Aux/IAA and clade B ARF interactions tested and that two Aux/IAAs directly interacted with at least one clade B ARF (ARF10) in our Y2H experiments (Supplemental Fig. S6D). Overall, these results suggest that additional complexity likely exists in the transcriptional control of auxin signaling.
Key TFs Are Misregulated in rel2 Tassels
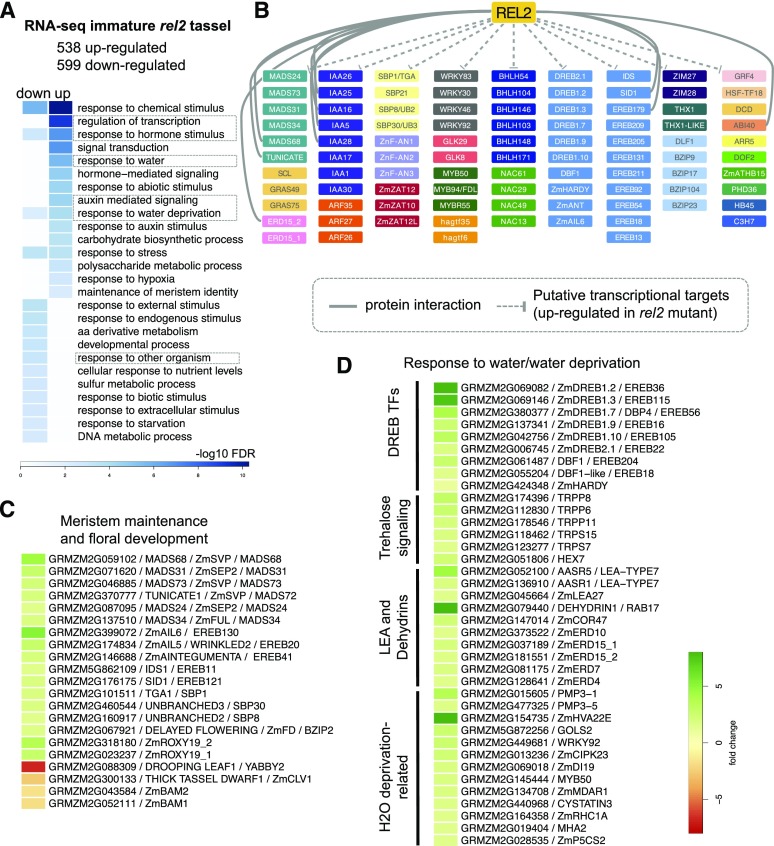
The pleiotropic phenotypes of rel2 mutants and the identification of many interacting TFs indicate that REL2 is involved in the transcriptional regulation of numerous pathways. To identify possible direct transcriptional targets of REL2-TF complexes, we profiled transcriptional changes caused by the rel2 mutation. We chose to focus on the meristem-rich structure of developing maize tassels given the fact that many rel2 phenotypes were related to meristem initiation and maintenance. Differential gene expression analysis from RNA-seq experiments performed on immature 2- to 3-mm wild-type and rel2-ref tassels (capturing the early steps of IM activity as well as AM initiation and development) identified 538 up- and 599 down-regulated genes (FDR < 0.05; Supplemental Table S3). Gene Ontology (GO) enrichment analysis of genes up-regulated in rel2 mutant tassels (genes likely to be directly repressed by REL2) showed strong enrichment for “transcriptional regulation” and included known negative regulators such as members of the AP2-EREB, MADS, SBP, and Aux/IAA families (Fig. 6, A and B).
Figure 6.
RNA-seq analysis of immature rel2 tassels shows widespread mis-regulation. A, GO enrichment analysis of differentially expressed genes. B, TFs up-regulated in rel2 mutant tassels. TFs that also physically interacted with REL2 are indicated with solid gray line. C, Differentially expressed genes involved in inflorescence development. D, Up-regulated genes involved in water response. FDR < 0.05; Scale bar indicates fold change.
Within these various families, up-regulated genes included those involved in meristem maintenance and floral organ identity such as IDS1, SID1, and TUNICATE1 (Chuck et al., 2008; Han et al., 2012), the inflorescence branching genes UB2 and UB3 (Chuck et al., 2014), and the FUSED LEAVES1 MYB TF involved in organ separation (La Rocca et al., 2015; Fig. 6, B and C). The increased expression of these genes in rel2 tassels suggests that their transcription is regulated by REL2-mediated repression and their mis-regulation could contribute to the branching and spikelet development defects observed in rel2 mutant tassels. Furthermore, up-regulation of multiple Aux/IAA genes may be responsible for the early patterning defects observed in young rel2 tassels, as previously reported in two semidominant aux/iaa mutants (Galli et al., 2015). Given the IM defects observed in rel2 mutants, we also examined the expression levels of genes known to function in the WUS/CLV pathway, the core regulatory pathway for meristem size. Among these, THICK TASSEL DWARF1, the maize ortholog of the LRR-kinase CLAVATA1 (Bommert et al., 2005), and two maize homologs of the related LRR-kinases BARELY ANY MERISTEM2 (GRMZM2G052111 and GRMZM2G043584; Deyoung and Clark, 2008; Nimchuk et al., 2015) were significantly down-regulated, suggesting a loss of tight control of the negative feedback loop regulation in the WUS/CLV pathway (Fig. 6C). A loss of negative regulation in this pathway may help explain the abnormal IM meristem development of rel2 tassels and the slight fasciation phenotype observed in A619 rel2 ears (Fig. 2, B–E).
GO enrichment analysis of genes up-regulated in rel2 tassels also unexpectedly showed enrichment for ‘response to water’ and ‘response to water deprivation.’ Up-regulated genes related to this process included known dehydration-induced genes such as DREB TFs (Liu et al., 2013), DEHYDRINS (Hanin et al., 2011; Hernández-Sánchez et al., 2017), EARLY RESPONSE TO DEHYDRATION (ERD), and six members of the trehalose-6 signaling pathway, which has been shown to impact maize yields underwater restrictive conditions by acting during the critical early maize inflorescence development period (Fig. 6D; Zhou et al., 2014; Nuccio et al., 2015). rel2 down-regulated genes also showed enrichment for ‘response to water deprivation,’ suggesting general misregulation of water allocation in rel2 mutant tassels.
The ‘response to hormones’ term identified by our GO enrichment analysis was also found in both up-regulated and down-regulated rel2 genes, likely reflecting the involvement of TPL family members in many different hormone-signaling pathways. Among the various hormones, auxin-related genes were most frequently found among the up-regulated genes (Supplemental Fig. S7A), while jasmonic acid (JA)-related genes, with the exception of two JAZ proteins (ZIM27 and ZIM28) that were up-regulated, were found in the down-regulated gene dataset. The latter included ZmbHLH91/GRMZM2G049229, an AtMYC4 homolog involved in JA signal transduction, and JA-biosynthesis pathway genes such as ALLELE OXIDE SYNTHASE and LIPOXYGENASE genes. The putative down-regulation of JA biosynthesis in rel2 mutants agrees with the tendency of rel2 mutant to show feminization of tassel florets, a known phenotype of the JA biosynthesis mutant tasselseed1/LIPOXYGENASE7 (Acosta et al., 2009). JA is also known to be involved in plant defense, and in general, rel2 mutants showed down-regulation of genes corresponding to categories associated with biotic stresses such as ‘response to other organism’ and ‘response to external stimulus,’ suggesting that REL2 may be involved in several defense-related pathways. These included genes involved in benzoxinoid biosynthesis and Phe ammonium lyase, both of which are important secondary metabolites that function in plant defense (Supplemental Fig. S7B). Our finding that REL2 may transcriptionally regulate many defense-related gene agrees with a recent study implicating REL2 in the response to the fungal pathogen Cochliobolus carbonum (Walley et al., 2018).
Interestingly, many of the up-regulated genes including SID1, TU1, and ERD15 directly interacted with REL2 in our Y2H screen (Chuck et al., 2008; Han et al., 2012), suggesting that they may repress their own transcription (Fig. 6B). Overall, 19% (14/74) of the directly interacting proteins identified in our Y2H screen were also misexpressed in our tassel RNA-seq dataset (11 up-regulated and three down-regulated), suggesting that many are subjected to feedback regulation. The auxin signaling pathway is known to respond to feedback inhibition, and indeed about one-third of Aux/IAA genes (11 out of 34 expressed) were mis-regulated in rel2 mutant tassels (eight up-regulated and three down-regulated; Supplemental Fig. S7A). We also noted that among the RELK genes, RELK1 expression was approximately four times higher in rel2 tassels (FDR = 0.002), suggesting REL2-TF complexes specifically repress RELK1 transcription in wild-type tassels (Supplemental Figure S7C). This finding indicates that the expression of REL2 and RELK1 is coordinated and supports a model where relatively higher levels of REL2 expression are important for proper tassel development.
DISCUSSION
The pleiotropic phenotypes and misregulation of diverse pathways in rel2 mutants indicate that REL2 functions in many developmental processes. Salient among these is the role of REL2 in controlling meristem development and overall plant and inflorescence architecture. These traits, in particular the increase in IM size and the moderate ear fasciation of the rel2 mutant in the A619 permissive background, suggest that modulation of REL2-mediated repression could be an effective means to increase kernel row number and grain production (Taguchi-Shiobara et al., 2001; Bommert et al., 2005, 2013a, 2013b; Xu et al., 2015). The core pathway that regulates meristem size is the well-established WUS/CLV negative feedback loop (Galli and Gallavotti, 2016; Somssich et al., 2016). WUS itself was reported to interact with TPL and TPR proteins (Kieffer et al., 2006). While we did not isolate a maize ortholog of WUS in our Y2H screen with REL2 (likely due to low expression levels), the rel2 ear phenotype may be due to a perturbation of the WUS/CLV or of functionally related pathways. The defects observed in rel2 tassels similarly point to REL2 as pivotal player in regulating male inflorescence architecture as well, a feature that affects pollen abundance, light exposure, and energy allocation (Xu et al., 2017). Furthermore, the observation that rel2 mutant tassels showed misregulation of many genes involved in water deprivation suggests that REL2 also operates during this critical stage of development.
The finding that REL2 behaves in a nongenetically redundant manner is distinct from the situation in Arabidopsis where single recessive mutations in TPL family members do not show a phenotype. REL2 has been shown to rescue the embryo defects of the dominant tpl-1 mutation in Arabidopsis, suggesting that it is capable of carrying out many of the same functions as TPL (Gallavotti et al., 2010). Whether the reciprocal is true however, i.e. TPL is able to rescue rel2 mutant phenotypes, is not known. However, our phylogenetic analysis of this highly conserved family placed REL2 and ASP1 in a closely related but separate clade with no obvious Arabidopsis ortholog (Fig. 3A), supporting unique functions for the REL2 clade that are yet to be determined.
Our finding that the WD40 domain of the REL2 C terminus binds to TFs containing RLFGV- and DLN-type motifs provides the first experimentally derived role for this domain of the TPL corepressor family. Given the high degree of conservation of both the REL2/TPL WD40 domain and these motifs across plant species, we propose that this function is central to all TPL family members. The single missense mutation present in the rel2-203 allele (G368D) is predicted to reside on the top face of WD40 β-propeller1 (Wang et al., 2015). This region is known to be a common protein interaction surface (Wu et al., 2012), and protein interaction assays with this mutation disrupt interactions with RLFGV- and DLN-type motifs, but not LxLxL motifs. An additional mutation in TPL (S578F; equivalent to S566F in REL2), which we predict also resides on the top face of the WD40 domain of TPL (Supplemental Fig. S5, B and C), was previously shown to rescue the dominant negative phenotype of the Arabidopsis tpl-1 mutant, providing further support that this region is likely important in all REL2/TPL family members (Long et al., 2006).
In general, the fact that REL2/TPL corepressors interact with different TFs using distinct N- and C-terminal domains suggests that the composition of these repressor complexes is likely complex. This finding may carry implications for many hormonal and developmental pathways, such as the auxin signaling pathway, whereby Aux/IAAs and clade A and clade B ARFs may come together in the same repressor complex to coordinate auxin transcriptional responses. We recently showed that clade A and clade B ARFs bind either identical or nearby sites at many loci in the maize genome (Galli et al., 2018), and we speculate that the formation of large complexes could potentially bridge nearby ARF binding sites and regulate auxin signaling targets. These findings are in line with the proposed TF-induced self-association of the TPL N terminus thought to mediate transcriptional repression through chromatin compaction (Ma et al., 2017). Although the role of the REL2/TPL WD40 domain in such a model remains unclear, it is tempting to speculate that bipartite TF-binding could contribute to this process.
As noted previously, TPL/REL2 proteins share several mechanistic and structural properties with other eukaryotic repressors (Long et al., 2006; Ma et al., 2017; Martin-Arevalillo et al., 2017). These similarities include an N terminus that mediates tetramerization, interaction with transcriptional regulators via hydrophobic peptides, HDAC recruitment, and nucleosome binding (Long et al., 2006; Krogan et al., 2012; Ma et al., 2017; Martin-Arevalillo et al., 2017). Our finding that the REL2 C-terminal WD40 domain binds to TFs via RLFGV and DLN-type motifs is highly analogous to the binding of WRPW/Y and LxIxxIL motifs by the WD40 domain of Groucho/TLE corepressors (Buscarlet and Stifani, 2007) and reveals an even deeper level of mechanistic convergence. Crystallization studies have shown that the WRPW/Y and LxIxxIL motifs bind the top face of the Gro/TLE WD40 domain via overlapping sites that span the central pore of the β-propeller (Jennings et al., 2006). Given the location of the REL2 G368 residue in a similar location and its ability to disrupt repression motif binding, a similar scenario for the binding of DLN-type and RLFGV motifs may occur. In contrast to other WD40 domain-containing corepressors, however, the REL2/TPL WD40 domain contains a tandem β-propeller. The open clamshell-shaped structure that is formed by such an arrangement has been proposed to accommodate large protein complexes and numerous interaction surfaces (Voegtli et al., 2003). It remains to be determined whether β-propeller2 has an independent function that is specific to plant corepressors or one that is mechanistically conserved across eukaryotes.
MATERIALS AND METHODS
Plant Materials and Phenotyping
Maize (Zea mays) plants segregating for the rel2-203 allele were obtained from the Maize Genetics Cooperation Stock Center (MGCSC) and originated from EMS mutagenesis performed by Gerry Neuffer in the inbred line B73 (03HI-B73GN-203). While unaware of the background-dependent earless phenotype, rel2-203 mutant plants were crossed to the inbred line Mo17 to generate an F2 mapping population. We performed bulked segregant analysis analysis using 25 mutant and 25 wild-type samples of the F2 population and used a high-throughput single nucleotide polymorphism genotyping platform to map the mutant locus to chromosome 10 (Gallavotti and Whipple, 2015). Subsequently, sequencing the REL2 gene revealed that earless plants carried a G-to-A transition at nucleotide position +1103 of the coding sequence that caused a missense mutation (G368D) in the first WD40-repeat domain. An allelism test was also performed by crossing rel2-203 plants to rel2-ref plants, and the resulting F1 generation failed to complement the rel2 phenotype. The rel2-341 (04HI-A632xOH43GN-341) and rel2-114 (04MO-A619xB73GN-114 or 04MO-A619xB73GN-139) alleles were similarly generated by Gerry Neuffer, mapped and sequenced, and are available from the MGCSC. The rel2-ref and rel2-SLO73 alleles used in this study were previously reported (Gallavotti et al., 2010).
All alleles were introgressed in both A619 and B73 inbred lines with a series of back-crosses. rel2-ref BC10F2 (A619) and BC9F2 (B73); rel2-SLO73 BC3F2 (A619) and BC4F2 (B73); and rel2-203 BC3F2 (A619) and BC2F2 (B73; using original stock 03HI-B73GN-203) were used for the data presented. Plant height was measured from the ground to the tip of the tassel. For tassel measurements, tassel length was considered as the distance from the base of the first tassel branch to the tip of the tassel and branch number as the number of branches with more than two spikelets. Ear shank length was measured as the distance from the base of the ear to the insertion on the main stem. Inflorescence meristem diameters were measured using a JMC-6000PLUS SEM. Immature ears of 4 to 6 mm were dissected for imaging. Kernel row number was determined using at least 50 ears from mature, open-pollinated cobs from each line.
For rel2 tb1 double mutant analysis, the tb1-117D mutant was obtained from the MGCSC and back-crossed twice in BC9F2 (B73) rel2-ref before being self-crossed. The tb1 (117D) allele was distinguished using simple sequence repeats marker umc1082, which is tightly linked to the tb1-117D allele (∼200 bp from ATG), using primers 5′-GCCTGCATAGAGAGGTGGTATGAT-3′ and 5′-CCGACCATGCATAAGGTCTAGG-3′. The genotype of rel2-ref was determined using a dCAPS marker developed on the causative point mutation, using primers 5′-AATGTTTGATACGTAAAGGTTCTG-3′ and 5′-GTCAACTCTTGAGCCCAAGCAAGC-3′ for amplification followed by a digestion using HindIII (only the mutant allele is cut). Student’s t test was used to determine statistical significance in all measurements with a cutoff for significance set at P ≤ 0.01.
SEM and Histology
For SEMs both freshly dissected or paraformaldehyde-fixed immature inflorescences were used. Fresh inflorescences (Fig. 3B) were imaged using a JMC-6000PLUS SEM. Fixed inflorescences (Fig. 3A) were treated with 4% paraformaldehyde in phosphate-buffered saline buffer. After dehydration through a graded ethanol series, samples were dried using a CPD 020 Balzers critical point dryer, mounted onto adhesive-coated aluminum stubs, sputter coated with palladium/gold alloy, and imaged using an Amray 1830 I SEM.
Phylogenetic Analysis
Full-length TPL family amino acid sequences were obtained from Phytozome (Goodstein et al., 2012), except in the case of RELK1-3, the sequences of which were cloned from cDNA based on publicly available aggregated RNA-seq data (see NCBI accessions below). Alignments were conducted with protein-coding sequences using ClustalW 2.0 software. The alignment file was then used to generate a neighbor-joining rooted tree with MEGA 5.0, applying the Bootstrap method and 1,000 bootstrap replications.
Expression and RNA-seq Analysis
For RT-qPCR analysis, total RNA was extracted from different tissues obtained from pools of three or more B73 plants using standard procedures. These include embryos and endosperm at 10 d after pollination, mature leaf blades, seedlings, roots, silks, and 6-mm tassels and ears. Primary root tips of rel2-SLO73 and rel2-203 B73 segregating populations were used in Supplemental Figure S2B. cDNA was obtained using the qScript cDNA synthesis kit and amplified with PerfeCTa SYBR Green Fast Mix (Quanta Biosciences). RT-qPCR was performed on two biological replicates using specific primer pairs for the REL2 and RELK genes and for UBIQUITIN as a control (Supplemental Table S4). All reactions were carried out on an Illumina Eco Real-Time PCR System and quantified using the Eco Real-Time PCR System Softwarev4 (Illumina).
For RNA-seq, immature tassels at the 2- to 3-mm stage were collected from rel2-ref plants and phenotypically wild-type siblings (BC7 in B73). Five tassels were bulked per sample and three biological replicates were prepared per phenotype. Libraries were prepared from 1.33 μg of total RNA using a TrueSeq kit (Illumina). Sequencing was performed on an Illumina NextSeq500 with single-end 150-bp reads. Sequenced reads were mapped to the B73v3 genome using Tophat, and differential gene expression was determined using CuffDiff (Trapnell et al., 2012).
Protein Structural Prediction
Structural modeling of the REL2 WD40 domain was performed using Phyre2 (Kelley et al., 2015). WD40 annotation was performed using the WD Structure Predictor (WDSP) web server, which predicts fourteen WD40 repeats in the REL2 C terminus (Wang et al., 2015). WDSP predicts the G368 residue that is mutated in rel2-203 resides in loop bc of WD40 blade 1 comprised of amino acids 366 to 384.
Constructs
To generate the pDEST-DB-REL2, pDEST-DB-RELK1, pDEST-DB-RELK2, and pDEST-DB-RELK3 constructs, the full-length coding sequences of REL2 cloned into pENTR223-Sfi and RELK1, RELK2, and RELK3 cloned into pDONR207 were recombined into pDEST-DB using LR clonase II (Life Technologies). To generate the pDEST-AD-REL2, pDEST-AD-RELK1, pDEST-AD-RELK2, and pDEST-AD-RELK3 constructs, the pENTR clones were similarly recombined into pDEST-AD (Arabidopsis Interactome Mapping, 2011).
For pDEST-AD-IAA27, pDEST-AD-MADS68 (amino acids, aa 96–236), pDEST-AD-NF-Y (aa 61–290), pDEST-AD-EREB179 (aa 29–303), pDEST-AD-ARF28 (aa 405–827), and pDEST-AD-ABI40 (aa 1–308, full length), constructs were regenerated from Y2H library screen clones. To generate the pDEST-AD-mIAA27, pDEST-AD-mMADS68, pDEST-AD-mNF-Y, pDEST-AD-mEREB179, pDEST-AD-mARF28, and pDEST-AD-mABI40 constructs, the above constructs were converted to pENTR clones using Gateway BP Clonase Enzyme Mix (Invitrogen). The subsequent pENTR plasmids were then used as templates for introducing specific site mutations using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene). Resulting pENTR clones were then recombined in the pDEST-AD vector. The respective pairs of primers for specific site mutations are shown in Supplemental Table S4.
To generate pDEST-DB-REL2-N (aa 1–229), pDEST-DB-REL2-C (aa 230–1,141), pDEST-DB-REL2-∆WD40-2 (aa 230–746), and pDEST-DB-REL2-C1 (aa 287–1,141) constructs, fragments were PCR amplified and cloned into the Sfi sites of pENTR223-Sfi. The REL2-G368D construct was generated using Gibson assembly with primers that introduced the G368 to D site mutation and cloned into the Sfi sites of pENTR223-Sfi. The REL2-N176H clone (Gallavotti et al., 2010) was PCR amplified and cloned into the Sfi sites of pENTR223-Sfi. All cloning primers are shown in Supplemental Table S4. All REL2 pENTR clones were recombined into the pDEST-DB vector using LR Clonase II.
To generate the pDEST-DB-TraM+REL2-C construct, TraM (PDB code, 2G7O) cDNA fused to the 5′ end of REL2-C was cloned into pENTR223-Sfi and recombined into pDEST-DB.
For pBridge-IAA27 and pBridge-IAA20, the full-length coding sequences of IAA27/BIF1 and IAA20/BIF4 were subcloned into the EcoRI and BamHI sites of multiple cloning site1 (MCS1) of the GAL4 binding domain vector pBridge (Clontech). To construct pBridge-IAA27/REL2, pBridge-IAA20/REL2, pBridge-IAA27/REL2G368D, and pBridge-IAA20/REL2G368D, the full-length coding sequences of IAA27 and IAA20 were subcloned into the EcoRI and BamHI sites of MCS1 and REL2 and REL2G368D were subcloned into the NotI and BglII sites of MCS2 of pBridge. To generate pBridge-empty/REL2, the full-length coding sequence of REL2 was cloned into the NotI and BglII sites of MCS2 of the pBridge vector. Coding sequences were PCR amplified, with the respective pairs of primers shown in Supplemental Table S4. For pGAD-ARF36, pGAD-ARF39, and pGAD-ABI40 constructs, plasmids pDEST-AD-ARF36, pDEST-AD-ARF39, and pDEST-AD-ABI40 were converted to pENTR clones using Gateway BP Clonase and recombined into pGAD-T7 with LR clonase II. To create the YFP-REL2-N and YFP-REL2-C constructs for transient expression in tobacco (Nicotiana benthamiana), pENTR-REL2-N and -REL2-C were recombined into pEarlyGate104 (Earley et al., 2006) using LR clonase II. To create the HA-mCHERRY-IAA27, -mIAA27, -EREB179, -mEREB179, -ABI40, and -mABI40 constructs, the corresponding pENTR clones were recombined into pEarlyGate-HA-mCHERRY (Gutierrez et al., 2009) using LR clonase II.
Y2H Library Screens
A cDNA library was prepared using approximately 200 µg of total RNA isolated from immature maize tassels (2 mm up to 10 mm) and ears (3 mm up to 10 mm) with a Qiagen RNAeasy kit according to manufacturer’s instructions. RNA was pooled, and mRNA was isolated with an Oligotex mRNA Mini Kit from Qiagen according to the manufacturer’s instructions. One microgram of mRNA was used for cDNA library construction. The CloneMiner cDNA Library Construction Kit from Invitrogen was used to obtain a high-quality cDNA library. To perform the LR library transfer reaction, 0.5 to 1 × 107 cfu (colony forming units) were inoculated in 50 mL LB media. Plasmid isolation was performed with the Qiagen Plasmid Midi Kit and used for an LR reaction with pDEST-AD as described in the CloneMiner manual.
The REL2 full-length open reading frame in pENTR223-Sfi was recombined into pDEST-DB and transformed into the yeast (Saccharomyces cerevisiae) strain Y8930 (Arabidopsis Interactome Mapping Consortium, 2011) using a standard lithium acetate (LiAc) transformation protocol. Competent cells were prepared with the pDEST-DB-REL2 strain and transformed with 10 µg library plasmid DNA using the LiAc transformation method. Positive interactors were selected on SD media with 1 mm 3AT -Leu/-Trp/-His. Positive colonies were picked after 7 and 14 d incubation at 30°C and analyzed by colony PCR and sequencing.
For the mixed tissue (Soderlund et al., 2009) and second inflorescence library screens, a mating-based strategy was employed. pENTR clones were recombined into pDEST-AD using LR clonase II and transformed into mating compatible yeast strain Y8800. pDEST-DB-REL2 was transformed into yeast strain Y8930. Library screens were performed by yeast mating between the bait and library strains according to standard procedures. In each case, over 1 million diploids were screened for interactions, and putative positives were isolated on -Leu/-Trp/-His +1 mm 3AT media plates.
Direct Y2H and Y3H Assays
For targeted Y2H assays, pDEST-AD and pDEST-DB clones were transformed into mating compatible yeast strains Y8800 and Y8930, respectively, using the LiAc transformation method. Mating was carried out according to standard procedures. Reporter gene activation was determined by assessing growth on -Leu/-Trp/-His +1 mm 3AT media (unless otherwise indicated) after 3 to 5 d at 28°C to 30°C.
For Y3H assays, clones in pGAD-T7 and pBridge were transformed into the Gold yeast strain using the LiAc transformation method. Reporter gene activation was determined by assessing growth on -Leu/-Trp/-His +5 mm 3AT after 3 d at 30°C.
Co-IP Experiments
YFP, YFP-REL2-N, YFP-REL2-C, HA-mCHERRY-IAA27, HA-mCHERRY-IAA27mEAR, HA-mCHERRY-EREB179, HA-mCHERRY-EREB179mDLN, HA-mCHERRY-ABI40, and HA-mCHERRY-ABI40mRLFGV were transiently expressed in Nicotiana benthamiana using Agrobacterium tumefaciens leaf injection. After 3 d, proteins were extracted from leaves with extraction buffer (50 mm Tris-HCL, pH 7.4, 150 mm NaCl, 10% [v/v] glycerol, 10 mm EDTA, 5 mm dithiothreitol, 0.5% [v/v] Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail Roche Complete Minitablets). Co-IP with GFP antibodies (Invitrogen) was performed at 4°C. GFP antibodies were incubated with Dynabeads-proteinG (Life Technologies) for 1 h. Dynabeads were washed once with 1× phosphate-buffered saline containing 0.02% Tween 20. Dynabeads with GFP antibodies were incubated with total extracted plant protein for 3 h at 4°C. Finally, the beads were washed four times with extraction buffer containing 0.2% (v/v) Nonidet P-40 and then eluted with 30 mL of SDS-PAGE sample buffer for immunoblot analysis. YFP, YFP-REL2-N, and YFP-REL2-C were detected by mouse anti-GFP monoclonal antibody (Clontech; 1:5,000 dilution). HA-mCHERRY fusion proteins were detected by mouse anti-HA monoclonal antibody (Covance; 1:5,000 dilution).
Accession Numbers
RNA-seq data have been deposited in GEO, accession number GSE115498. RELK experimentally determined coding sequences that differ from incorrectly annotated B73v3 gene models are deposited at NCBI with the following accession numbers: RELK1/ GRMZM2G316967/Zm00001d040279 MH230891, RELK2/GRMZM2G030422/Zm00001d028481 MH230892, RELK3/ GRMZM2G550865/Zm00001d047897 MH230893. Additional gene identification numbers for genes described in this study can be found in Supplemental Tables S2 and S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quantification of the rel2 phenotype
Supplemental Figure S2. REL2 expression in different rel2 alleles
Supplemental Figure S3. Expression and protein interaction analysis of REL2 and RELK genes
Supplemental Figure S4. Conservation of repression motifs across species
Supplemental Figure S5. Structural modeling of the REL2 WD40 domain
Supplemental Figure S6. REL2 interaction details and repression motif comparisons
Supplemental Figure S7. RNA-seq analysis of immature rel2 tassels shows widespread misregulation
Supplemental Table S1. Quantification of rel2 allele phenotypes
Supplemental Table S2. List of REL2-interacting proteins from Y2H screens
Supplemental Table S3. List of differentially expressed genes from RNA-seq analysis
Supplemental Table S4. List of primers used for genotyping, expression analysis, and construct preparation
Acknowledgments
The authors are grateful to the MGCSC for seeds; Gerald Neuffer for EMS mutagenesis; Robert Schmitz for RNA-seq library preparation and the Georgia University Genetic Department for sequencing; and Marc Probasco and Joshua Gager for greenhouse and field management.
Footnotes
I.C. was supported by the Waksman Charles and Johanna Busch Fellowship. This research was supported by grants from the National Science Foundation (IOS-1456950 and IOS-1546873 to A.G.).
Articles can be viewed without a subscription.
Senior author.
References
- Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL (2009) tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323: 262–265 [DOI] [PubMed] [Google Scholar]
- Alter P, Bircheneder S, Zhou LZ, Schlüter U, Gahrtz M, Sonnewald U, Dresselhaus T (2016) Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1. Plant Physiol 172: 389–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves MS, Reis PA, Dadalto SP, Faria JA, Fontes EP, Fietto LG (2011) A novel transcription factor, ERD15 (Early Responsive to Dehydration 15), connects endoplasmic reticulum stress with an osmotic stress-induced cell death signal. J Biol Chem 286: 20020–20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W (2005) thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldshmidt A, Jackson D (2013a) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Bommert P, Nagasawa NS, Jackson D (2013b) Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet 45: 334–337 [DOI] [PubMed] [Google Scholar]
- Buscarlet M, Stifani S (2007) The ‘Marx’ of Groucho on development and disease. Trends Cell Biol 17: 353–361 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B (2012a) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Lloyd J, Stevens L, Davies B (2012b) TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal Behav 7: 325–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019 [DOI] [PubMed] [Google Scholar]
- Chuck GS, Brown PJ, Meeley R, Hake S (2014) Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc Natl Acad Sci USA 111: 18775–18780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyoung BJ, Clark SE (2008) BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Flores-Sandoval E, Eklund DM, Bowman JL (2015) A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort Marchantia polymorpha. PLoS Genet 11: e1005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Whipple CJ (2015) Positional cloning in maize (Zea mays subsp. mays, Poaceae). Appl Plant Sci 3: apps1400092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Long JA, Stanfield S, Yang X, Jackson D, Vollbrecht E, Schmidt RJ (2010) The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Gallavotti A (2016) Expanding the Regulatory Network for Meristem Size in Plants. Trends Genet 32: 372–383 [DOI] [PubMed] [Google Scholar]
- Galli M, Liu Q, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL, et al. (2015) Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci USA 112: 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Khakhar A, Lu Z, Chen Z, Sen S, Joshi T, Nemhauser JL, Schmitz RJ, Gallavotti A (2018) The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat Commun 9: 4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Han JJ, Jackson D, Martienssen R (2012) Pod corn is caused by rearrangement at the Tunicate1 locus. Plant Cell 24: 2733–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin M, Brini F, Ebel C, Toda Y, Takeda S, Masmoudi K (2011) Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms. Plant Signal Behav 6: 1503–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Wang X, Li X, Bassa C, Mila I, Audran C, Maza E, Li Z, Bouzayen M, van der Rest B, et al. (2014) Genome-wide identification, phylogenetic analysis, expression profiling, and protein-protein interaction properties of TOPLESS gene family members in tomato. J Exp Bot 65: 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Sánchez IE, Maruri-López I, Graether SP, Jiménez-Bremont JF (2017) In vivo evidence for homo- and heterodimeric interactions of Arabidopsis thaliana dehydrins AtCOR47, AtERD10, and AtRAB18. Sci Rep 7: 17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ohme-Takagi M (2009) A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D (2006) Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell 22: 645–655 [DOI] [PubMed] [Google Scholar]
- Ke J, Ma H, Gu X, Thelen A, Brunzelle JS, Li J, Xu HE, Melcher K (2015) Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci Adv 1: e1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar A, Leydon AR, Lemmex AC, Klavins E, Nemhauser JL (2018) Synthetic hormone-responsive transcription factors can monitor and re-program plant development. eLife 7: e34702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B (2006) Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18: 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Long JA (2009) Why so repressed? Turning off transcription during plant growth and development. Curr Opin Plant Biol 12: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA (2012) APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca N, Manzotti PS, Cavaiuolo M, Barbante A, Dalla Vecchia F, Gabotti D, Gendrot G, Horner DS, Krstajic J, Persico M, et al. (2015) The maize fused leaves1 (fdl1) gene controls organ separation in the embryo and seedling shoot and promotes coleoptile opening. J Exp Bot 66: 5753–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Golz JF (2012) Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal Behav 7: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Huang KF, Du HM, Wang HL, Chen X, Gao SB, Liu HL, Cao MJ, Lu YL, Rong TZ, et al. (2016) Genome-wide analysis of Gro/Tup1 family corepressors and their responses to hormones and abiotic stresses in maize. J Plant Biol 59: 603–615 [Google Scholar]
- Liu Z, Karmarkar V (2008) Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci 13: 137–144 [DOI] [PubMed] [Google Scholar]
- Liu S, Wang X, Wang H, Xin H, Yang X, Yan J, Li J, Tran LS, Shinozaki K, Yamaguchi-Shinozaki K, et al. (2013) Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet 9: e1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK (2002) Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129: 2797–2806 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lu J, Edwards RA, Wong JJ, Manchak J, Scott PG, Frost LS, Glover JN (2006) Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM. EMBO J 25: 2930–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Duan J, Ke J, He Y, Gu X, Xu TH, Yu H, Wang Y, Brunzelle JS, Jiang Y, et al. (2017) A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci Adv 3: e1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, Bangarusamy DK, Zhu JK (2012) Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol 78: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Arevalillo R, Nanao MH, Larrieu A, Vinos-Poyo T, Mast D, Galvan-Ampudia C, Brunoud G, Vernoux T, Dumas R, Parcy F (2017) Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression. Proc Natl Acad Sci USA 114: 8107–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Ji J, Werr W, Scanlon MJ (2004) The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131: 2827–2839 [DOI] [PubMed] [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM (2015) Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio ML, Wu J, Mowers R, Zhou HP, Meghji M, Primavesi LF, Paul MJ, Chen X, Gao Y, Haque E, et al. (2015) Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat Biotechnol 33: 862–869 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payankaulam S, Li LM, Arnosti DN (2010) Transcriptional repression: conserved and evolved features. Curr Biol 20: R764–R771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles LM, Roe SM, Hemingway EJ, Stifani S, Pearl LH (2002) Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure 10: 751–761 [DOI] [PubMed] [Google Scholar]
- Reis PA, Carpinetti PA, Freitas PP, Santos EG, Camargos LF, Oliveira IH, Silva JC, Carvalho HH, Dal-Bianco M, Soares-Ramos JR, Fontes EP (2016) Functional and regulatory conservation of the soybean ER stress-induced DCD/NRP-mediated cell death signaling in plants. BMC Plant Biol 16: 156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, Currie J, Angelova A, Collura K, Wissotski M, Ashley E, et al. (2009) Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet 5: e1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D (2016) CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143: 3238–3248 [DOI] [PubMed] [Google Scholar]
- Sun B, Xu Y, Ng KH, Ito T (2009) A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev 23: 1791–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guédon Y, Armitage L, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegtli WC, Madrona AY, Wilson DK (2003) The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization. J Biol Chem 278: 34373–34379 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES IV, Martienssen R (2005) Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126 [DOI] [PubMed] [Google Scholar]
- Walley JW, Sartor RC, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, Briggs SP (2016) Integration of omic networks in a developmental atlas of maize. Science 353: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Shen Z, McReynolds MR, Schmelz EA, Briggs SP (2018) Fungal-induced protein hyperacetylation in maize identified by acetylome profiling. Proc Natl Acad Sci USA 115: 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu XJ, Zou XD, Wu XH, Ye ZQ, Wu YD (2015) WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res 43(D1): D339–D344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XH, Wang Y, Zhuo Z, Jiang F, Wu YD (2012) Identifying the hotspots on the top faces of WD40-repeat proteins from their primary sequences by β-bulges and DHSW tetrads. PLoS One 7: e43005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liberatore KL, MacAlister CA, Huang Z, Chu YH, Jiang K, Brooks C, Ogawa-Ohnishi M, Xiong G, Pauly M, Van Eck J, Matsubayashi Y, et al. (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet 47: 784–792 [DOI] [PubMed] [Google Scholar]
- Xu G, Wang X, Huang C, Xu D, Li D, Tian J, Chen Q, Wang C, Liang Y, Wu Y, Yang X, Tian F (2017) Complex genetic architecture underlies maize tassel domestication. New Phytol 214: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano HY (2012) Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J 70: 327–339 [DOI] [PubMed] [Google Scholar]
- Zhou M-L, Zhang Q, Sun Z-M, Chen L-H, Liu B-X, Zhang K-X, Zhu X-M, Shao J-R, Tang Y-X, Wu Y-M (2014) Trehalose metabolism-related genes in maize. J Plant Growth Regul 33: 256–271 [Google Scholar]