OHP1, OHP2, and HCF244 form a transient PSII reaction center (RC)-like complex that promotes the assembly and repair of the PSII complex.
Abstract
The reaction center (RC) of photosystem II (PSII), which is composed of D1, D2, PsbI, and cytochrome b559 subunits, forms at an early stage of PSII biogenesis. However, it is largely unclear how these components assemble to form a functional unit. In this work, we show that synthesis of the PSII core proteins D1/D2 and formation of the PSII RC is blocked specifically in the absence of ONE-HELIX PROTEIN1 (OHP1) and OHP2 proteins in Arabidopsis (Arabidopsis thaliana), indicating that OHP1 and OHP2 are essential for the formation of the PSII RC. Mutagenesis of the chlorophyll-binding residues in OHP proteins impairs their function and/or stability, suggesting that they may function in the binding of chlorophyll in vivo. We further show that OHP1, OHP2, and HIGH CHLOROPHYLL FLUORESCENCE244 (HCF244), together with D1, D2, PsbI, and cytochrome b559, form a complex. We designated this complex the PSII RC-like complex to distinguish it from the RC subcomplex in the intact PSII complex. Our data imply that OHP1, OHP2, and HCF244 are present in this PSII RC-like complex for a limited time at an early stage of PSII de novo assembly and of PSII repair under high-light conditions. In a subsequent stage of PSII biogenesis, OHP1, OHP2, and HCF244 are released from the PSII RC-like complex and replaced by the other PSII subunits. Together with previous reports on the cyanobacterium Synechocystis, our results demonstrate that the process of PSII RC assembly is highly conserved among photosynthetic species.
PSII is a multisubunit pigment-protein complex present in the thylakoid membranes of oxygenic photosynthetic organisms (Nelson and Yocum, 2006; Nelson and Junge, 2015). It captures light energy to extract electrons from water and drive the transfer of electrons to plastoquinone, resulting in the production of oxygen (Nelson and Junge, 2015). A series of structural analyses revealed that the PSII structure is highly conserved among cyanobacteria and chloroplasts, except for the light-harvesting system (Umena et al., 2011; Su et al., 2017). The photochemical reaction center (RC) is the smallest unit having photochemical activity in PSII and is composed of D1, D2, PsbI, as well as the cytochrome (Cyt) b559 α- and β-subunits (PsbE and PsbF, respectively). In addition, the PSII RC contains several cofactors, including six chlorophylls, two pheophytins a, the plastoquinones QA and QB, one heme, and one β-carotene molecule (Nelson and Yocum, 2006), which are required for energy transfer, charge separation, and electron transfer. Surrounding the PSII RC are the CP47 and CP43 subunits, which bind chlorophyll a and β-carotene molecules. CP43 and D1 also provide ligands for the CaMn4 cluster that is essential for water oxidation in the oxygen-evolving complex of PSII (Shen, 2015). In addition to these core subunits, PSII contains at least 11 subunits with low molecular mass (Shi et al., 2012). The phycobilisomes and chlorophyll a/b-binding complex (LHCII) associated with PSII core complexes in cyanobacteria and chloroplasts, respectively, act as light-harvesting systems.
In the past two decades, researchers paid great attention to the molecular mechanisms underlying de novo PSII biogenesis and showed that assembly of the PSII complex occurs in a step-wise manner (Baena-González and Aro, 2002; Nixon et al., 2010; Komenda et al., 2012; Nickelsen and Rengstl, 2013; Heinz et al., 2016). The Cyt b559 subunits are initially synthesized and used for the integration of D2 to form the D2-Cyt b559 subcomplex (Komenda et al., 2004). D1 protein, together with the PsbI subunit, is then cotranslationally assembled into the D2-Cyt b559 subcomplex to form the PSII RC. The PSII RC subcomplex serves as a platform for the incorporation of CP47 and several low-molecular-mass subunits to generate a CP43-less PSII monomer. Subsequently, CP43-PsbK is added to form the PSII core monomer. After association of the oxygen-evolving complex, the monomeric PSII complex is generated. In chloroplasts, PSII monomers in turn form PSII dimers and PSII supercomplexes after association of the LHC complexes (Rokka et al., 2005; Nickelsen and Rengstl, 2013). During the biogenesis of PSII, D1 protein is synthesized as a precursor (pD1). After insertion of pD1 into the PSII RC, nine to 16 residues of the C terminus of pD1 are cleaved by the CtpA peptidase in the thylakoid lumen (Shestakov et al., 1994). This process appears to be a prerequisite for the assembly of the Mn4CaO5 cluster (Roose and Pakrasi, 2004).
During PSII assembly, at least two dozen facilitating factors were identified by genetics approaches (Pagliano et al., 2013; Järvi et al., 2015; Lu, 2016; Plöchinger et al., 2016). They are involved at various stages of biogenesis through specific interactions with distinct subunits or assembly intermediates. At an early stage of PSII assembly, efficient formation of the PSII RC is one of the initial critical steps for PSII assembly (Komenda et al., 2004; Rokka et al., 2005), which requires several biogenesis factors. High Chlorophyll Fluorescence136 (HCF136) was identified in the screening of the Arabidopsis (Arabidopsis thaliana) hcf mutants (Meurer et al., 1998). It localizes to the thylakoid lumen and facilitates PSII RC formation by interacting with the D2-Cyt b559 subcomplex (Plücken et al., 2002). The HCF136 homolog in the cyanobacterium Synechocystis, YCF48, has been shown to interact with pD1 protein and associate with the PSII RC complex (Komenda et al., 2008). Chloroplast-encoded PsbN is not an integral subunit of PSII, but it is required for the efficient formation of the PSII RC, although the exact molecular mechanism is still unclear (Torabi et al., 2014). HCF244 and HCF173 contain one and two atypical short-chain dehydrogenase/reductase domains, respectively, and both proteins have been proposed to be required for translation initiation of the psbA mRNA (Schult et al., 2007; Link et al., 2012). Recently, HCF173 was shown to interact with the pentatricopeptide repeat protein LOW PHOTOSYNTHETIC EFFICIENCY1, and both proteins are jointly involved in the light-induced synthesis of D1 in Arabidopsis (Jin et al., 2018).
The incorporation of cofactors such as chlorophyll into the PSII subunits is an essential step in the formation of the PSII RC. These processes also may be essential for the protection of newly assembled intermediates from photodamage during PSII biogenesis (Eichacker et al., 1996). In Synechocystis, two High-light-inducible protein family proteins, HliC and HliD, both of which contain one light-harvesting chlorophyll a/b-binding (LHC) domain, and the short-chain dehydrogenase/reductase protein Ycf39, the homolog of HCF244 in chloroplasts, were found to be associated with the RC complex (Knoppová et al., 2014). In addition, chlorophyll and β-carotene were copurified in the Ycf39-HliD complex (Knoppová et al., 2014). Moreover, Chidgey et al. (2014) discovered that the chlorophyll biosynthesis enzyme, chlorophyll synthase, can be coimmunoprecipitated with Ycf39 and HliD. These results suggest that HliC/HliD proteins may deliver chlorophyll to the D1/D2 subunits during the biogenesis of the PSII RC. In higher plants, ONE-HELIX PROTEIN1 (OHP1) was proposed recently to deliver chlorophyll to the PSII RC (Myouga et al., 2018). OHP1 and another one-helix protein, OHP2, contain one LHC domain (Jansson et al., 2000; Andersson et al., 2003). In the absence of OHP1 or OHP2, the plants showed pale-green phenotypes and were impaired in the normal accumulation of photosystems (Beck et al., 2017; Myouga et al., 2018). Further analyses showed that, together with D1/D2 protein, HCF136, HCF244, HCF173, and several other proteins can be copurified with OHP1 (Myouga et al., 2018). More recently, Hey and Grimm (2018) proposed that HCF244 can bind to the N-terminal end of OHP2 and then forms a heterotrimer with OHP1. However, it is unclear how these proteins promote the assembly of the PSII complex.
In this study, we found that OHP1, OHP2, and HCF244 form a transient PSII RC-like complex with D1, D2, PsbI, and Cyt b559 subunits in thylakoids at an early stage of PSII biogenesis as well as during PSII repair. Together with the discovery of the counterpart Ycf39-HliD complex in Synechocystis (Chidgey et al., 2014; Knoppová et al., 2014), our results imply that the mechanisms underlying PSII RC assembly are highly conserved among photosynthetic organisms.
RESULTS
Both ohp1 and ohp2 Mutants Are Defective in PSII Activity
To investigate the function of OHP1 and OHP2, we obtained the T-DNA insertion mutants ohp1 and ohp2 (Supplemental Fig. S1, A and B). As reported earlier (Beck et al., 2017; Hey and Grimm, 2018; Myouga et al., 2018), both ohp1 and ohp2 mutants display a pale-green phenotype when the plants are grown on Murashige and Skoog (MS) medium (Supplemental Fig. S1B). They cannot grow photoautotrophically on soil and show a high chlorophyll fluorescence phenotype (Supplemental Fig. S1B), indicating severe defects in photosynthesis. Immunoblot analysis detected the OHP1 and OHP2 signals with molecular masses of less than 10 and 15 kD, respectively (calculated molecular masses of mature OHP1 and OHP2 are 7.8 and 14.3 kD, respectively) in the thylakoid membrane of wild-type and complemented plants but absent in the ohp1 or ohp2 thylakoids and stromal fraction of wild-type plants (Supplemental Fig. S1, C and D), confirming that both OHP1 and OHP2 are localized in the thylakoid membrane.
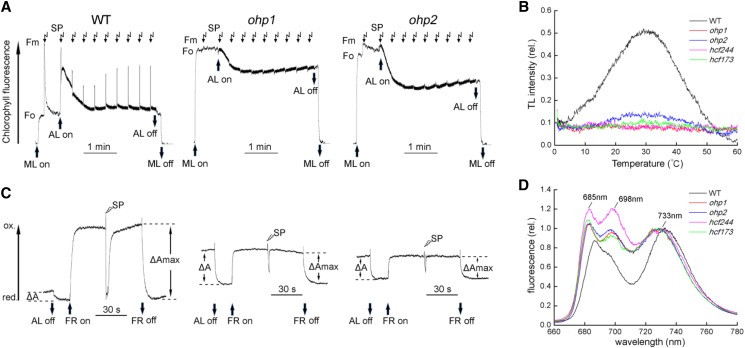
Noninvasive fluorimetric and spectroscopic analyses of chlorophyll a fluorescence revealed that minimum chlorophyll a fluorescence was increased considerably in the ohp1 and ohp2 mutants (Fig. 1A). As shown by Beck et al. (2017), in the ohp1 mutant, no increase of fluorescence was observed when saturating pulses were applied to most of the leaves, revealing the absence of variable chlorophyll fluorescence. In some of the immature ohp1 leaves, a slight increase of fluorescence could be detected, indicating that PSII activity is retained to some extent in the immature ohp1 leaves and dropped to undetectable levels when the leaves developed further. In the ohp2 mutant, however, a small increase of fluorescence could be detected when saturating pulses were applied to most of the leaves, resulting in a quantum yield of PSII (Fv/Fm) of only about 0.1 to 0.2. This indicates that PSII activity in the ohp2 mutant is higher than that in the ohp1 mutant. We also measured the induction of chlorophyll a fluorescence in the hcf173 and hcf244 mutants. Consistent with earlier reports (Schult et al., 2007; Link et al., 2012), only a small and no variable chlorophyll fluorescence were detected in the hcf173 and hcf244 leaves, respectively (Supplemental Fig. S2).
Figure 1.
Spectroscopic analysis of the ohp1 and ohp2 mutants. A, Chlorophyll a fluorescence induction analysis. Plants were dark adapted for at least 20 min, and the minimal level of fluorescence (Fo) was determined by turning on measuring light (ML). After application of a saturating pulse (SP) to determine the maximum level of fluorescence (Fm), the leaves were illuminated with actinic light (AL; 100 µmol photons m−2 s−1) for 3 min. During the illumination, a set of saturating pulses was applied every 20 s. B, TL curves in the wild type (WT) and mutants as indicated. Leaf segments with similar area were cooled to 0°C in the TL apparatus for 120 s and then excited by a single actinic flash. The TL signal was recoded when the leaves were heated from 0°C to 60°C. C, P700 redox kinetics. Dark-adapted plants were illuminated with actinic light for 2 min. After illumination, absorbance changes of P700 at 830 nm induced by actinic light (ΔA) were determined. Then, far-red light (FR) was turned on to oxidize all of P700 (ΔAmax). During the illumination with far-red light, a saturating pulse was applied to induce the rereduction of P700+. The absorbance change at 830 nm in the mutants after turning on far-red light or applying a saturating pulse indicates a functional PSI complex in the ohp1 and ohp2 mutants. D, Chlorophyll fluorescence emission at 77K. Thylakoid membranes isolated from ohp1, ohp2, hcf244, hcf173, and wild-type plants were excited at 436 nm, and the fluorescence was normalized to the emission at the maximum level of fluorescence of PSI (around 733 nm).
Thermoluminescence (TL) emission was measured to investigate the charge recombination of S2QB, which is reflected by the B band of the TL signal at about 30°C (Ducruet and Vass, 2009). As shown in Figure 1B, the TL intensity was reduced drastically in all of the PSII mutants, especially in the ohp1 and hcf244 mutants, in which the B band was almost completely absent (Fig. 1B). In the ohp2 and hcf173 mutants, a slight increase of the B band was detected (Fig. 1B), indicating that these two mutants still retain a small amount of PSII activity, as shown by the fluorescence induction analysis (Fig. 1A; Supplemental Fig. S2).
Spectroscopic analysis of the redox kinetics of PSI showed that application of far-red light results in the oxidation of P700 in both mutants and wild-type plants (Fig. 1C). Also, under a far-red light background, saturating pulses induced the rereduction of P700+ (Fig. 1C). These results indicate that the PSI complex is at least partially functional in the ohp1 and ohp2 mutants.
Measurements of 77K fluorescence emission spectra were performed to probe the coupling of antenna complexes to the RCs of photosystems. As shown in Figure 1D, three major fluorescence peaks, F685, F698, and F733, originating from CP43, CP47, and LHCI, respectively, were obtained with wild-type thylakoid membranes. However, in the ohp1 and ohp2 mutants as well as in the hcf173 and hcf244 mutants, the levels of F685 and F698 were enhanced significantly when the fluorescence was normalized to the level of F733, suggesting that the energy transfer from CP43 and CP47 to the PSII RC is impaired (Fig. 1D). In addition, we detected a blue shift of the PSI fluorescence (F733) in all of the mutants (Fig. 1D). This blue shift might be explained by high levels of free LHCI proteins uncoupled from the PSI RC and also was observed in PSII-deficient mutants such as hcf136 (Meurer et al., 1998).
Taken together, our fluorimetric and spectroscopic analyses indicate that the activity of PSII is reduced significantly in the ohp2 mutant and more so in the ohp1 mutant. The defects in PSII activity in the mutants may represent the primary lesion, which indirectly further affects the function of PSI and photoautotrophic growth.
Accumulation of PSII Is Severely Affected in the ohp1 and ohp2 Mutants
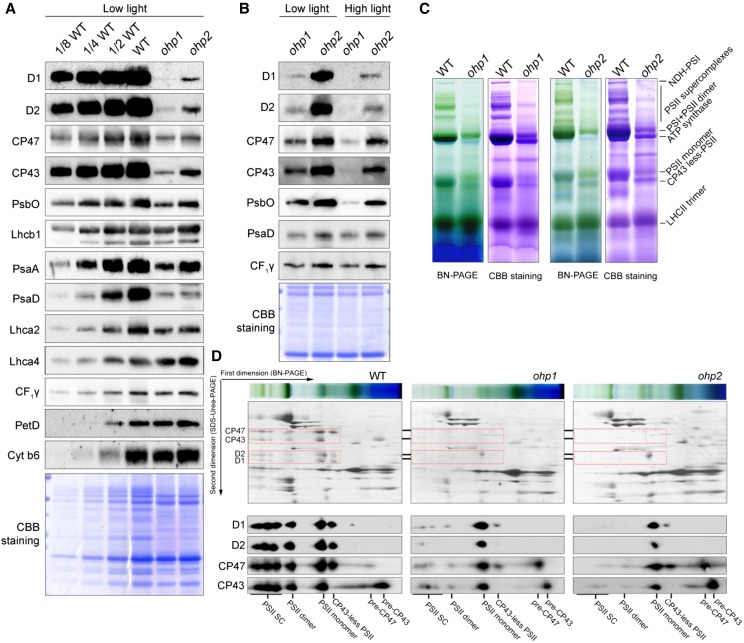
The levels of thylakoid membrane proteins in the ohp1 and ohp2 mutants were determined by immunoblot analyses using a series of antibodies. When protein samples were loaded on an equal protein level, the levels of the PSII core proteins D1, D2, CP43, and CP47 in the ohp1 and ohp2 mutants were reduced to less than one-eighth in comparison with wild-type plants (Fig. 2A). The PsbO level in both mutants was reduced one-fourth to one-half compared with the wild type. The levels of PSII subunits in the ohp2 mutant are higher than those in ohp1, which may explain the higher PSII activity detected in the ohp2 mutant (Fig. 1). By contrast, the PSI complex was less affected than PSII in both mutants. While the levels of PsaA and PsaD were reduced to about one-fourth of wild-type levels, Lhca2 and Lhca4 levels were reduced to more than one-half and comparable with wild-type levels, respectively (Fig. 2A), indicating that more LHCI proteins accumulate in thylakoids and are uncoupled from the PSI core in the ohp1 and ohp2 mutants. This result also explains the blue shift of the PSI fluorescence in these mutants (Fig. 1D). The levels of CF1γ (a subunit of chloroplast ATP synthase) as well as PetD and Cyt b6 (subunits of the Cyt b6f complex) in the ohp1 and ohp2 mutants are comparable to those of the wild type (Fig. 2A), suggesting that the absence of OHP1 and OHP2 does not affect the accumulation of ATP synthase and the Cyt b6f complex.
Figure 2.
Accumulation of thylakoid protein complexes in the ohp1 and ohp2 mutants. A, Immunoblot analysis of representative thylakoid proteins. Thylakoid membranes isolated from wild-type (WT), ohp1, and ohp2 plants were separated by 15% (w/v) SDS-urea-PAGE and then probed with antibodies as indicated. Proteins were loaded on an equal protein level. B, Immunoblot analysis of thylakoid proteins in the ohp1 and ohp2 mutants. The mutants grown under low-light conditions (50 μmol photons m−2 s−1) were shifted to high-light conditions (300 μmol photons m−2 s−1) for 2 d. Thylakoids were isolated for immunoblot analysis. Proteins were loaded on an equal protein level. C, BN-PAGE analysis of thylakoid protein complexes. Thylakoids corresponding to 10 µg of chlorophyll were solubilized with 1% (w/v) dodecyl-β-d-maltopyranoside and separated by 5% to 12% (w/v) BN-PAGE. The gels were stained with Coomassie Brilliant Blue (CBB) for visualization of the proteins. D, 2D BN/SDS-urea-PAGE analysis of the thylakoid membrane complexes. Thylakoid complexes were separated by BN-PAGE (C) and further subjected to 2D SDS-urea-PAGE. The gels were stained with Coomassie Brilliant Blue or probed with antibodies against D1, D2, CP43, and CP47. The positions of D1, D2, CP43, and CP47 in various PSII complexes are framed with red dotted boxes. SC, Supercomplexes.
We also transferred the mutant plants grown under low-light conditions (50 μmol photons m−2 s−1) to high-light conditions (300 μmol photons m−2 s−1) for 2 d and then measured the accumulation of PSII proteins. The results showed that the levels of D1, D2, CP43, CP47, and PsbO were reduced in both mutants treated with high light as compared with when they were grown under low-light conditions (Fig. 2B). These results indicate that the retained PSII complex in the ohp1 and ohp2 mutants is sensitive to high light. By contrast, PSI and ATP synthase are more stable in both mutants (Fig. 2B).
To investigate the accumulation of protein complexes in thylakoids in the ohp1 and ohp2 mutants, thylakoid membranes isolated from mutants and wild-type plants were solubilized with 1% (w/v) dodecyl-β-d-maltopyranoside and thylakoid protein complexes were separated by blue native (BN)-PAGE. The BN gel was further stained with Coomassie Brilliant Blue or subjected to 2D SDS-urea-PAGE for visualization of the individual subunits (Fig. 2, C and D). As shown in Figure 2C, several bands corresponding to PSI-NDH supercomplex, PSII supercomplexes, PSI monomer, PSII dimer, chloroplast ATP synthase, PSII monomer, CP43-less PSII, and trimeric LHCII were resolved clearly in the wild-type samples. Expectedly, PSII supercomplexes, dimeric and monomeric PSII, as well as the CP43-less PSII complexes were barely detectable in the ohp1 and ohp2 mutants. In contrast, PSI subunits PsaA/B were assembled in the intact complex (Fig. 2D), in agreement with the finding that the PSI complex is still partially functional in the ohp1 and ohp2 mutants (Fig. 1C).
Immunoblot analyses showed that the ohp1 and ohp2 mutants accumulate less than one-eighth of PSII compared with the wild type (Fig. 2A). To investigate the levels of the subunits in the remaining PSII in the ohp1 and ohp2 mutants, thylakoid proteins were separated by 2D BN/SDS-PAGE and subsequently subjected to immunoblot assays using the antibodies against PSII core subunits D1, D2, CP43, and CP47. As shown in Figure 2D, the PSII core subunits were detected in the various PSII complexes in the wild type. However, the PSII subunits were prominently detected in the PSII monomer in both ohp1 and ohp2 mutants (Fig. 2D). In addition, in the two mutants, more than half of CP43 and CP47 were detected in the low molecular positions, which may represent the PSII assembly intermediates pre-CP43 and pre-CP47. The levels of PSII supercomplexes and PSII dimer were extremely low in both mutants (Fig. 2D). These data indicate that the efficient biogenesis and/or stability of PSII complexes are impaired in the absence of OHP1 or OHP2.
Formation of the PSII RC Is Blocked in the ohp1 and ohp2 Mutants
To investigate how OHP1 and OHP2 are involved in the normal accumulation of PSII, we first investigated whether OHP1 and OHP2 are required for the transcription of the psbA and psbD genes, which encode the PSII core subunits D1 and D2, respectively, by RNA-blot analysis. As shown in Supplemental Figure S3A, the levels of the psbA and psbD transcripts were comparable to those in wild-type plants. We next analyzed the association of polysomes with psbA and psbD transcripts by Suc density gradient centrifugation. The results showed that the polysome profiles of psbA and psbD RNA were almost identical in ohp1, ohp2, and wild-type plants (Supplemental Fig. S3B). These results suggest that OHP1 and OHP2 are unlikely to be involved in the transcription or translation initiation of the D1 and D2 subunits.
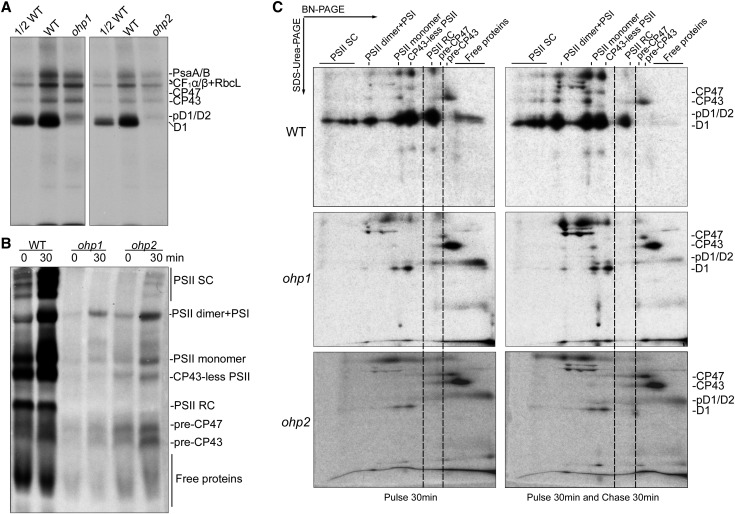
To investigate the possibility that OHP1 and OHP2 are required for protein synthesis and/or PSII complex assembly, the de novo synthesis of chloroplast-encoded proteins was studied by in vivo pulse labeling with [35S]Met (Fig. 3). Primary leaves of 12-d-old seedlings were first incubated with cycloheximide to inhibit the translation of nucleus-encoded proteins and then labeled with [35S]Met for 30 min. After pulse labeling, thylakoid proteins were isolated from equal amounts of leaves and fractionated by SDS-urea-PAGE. Newly synthesized chloroplast-encoded proteins were visualized by autoradiography. As shown in Figure 3A, the levels of radiolabeled PSII core proteins CP43 and CP47, the PSI core proteins PsaA and PsaB, the α- and β-subunits of the chloroplast ATP synthase, as well as RbcL, known to bind to thylakoid membranes, in the ohp1 and ohp2 mutants were reduced less than 2-fold compared with the wild type (Fig. 3A; Zhang et al., 2018). However, the amount of newly synthesized pD1/D1/D2 proteins in both mutants was reduced drastically in comparison with wild-type plants (Fig. 3A). This result indicates that the de novo synthesis of D1 and D2, especially the D1 protein, is strongly impaired in the absence of OHP1 and OHP2. Reduced synthesis of other chloroplast-encoded proteins may be a secondary effect due to a deficiency in PSII accumulation and/or to other causes linked to the loss of OHP1 and OHP2.
Figure 3.
Thylakoid protein synthesis and assembly of PSII complexes in ohp1, ohp2, and wild-type (WT) plants. A, In vivo pulse labeling of plastid-encoded thylakoid proteins of wild-type, ohp1, and ohp2 plants. Primary leaves were first incubated with cycloheximide for 30 min to inhibit the synthesis of nucleus-encoded proteins and then were pulse labeled with [35S]Met for 30 min. Thylakoid membrane proteins were isolated and fractionated by SDS-urea-PAGE. Radiolabeled proteins were visualized by autoradiography. B, Assembly kinetics of the PSII complexes from the ohp1, ohp2, and wild-type plants. After labeling for 30 min, primary leaves were further incubated with excess cold Met (1,000 times higher than [35S]Met) for a 30-min chase. Thylakoid protein complexes were separated by BN-PAGE, and newly assembled complexes were visualized by autoradiography. C, 2D BN/SDS-urea-PAGE analysis of the assembly of the thylakoid protein complexes. Protein complexes were pulse labeled and chased as in B and then separated by 2D BN/SDS-urea-PAGE, and the labeled proteins were detected by autoradiography. SC, Supercomplexes.
To investigate the assembly kinetics of PSII complexes in the ohp1 and ohp2 mutants, radiolabeled leaves were further chased for an additional 30 min with unlabeled Met. After labeling and chase, thylakoid protein complexes were separated by BN-PAGE and 2D SDS-urea-PAGE (Fig. 3, B and C). Newly synthesized PSII subunits were incorporated into various PSII complexes, including the PSII RC, CP43-less PSII, PSII monomer, PSII dimer, and PSII supercomplexes, after labeling for 30 min in the wild-type plants. During the 30-min chase, free proteins including pD1/D2 and the PSII RC were incorporated progressively in the large PSII complexes in the wild-type plants (Fig. 3, B and C). However, in both the ohp1 and ohp2 mutants, PSII RC formation was impaired. Most of the newly synthesized PSII subunits were present in the pre-CP47 and pre-CP43 complexes as well as in the form of free pD1/D2 proteins after labeling and chase (Fig. 3, B and C). In addition, a small amount of PSII subunits was directly assembled into PSII dimer, PSII monomer, and CP43-less complexes without formation of the PSII RC in both mutants (Fig. 3, B and C). These results indicate that OHP1 and OHP2 are essential for the assembly of the PSII RC during the early stages of PSII biogenesis.
In summary, our radiolabeling and chase experiments demonstrate that OHP1 and OHP2 are jointly required for the synthesis of D1 and D2 and more essentially for the assembly of the PSII RC.
OHP1, OHP2, and HCF244 Are Present in the PSII RC-Like Complex at Early Stages of PSII Biogenesis
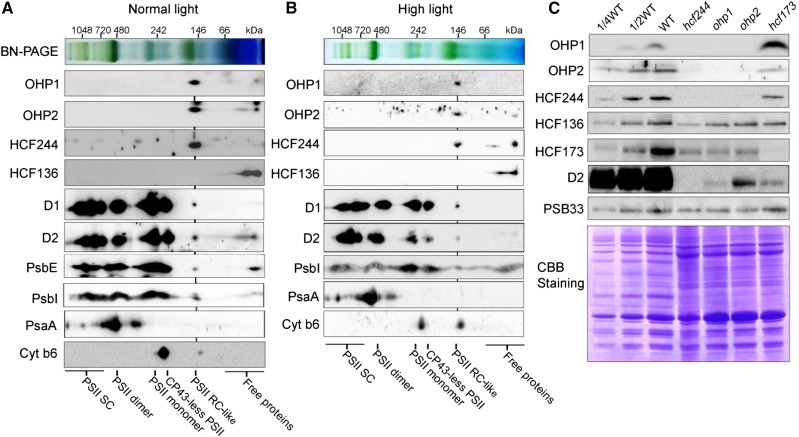
Since OHP1 and OHP2 are essential for PSII RC formation (Fig. 3), they may function as components of this newly synthesized RC complex. To investigate this possibility, thylakoid membranes were solubilized with 1% (w/v) dodecyl-β-d-maltopyranoside and protein complexes were separated by 2D BN/SDS-urea-PAGE. Subsequent immunoblots using antibodies against OHP1 and OHP2 showed that both proteins are present in a putative complex with a molecular mass of about 150 kD (Fig. 4A). Our results indicate that HCF244, but not HCF136, also comigrates with OHP1 and OHP2 in the BN-PAGE, suggesting the association of HCF244 with the two OHP proteins (Fig. 4A), which is consistent with previous reports showing that HCF244 can be copurified with OHP1 and OHP2 (Hey and Grimm, 2018; Myouga et al., 2018). A trace amount of D1, D2, PsbE, and PsbI also was detected at the same position in the BN-PAGE (Fig. 4A), suggesting that this complex contains OHP1, OHP2, HCF244, D1, D2, PsbE, and PsbI. Although we have no evidence for the presence of PsbF in this complex, PsbF is likely one of the components because PsbE and PsbF form a Cyt b559 heterodimer in thylakoids. To distinguish the RC, which contains D1/D2/PsbI/Cyt b559, in the final PSII complex from this complex, which is formed during the PSII biogenesis process, we designated the latter as the PSII RC-like complex.
Figure 4.
Analysis of the putative complex containing OHP1, OHP2, and HCF244. A, BN-PAGE analysis of the PSII RC-like complex containing OHP1, OHP2, and HCF244. Thylakoid protein complexes separated by 2D BN/SDS-urea-PAGE were detected by immunoblot analysis using the indicated antibodies. The positions of the various PSII complexes are shown. B, BN-PAGE analysis of the PSII RC-like complex from mature leaves treated with high light (1,200 μmol photons m−2 s−1 for 8 h). C, Immunoblot analysis of OHP1, OHP2, HCF244, HCF173, PSB33, and HCF136 from various mutants as indicated. Freshly isolated thylakoid membranes from hcf244, ohp1, ohp2, hcf173, and wild-type (WT) plants were separated by SDS-PAGE and immunoblotted with the indicated antibodies. A portion of the gel stained with Coomassie Brilliant Blue (CBB) is shown as a loading control. SC, Supercomplexes.
We also investigated the distribution of OHP1, OHP2, and HCF244 in the thylakoid membrane from mature leaves treated with high light. Under these conditions, PSII repair rather than PSII de novo synthesis occurs. Our results show that most of the OHP1, OHP2, and HCF244 proteins are still present at the position of the PSII RC-like complex (Fig. 4B). These results indicate that OHP1, OHP2, and HCF244 are present in the PSII RC-like complex not only during the de novo synthesis of PSII but also during the repair of PSII in the mature leaves.
We further determined their accumulation in the ohp1, ohp2, hcf244, and hcf173 mutants by immunoblotting (Fig. 4C). The results show that OHP1, OHP2, and HCF244 are barely detectable in the ohp1, ohp2, and hcf244 mutants but accumulate normally in the hcf173 mutant (Fig. 4C). It is interesting that OHP1 protein is overaccumulated in the hcf173 mutant compared with wild-type plants (Fig. 4C), suggesting that there are some unknown interaction links between HCF173 with OHP1 but not with OHP2 and HCF244. In a reciprocal way, HCF173 protein level in the ohp1, ohp2, and hcf244 mutants was reduced to half of the wild type level (Fig. 4C). The levels of HCF136 and PHOTOSYSTEM II PROTEIN33 (PSB33) were almost not changed in all mutants compared with the wild type (Fig. 4C; Fristedt et al., 2015). These results indicate that the stable accumulation of OHP1, OHP2, and HCF244 is dependent on the presence of the others, explaining why the newly synthesized PSII subunits could not be assembled into the PSII RC in the ohp1 and ohp2 mutants (Fig. 3). Thus, we conclude that the PSII RC-like complex contains OHP1, OHP2, and HCF244 as well as D1/D2/PsbI/Cyt b559 and that this complex forms at early stages during PSII assembly.
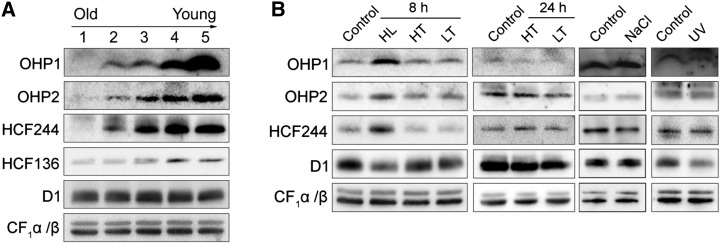
The expression of OHP1, OHP2, and HCF244 at different leaf development stages and under various environmental conditions was investigated further by immunoblot analyses (Fig. 5). The results showed that their amounts decreased gradually with the development of the leaves (Fig. 5A; Supplemental Fig. S4). The PSII assembly factor HCF136 also exhibits a similar accumulation pattern (Fig. 5A). The high levels of these proteins in immature leaves coincide with a highly active biogenesis of the PSII complex. Furthermore, the accumulation of OHP1, OHP2, and HCF244 was enhanced under high-light conditions, in which the level of PSII was decreased and the PSII complex needed to be repaired (Fig. 5B). No change of OHP1, OHP2, and HCF244 level was found under other stress conditions, such as high temperature, low temperature, salt treatment, and UV light illumination (Fig. 5B). These results imply that the induction of OHP1, OHP2, and HCF244 expression is specific to high light.
Figure 5.
OHP1, OHP2, and HCF244 exhibit a leaf age-dependent reduction and are induced by high light. A, Immunodetection of the OHP1, OHP2, and HCF244 proteins during leaf development. Five-week-old wild-type plants were used in this study. The leaves corresponding to lanes 1 to 5, representing the leaves from old to young, are shown in Supplemental Figure S4. Thylakoid membranes were loaded on an equal protein level and probed with the indicated antibodies. B, Immunodetection of OHP1, OHP2, and HCF244 under stress conditions. HL, HT, and LT represent high light, high temperature, and low temperature, respectively. After treatment, thylakoids were isolated and subjected to SDS-urea-PAGE analysis. The proteins were probed with the indicated antibodies. An immunoblot with an antibody against CF1α/β was used as a loading control.
OHP1 and OHP2 Are Tightly Integrated in the Stromal Thylakoid Membranes
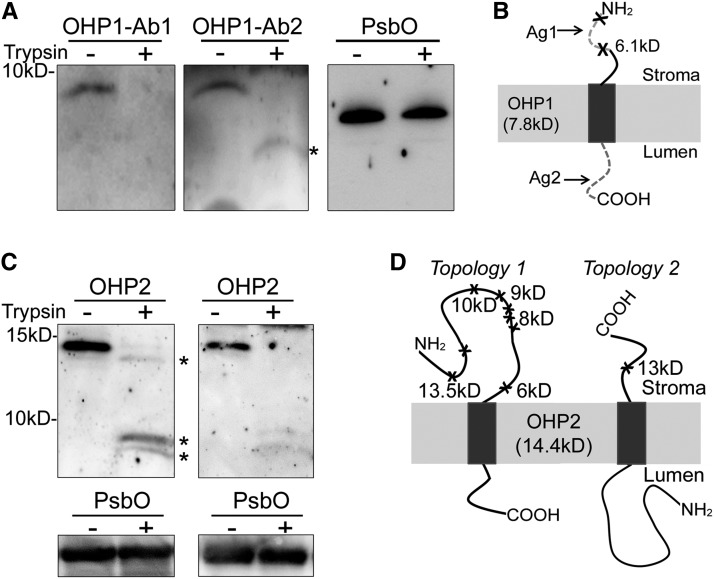
Both OHP1 and OHP2 contain an LHC domain, suggesting that they are integrated into thylakoid membranes (Andersson et al., 2003). To better understand the function of OHP1 and OHP2, we determined their topology in the thylakoids. Freshly isolated thylakoids were subjected to digestion with trypsin, which only had access to the stroma-exposed face. For OHP1, we produced two antibodies against the N- and C-terminal peptides of OHP1 (Fig. 6, A and B). After the digestion of thylakoids with trypsin, the antibody against the C-terminal peptides of OHP1 (Ab2) recognized an OHP1 fragment with a predicted molecular mass of 6.1 kD, but immunoblotting with the antibody against the N-terminal peptide of OHP1 (Ab1) did not detect any fragment (Fig. 6A), indicating that the N terminus of OHP1 was digested by trypsin. Thus, we conclude that the OHP1 N terminus is located on the stromal side of the thylakoids while its C terminus is in the thylakoid lumen (Fig. 6B).
Figure 6.
Topologies of OHP1 and OHP2. A, Immunoblot analyses of wild-type thylakoids treated with (+) or without (−) 10 µg mL−1 trypsin for 5 min. After treatment, proteins were separated and probed using two antibodies against OHP1 (Ab1 and Ab2) and the antibody against PsbO. PsbO located on the lumenal side of thylakoids was used as a control. * Indicates degradation fragment of OHP1. B and D, Schematic representation of the OHP1 topology (B) and two possible OHP2 topologies (D). Ag1 and Ag2 represent the peptides used to produce the OHP1-Ab1 and OHP1-Ab2 antibodies (used in A), respectively. The cleavage sites for trypsin are marked with crosses. The molecular sizes close to the marked crosses indicate the sizes of the remaining OHP fragments in thylakoids after cleavage at these sites with trypsin. C, Immunoblot analyses of wild-type thylakoids treated with (+) or without (−) 1 µg mL−1 (left) or 10 µg mL−1 (right) trypsin for 5 min. A concentration of 1 µg mL−1 trypsin was used for the mild digestion of thylakoids to detect the fragments resulting from partial digestion. * Indicates degradation fragments of OHP2. Immunoblots with PsbO antibody were used as controls.
For OHP2, we produced an antibody against the whole mature OHP2 protein. If the N terminus of OHP2 faces the chloroplast stroma, trypsin digestion will result in several OHP2 fragments with molecular masses ranging from 6 to 13.5 kD (Topology 1 in Fig. 6D); in the other case, only a 13-kD fragment of OHP2 will be produced (Topology 2 in Fig. 6D). Our immunoblots showed that two fragments below 10 kD were detected after digestion with trypsin at a concentration of 10 µg mL−1 (right gel in Fig. 6C). When thylakoids were treated with a lower concentration of trypsin (1 µg mL−1), one additional band with a molecular mass slightly less than that of OHP2 was detected (left gel in Fig. 6C). Detection of these proteolytic fragments after trypsin treatment indicates that, similar to OHP1, the N terminus of OHP2 faces the chloroplast stroma (Topology 1 in Fig. 6D). Taken together, our results demonstrate that both OHP proteins insert their LHC motifs into the thylakoid membrane with the same orientation as helix 1 and helix 3 in Lhcb subunits (Liu et al., 2004; Büchel, 2015).
The LHC Domain Is Important for the Function of OHP1 and OHP2
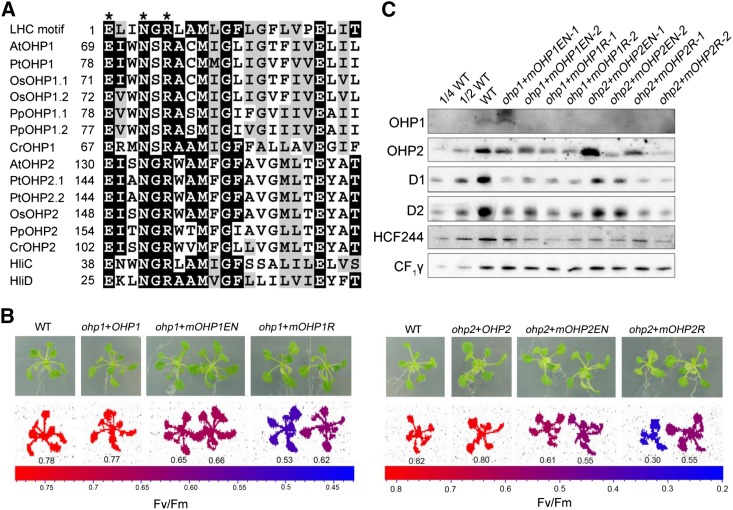
Both OHP1 and OHP2 proteins contain one characteristic LHC motif. Protein alignment analysis showed that the LHC motifs of OHP1 and OHP2 homologs in photosynthetic eukaryotes are well conserved, especially the conserved pigment-binding motif E-X-X-H/N-X-R (Fig. 7A), in which the residues E and H/N are responsible for the binding of the central Mg2+ of chlorophyll (Hoober and Eggink, 1999; Eggink and Hoober, 2000). The Glu residue from one LHC motif and the Arg residue from another LHC motif also form a salt bridge, which is important for chlorophyll binding as well as for protein folding and stability (Kühlbrandt et al., 1994). Conservation of these residues in OHP1 and OHP2 suggests that they may bind chlorophyll in vivo. To test this possibility, we mutated the Glu (E) and Asn (N) residues of the LHC motif to Ala (mOHP1-EN and mOHP2-EN, respectively) and also the Arg (R) to Ala (mOHP1-R and mOHP2-R, respectively; Fig. 7B). The mutated genes were introduced into ohp1 and ohp2 mutants by transformation to test the importance of these residues. The results showed that the Fv/Fm ratio was increased to about 0.5 to 0.6 in the transformed plants, significantly lower than in the wild type (Fig. 7B), which is consistent with the levels of D1 and D2 proteins (Fig. 7C).
Figure 7.
Mutagenesis of the LHC motif in OHP1 and OHP2. A, Sequence comparison of the LHC motif in OHP1 and OHP2. At, Arabidopsis; Pt, Populus trichocarpa; Os, Oryza sativa; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii; HliC and HliD are from Synechocystis 6803. Three residues important for chlorophyll binding are labeled with asterisks at the top of the protein sequences. The generic LHC motif defined by Jansson (1999) is shown in the top line of the alignment. While identical residues are indicated on black backgrounds, similar residues are shown as black letters on gray backgrounds. B, Phenotypes of ohp1 and ohp2 plants complemented with mutated OHP1 and OHP2 proteins. Plants were grown in MS medium for 3 weeks. Fv/Fm was measured using a closed FluorCam imaging system as described in “Materials and Methods.” ohp1+OHP1 and ohp2+OHP2, ohp1 and ohp2 mutants transformed with genomic sequences of OHP1 and OHP2, respectively; ohp1+mOHP1EN and ohp2+mOHP2EN, ohp1 and ohp2 mutants transformed with mutated OHP1 and OHP2 protein, in which Glu (E) and Asn (N) in the LHC motif were changed to Ala; ohp1+mOHP1R and ohp2+mOHP2R, ohp1 and ohp2 mutants transformed with mutated OHP1 and OHP2 protein, in which Arg (R) in the LHC motif was changed to Ala. The Fv/Fm ratio is indicated according to the color scale. C, Accumulation of the mutated OHP1 and OHP2 proteins in various genotypes. Two different lines of the ohp1 and ohp2 plants complemented with mutated OHP1 and OHP2 proteins were selected for immunoblot analyses using the indicated antibodies. An equal amount of protein was loaded, and detection of CF1γ was used as a loading control. WT, Wild type.
Immunoblot analyses showed that the mutated OHP1 and OHP2 levels were reduced in most of the transgenic plants even when the cauliflower mosaic virus 35S promoter was used to drive the expression of the mutated genes in the complemented plants (Fig. 7C). Although the levels of the mutated OHP1 and OHP2 in the two lines ohp1+mOHP1EN-1 and ohp2+mOHP2EN-1, respectively, were identical or higher than those in wild-type plants, the accumulation of their corresponding interaction partner (HCF244, OHP1, or OHP2) in thylakoids was reduced (Fig. 7C). These results indicate that the Glu, Asn, and Arg residues in the conserved LHC motif of OHP1 and OHP2 are important for the stabilization of individual proteins as well as for the formation and/or stabilization of the PSII RC-like complex. In the ohp1+mOHP1EN-1 line, levels of the HCF244, OHP1, or OHP2 proteins were more than one-half of wild-type levels. However, D1 and D2 protein amounts were only about one-fourth of wild-type levels (Fig. 7C). These results suggest that the Glu and Asn residues in the conserved LCH motif are essential for the function of OHP and that they might be involved in chlorophyll binding in vivo.
DISCUSSION
Formation of the PSII RC is the first step of PSII assembly (Nixon et al., 2010; Nickelsen and Rengstl, 2013). By radiolabeling of chloroplast proteins in combination with BN-PAGE or Suc density gradient centrifugation, pioneering studies demonstrated that the PSII RC complex is assembled with a molecular mass of about 150 kD during the biogenesis of PSII (Müller and Eichacker, 1999; Komenda et al., 2004; Rokka et al., 2005). Although this complex has been described for more than 20 years, its detailed components are still unclear.
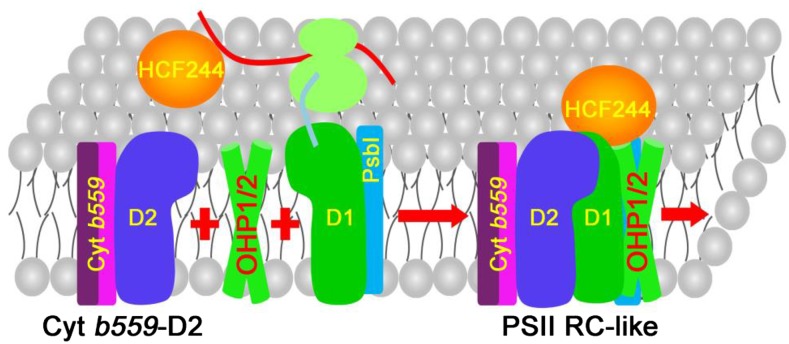
In this work, we provide evidence that this newly synthesized PSII RC complex in BN-PAGE is different from the PSII RC in the final functional PSII complex. Therefore, we have designated this complex formed during PSII biogenesis the PSII RC-like complex to make it clear that its composition is different from that of the PSII RC of the mature PSII complex. BN-PAGE and immunoblot analyses show that, in addition to D1, D2, Cyt b559, and PsbI, the PSII RC-like complex is composed of HCF244, OHP1, and OHP2 (Figs. 4 and 8). Our conclusion is supported by the following results or previous reports. (1) In the absence of OHP1 and OHP2, formation of the PSII RC-like complex is impaired in immature leaves (Fig. 3). (2) OHP1, OHP2, and HCF244 comigrate with the PSII RC-like complex in mature leaves or mature leaves treated with high light, in which the PSII repair cycle occurs (Fig. 4, A and B). (3) The stabilities of OHP1, OHP2, and HCF244 are dependent on each other (Fig. 4C). (4) A previous report indicates that HCF244 interacts with the stromal N terminus of OHP2 (Hey and Grimm, 2018). (5) Copurification of HCF244, OHP1, and OHP2 proteins can be achieved by immunoprecipitation (Myouga et al., 2018).
Figure 8.
Model for PSII RC-like complex formation. The D2-Cyt b559 precomplex functions as a target complex to accept the newly synthesized D1 protein. HCF244 and HCF173 may be involved in D1 translation and guide the proper insertion of newly synthesized D1 into the precomplex. During the cotranslational assembly of D1, OHP1 and OHP2 may form a heterodimer complex and deliver the chlorophyll and/or other cofactors to the D1/D2 subunits. HCF244 interacts with the stromal N terminus of OHP2 protein (Hey and Grimm, 2018). After assembly, OHP1, OHP2, and HCF244 remain in the PSII RC-like complex until CP47 and CP43 associate with the PSII RC. HCF173, HCF136, and PsbN are likely to act upstream of OHP1, OHP2, and HCF244 during the formation of the PSII RC and are not included in this model.
The PSII RC-like complex has a molecular mass about 150 kD. If only one copy of HCF244, OHP1, OHP2, D1, D2, Cyt b559, and PsbI is included in the complex, the calculated molecular mass of the complex should be about 140 kD, which is close to the molecular mass of the PSII RC detected in our BN-PAGE (about 150 kD; Fig. 4A). Thus, it is likely that most of the components of the PSII RC-like complex have been discovered.
Although the loss of OHP1, OHP2, and HCF244 results in a strong decrease of PSII, the extent of this decrease is slightly different in the three corresponding mutants. More PSII complex accumulates in ohp2 than in the ohp1 and hcf244 mutants (Figs. 2A and 4C), resulting in higher PSII activity (Fig. 1, A and B; Supplemental Fig. S2). To explain this difference, we have checked the OHP2 mRNA level of the mutant carefully, since the T-DNA was inserted in the intron of the OHP2 gene (Supplemental Fig. S1). We have repeated reverse transcription-PCR experiments twice, and a trace amount of OHP2 transcripts always could be detected (Supplemental Fig. S5). This raises the possibility that the leaky expression of OHP2 results in a higher level of PSII in the ohp2 mutant than in the other two mutants. It is also possible that OHP1, OHP2, and HCF244 play different roles during the formation of the PSII RC-like complex and that the absence of any of these proteins may have different effects. Overaccumulation of OHP1, but not of OHP2 and HCF244, in the hcf173 mutant supports this hypothesis. It is possible that OHP1 interacts specifically with HCF173 and is jointly involved in the translation initiation of the D1 protein (Fig. 4C; Schult et al., 2007).
What is the function of OHP1, OHP2, and HCF244 during PSII RC-like complex formation? OHP1 and OHP2 are likely to be involved in the incorporation of cofactors into the PSII RC during its biogenesis, like the related proteins HliC and HliD in Synechocystis (Chidgey et al., 2014; Knoppová et al., 2014). Although we have no direct evidence that they bind chlorophyll, several lines of evidence support this notion. Our results show that both OHP1 and OHP2 are inserted into the thylakoid membrane with the same topology as helices 1 and 3 of Lhcb proteins, in which their N and C termini face the stroma and lumen, respectively (Fig. 6; Kühlbrandt et al., 1994; Liu et al., 2004). In the Lhcb proteins, three transmembrane helices are integrated into thylakoids. While helices 1 and 3 intertwine in a left-handed supercoil, helix 2 is present in the peripheral part of Lhcb (Kühlbrandt et al., 1994). It has been proposed that the first residue, Glu, in the LHC motif functions not only in chlorophyll a binding but also in locking together two helices. Structure analysis also revealed that the fourth residue, Asn, and the sixth residue, Arg, are required for the binding of chlorophyll a (Kühlbrandt et al., 1994). Site-directed mutagenesis of these residues in OHP1 and OHP2 results in a partial loss of stabilization (Fig. 7). Although some transgenic lines accumulate more mutant OHP1 and OHP2 proteins, their PSII levels are reduced compared with wild-type plants (Fig. 7C), suggesting that OHP1 and OHP2 bind and deliver chlorophyll a in vivo.
The reduced stability of OHP1 and OHP2 with changes of the conserved residue Arg suggests that OHP1 and OHP2 may intertwine, forming a heterodimer structure similar to the corresponding structure of helices 1 and 3 of Lhcb proteins. While helices 1 and 3 of Lhcb are required for the binding of chlorophyll a, helix 2 of Lhcb binds chlorophylls a and b. However, chlorophyll b is not present in the PSII RC complex. This may be an important reason why photosynthetic organisms utilize two one-helix proteins (OHP1/OHP2 in photosynthetic eukaryotes and HliC/HliD in Synechocystis), but not three-helix proteins, for the delivery of chlorophyll a, perhaps also for other specific cofactors of the D1/D2 proteins in the PSII RC.
The recently characterized protein PSB33 also contains a LHC motif, and the formation of PSII supercomplexes is partially impaired in the psb33 mutant (Fristedt et al., 2015, 2017). Our results show that PSB33 protein stably accumulates in the ohp1, ohp2, hcf244, and hcf173 mutants (Fig. 4C), suggesting that PSB33 is involved in a different PSII assembly step than OHP1, OHP2, HCF244, and HCF173. This conclusion is consistent with previous reports that PSB33 is an integral membrane protein located in the vicinity of LHCII and the PSII CP43 RC protein (Fristedt et al., 2015, 2017).
OHP1 and OHP2 may play essential roles during the biogenesis of the PSII RC-like complex. Our pulse-chase labeling results indicate that the rate of synthesis of D1 and D2 is reduced significantly in these two mutants (Fig. 3A), which may be due to two possible causes. The first is that the translation elongation of D1 is affected in the absence of OHP1 and OHP2. It has been shown that the translation elongation of D1 is tightly coupled to its membrane insertion into the precomplex containing D2 and Cyt b559 (Zhang et al., 2000). Without OHP1 and OHP2, the insertion of D1 might be blocked at a certain stage, resulting in an impairment of the translation elongation of D1 and, consequently, in a reduced rate of synthesis of D1 (Fig. 3A). The second possibility is that the newly synthesized D1/D2 proteins are unstable, because failure of inserting cofactors into the D1/D2 proteins by OHP1 and OHP2 results in a rapid degradation before assembly into the PSII RC complex. In the ohp1 and ohp2 mutants, most of the newly synthesized D1/D2 proteins are present as free proteins in thylakoids without insertion into the PSII RC complex (Fig. 3C). This clearly implies that OHP1 and OHP2 are essential for the formation of the PSII RC complex. The increase in D1/D2 turnover rate also was observed in other mutants defective in PSII assembly, like lpa1 (Peng et al., 2006). The impairment of the de novo synthesis of the PSII RC is likely in the hcf244 mutant, because OHP1 and OHP2 are missing in this mutant (Fig. 4C).
OHP1, OHP2, and HCF244 form a stable complex with PSII RC subunits, strongly suggesting that they are not only involved in the biogenesis of the PSII RC complex but also play essential roles in the stabilization of the PSII RC-like complex after it is formed. By contrast, although the PSII assembly factors HCF136 and PsbN also are required for the formation of the PSII RC-like complex, they are unlikely to be present in the PSII RC-like complex (Fig. 4A; Plücken et al., 2002; Torabi et al., 2014), suggesting that they are not required for the stabilization of the PSII RC-like complex once the latter is assembled. While OHP1, OHP2, and HCF244 are part of PSII RC-like complex, they are not detected in the intact PSII complex (Fig. 4; Umena et al., 2011), suggesting that they are released after the association of CP47 and CP43 to the PSII RC complex (Fig. 8). OHP1, OHP2, and HCF244 also may function in the association of CP47, CP43, and other subunits with low molecular mass into PSII or in the delivery of cofactors to CP47 and CP43 during the subsequent assembly process of PSII.
In conclusion, we propose a model for PSII RC biogenesis in higher plant chloroplasts (Fig. 8). At early stages of assembly, D2-Cyt b559, OHP1, OHP2, and HCF244 may participate jointly in the integration of the D1 protein. During the cotranslational assembly process of D1, OHP1 and OHP2 may deliver chlorophyll and/or other cofactors to the D1/D2 heterodimer. After formation of the PSII RC, HCF244, OHP1, and OHP2 remain in the complex for a limited time that is essential for the efficient assembly of PSII. After the binding of CP47, CP43, and other low-molecular-mass subunits, OHP1, OHP2, and HCF244 are released from the PSII RC (Fig. 8). Our results also reveal that PSII RC formation is highly conserved among photosynthetic species. In Synechocystis, HliC/D and Ycf39 are likely to play similar roles to OHP1/2 and HCF244 in chloroplasts, respectively (Knoppová et al., 2014).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutants ohp1 (GK-362D02), ohp2 (GK-071E10), hcf173 (GK-246C02), and hcf244 (GK-088C04) were obtained from the European Arabidopsis Stock Centre. Homozygous and heterozygous mutants were identified by chlorophyll fluorescence analysis or PCR using gene-specific primers and T-DNA-specific primers (Supplemental Table S1). Sterilized seeds were planted on MS medium with 0.75% (w/v) agar and 3% (w/v) Suc. The seedlings were grown under greenhouse conditions (50 μmol photons m−2 s−1, 16-h-light/8-h-dark photoperiod, and 23°C). Heterozygous mutants were grown in soil in the greenhouse for seed production.
Complementation of the ohp1 and ohp2 Mutants
Genomic DNA of OHP1 and OHP2 was cloned into the plant expression vector pCAMBIA1301. The constructed vectors were introduced into the ohp1 and ohp2 heterozygous mutants, respectively, by the floral dip method. Transgenic seeds were selected on MS medium containing hygromycin and further confirmed by PCR. Site-directed mutagenesis was performed as described by Zhang et al. (2016), and the sequences were cloned into the pBI121 vector for plant transformation.
Chlorophyll Fluorescence Measurements and P700 Oxidation Measurements
Chlorophyll fluorescence of the plants was measured after illumination with actinic light (100 µmol photons m−2 s−1) for 1 min using a closed FluorCam imaging system (Photon Systems Instruments). Chlorophyll fluorescence induction kinetics was monitored with a mini-PAM chlorophyll fluorometer (Walz). Leaves were dark adapted for 30 min and then illuminated with low-intensity red light to determine minimum fluorescence. A saturating pulse of white light was applied to induce maximum fluorescence. The steady-state fluorescence then was recorded with actinic light (100 μmol photons m−2 s−1) for 4 min. P700 absorbance changes at 830 nm were measured with a PAM101 chlorophyll fluorometer (Walz) as described previously (Meurer et al., 1998). The 77K fluorescence emission spectrum was recorded using a spectrofluorometer (Hitachi F-4500). Thylakoid membranes with a chlorophyll concentration of 10 μg mL−1 were excited by 436-nm light, and the emission spectrum from 600 to 800 nm was recorded. The emission signals of fluorescence were normalized to the PSI emission peak around 733 nm.
TL Measurements
TL measurements were performed using the TL500/PMT system (Photon System Instruments). Arabidopsis leaves were adapted in the dark for 30 min at 22°C, and then the detached leaves were placed in the system. For better thermal contact, the leaves were covered with a layer of barbed wire (1 cm in diameter). After dropping to 0°C, the leaves were illuminated with a single actinic flash of 50 μs duration and heated to 60°C at a heating rate of 0.5°C s−1. The TL signal was recorded from 0°C to 60°C.
In Vivo Pulse-Chase Labeling of Chloroplast-Encoded Proteins
Chloroplast protein labeling and chasing were performed essentially as described previously (Meurer et al., 1998) with the following modifications (Zhang et al., 2018). For pulse radiolabeling, a total of 20 primary leaves of 12-d-old young seedlings were incubated in a buffer containing 20 µg mL−1 cycloheximide for 30 min to block the synthesis of nucleus-encoded proteins. Then, [35S]Met was added to a final concentration of 1 mCi mL−1. After labeling for 30 min under 80 μmol photons m−2 s−1 light conditions, the leaves were washed and frozen in liquid nitrogen or further chased in a buffer containing 1 mm unlabeled Met and 20 mg mL−1 cycloheximide for 30 min. After pulse labeling or chase, thylakoid membranes were isolated and the proteins were subjected to BN-PAGE or 2D BN/SDS-urea-PAGE. Radiolabeled proteins were visualized by autoradiography.
Antiserum Production
Antibodies against OHP1 protein were generated against the synthetic peptides AAKLPEGVIVPKAQPKSQC (Ab1 antibody in Fig. 6A) and CGILELIGVEIGKGLDLPL (Ab2 antibody in Fig. 6A) in the N and C terminus of OHP1, respectively. For generation of the OHP2 antibody, the sequence encoding the mature OHP2 protein (amino acids 80–172) was amplified from cDNA and then cloned into the pET28a expression vector. Expression of the recombinant proteins was induced in the host Escherichia coli strain BL21 (DE3), and the recombinant proteins were purified on an Ni-NTA agarose resin matrix under denaturing conditions. Purified protein was used to raise polyclonal antibodies in rabbits.
Thylakoid Membrane Preparation, SDS-Urea-PAGE, Tricine-SDS-PAGE, BN-PAGE, and Immunoblot Analysis
Thylakoid membranes were isolated as described previously (Zhang et al., 2016). For SDS-PAGE, thylakoid proteins were solubilized and separated by SDS-PAGE using 15% (w/v) acrylamide gels containing 6 m urea. For the detection of OHP1 as well as the proteolytic fragments of OHP1 and OHP2, Tricine-SDS-PAGE was used as described by Schägger (2006). BN-PAGE and immunoblot analysis were performed as described (Zhang et al., 2016). Antibodies against D1 (PHY0057), D2 (PHY0060), CP43 (PHY0318), CP47 (PHY0319), PsbO (PHY0344), PsaA (PHY0342), PsaD (PHY0343), CF1γ (PHY0313), CF1α (PHY0311), RbcL (PHY0346), HCF173 (PHY0326), HCF244 (PHY0327), PetD (PHY0354), Lhca2 (PHY0082S), Lhca4 (PHY0666S), PsbE (PHY0124A), PsbI (PHY0132A), and PSB33 (PHY0305) were obtained from PhytoAB.
Determination of OHP1 and OHP2 Topology
Intact chloroplasts isolated from wild-type plants were ruptured in a buffer containing 20 mm HEPES/KOH, pH 7.6. Thylakoids were resuspended in 20 mm HEPES/KOH, pH 7.6, 100 mm Suc, and 5 mm NaCl at a chlorophyll concentration of 0.1 mg mL−1. Trypsin then was added to the thylakoid samples at a final concentration of 1 or 10 μg mL−1. After incubation on ice for 5 min, thylakoids were precipitated by centrifugation for 5 min at 12,000g at 4°C, and protein gel-blot analysis was performed using specific antibodies.
Stress Treatment
For light stress, 4-week-old wild-type plants growing under greenhouse conditions (100 μmol photons m−2 s−1, 16-h-light/8-h-dark photoperiod, and 23°C) were illuminated with high light (1,200 µmol photons m−2 s −1) for 8 h. For heat and cold treatments, plants were incubated in a climate chamber at 42°C and 4°C, respectively, with a light intensity of 100 µmol photons m−2 s −1 for 8 and 24 h. For the UV-B treatment, plants were exposed to white light (100 μmol photons m−2 s−1) supplemented with UV-B for 5 h. UV-B was obtained from UVB-313 fluorescent tubes. For salt treatment, 3-week-old wild-type seedlings cultured on MS medium were transferred onto Whatman filter paper soaked with 300 mm NaCl for 8 h (100 μmol photons m−2 s−1 light).
Accession Numbers
Accession numbers are as follows: AtOHP1 (AT5G02120; Arabidopsis), AtOHP2 (AT1G34000; Arabidopsis), PtOHP1 (Potri.006G088200; Populus trichocarpa), OsOHP1.1 (Os05g22730; Oryza sativa), OsOHP1.2 (Os12g29570; O. sativa), PpOHP1.1 (Pp1s12_176V6; Physcomitrella patens), PpOHP1.2 (Pp1s33_127V6; P. patens), CrOHP1 (Cre02.g109950; Chlamydomonas reinhardtii), PtOHP2.1 (Potri.002G065000; P. trichocarpa), PtOHP2.2 (Potri.005G196100; P. trichocarpa), OsOHP2 (Os01g40710; O. sativa), PpOHP2 (Pp1s22_380V6; P. patens), CrOHP2 (Cre06.g251150; C. reinhardtii), HliC (ssl1633; Synechocystis 6803), HliD (ssr1789; Synechocystis 6803), AtHCF244 (AT4G35250; Arabidopsis), AtHCF173 (AT1G16720; Arabidopsis), AtHCF136 (AT5G23120; Arabidopsis), and AtPSB33(AT1G71500; Arabidopsis).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification and characterization of the ohp1 and ohp2 mutants.
Supplemental Figure S2. Chlorophyll a fluorescence induction analysis of the hcf173 and hcf244 mutants.
Supplemental Figure S3. RNA blot and polysome profile analysis of psbA and psbD transcripts in the ohp1 and ohp2 mutants.
Supplemental Figure S4. Selection of leaves with different ages in wild-type plants.
Supplemental Figure S5. Reverse transcription-PCR analysis of transcript levels of OHP2 in the ohp2 mutant.
Supplemental Table S1. Primers used in this work.
Acknowledgments
We thank the European Arabidopsis Stock Centre for providing the mutant seeds.
Footnotes
This work was supported by the National Natural Science Foundation of China (31470760), the National Natural Science Foundation for Outstanding Youth (31322007), and funds from the Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700).
Articles can be viewed without a subscription.
Senior authors.
References
- Andersson U, Heddad M, Adamska I (2003) Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol 132: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Aro EM (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Lohscheider JN, Albert S, Andersson U, Mendgen KW, Rojas-Stütz MC, Adamska I, Funck D (2017) Small one-helix proteins are essential for photosynthesis in Arabidopsis. Front Plant Sci 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchel C. (2015) Evolution and function of light harvesting proteins. J Plant Physiol 172: 62–75 [DOI] [PubMed] [Google Scholar]
- Chidgey JW, Linhartová M, Komenda J, Jackson PJ, Dickman MJ, Canniffe DP, Koník P, Pilný J, Hunter CN, Sobotka R (2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducruet JM, Vass I (2009) Thermoluminescence: Experimental. Photosynth Res 101: 195–204 [DOI] [PubMed] [Google Scholar]
- Eggink LL, Hoober JK (2000) Chlorophyll binding to peptide maquettes containing a retention motif. J Biol Chem 275: 9087–9090 [DOI] [PubMed] [Google Scholar]
- Eichacker LA, Helfrich M, Rüdiger W, Müller B (1996) Stabilization of chlorophyll a-binding apoproteins P700, CP47, CP43, D2, and D1 by chlorophyll a or Zn-pheophytin a. J Biol Chem 271: 32174–32179 [DOI] [PubMed] [Google Scholar]
- Fristedt R, Herdean A, Blaby-Haas CE, Mamedov F, Merchant SS, Last RL, Lundin B (2015) PHOTOSYSTEM II PROTEIN33, a protein conserved in the plastid lineage, is associated with the chloroplast thylakoid membrane and provides stability to photosystem II supercomplexes in Arabidopsis. Plant Physiol 167: 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Trotta A, Suorsa M, Nilsson AK, Croce R, Aro EM, Lundin B (2017) PSB33 sustains photosystem II D1 protein under fluctuating light conditions. J Exp Bot 68: 4281–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Liauw P, Nickelsen J, Nowaczyk M (2016) Analysis of photosystem II biogenesis in cyanobacteria. Biochim Biophys Acta 1857: 274–287 [DOI] [PubMed] [Google Scholar]
- Hey D, Grimm B (2018) ONE-HELIX PROTEIN2 (OHP2) is required for the stability of OHP1 and assembly factor HCF244 and is functionally linked to PSII biogenesis. Plant Physiol 177: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober JK, Eggink LL (1999) Assembly of light-harvesting complex II and biogenesis of thylakoid membranes in chloroplasts. Photosynth Res 61: 197–215 [Google Scholar]
- Jansson S. (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersson J, Kim SJ, Jackowski G (2000) An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible proteins. Plant Mol Biol 42: 345–351 [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Aro EM (2015) Photosystem II repair in plant chloroplasts: Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim Biophys Acta 1847: 900–909 [DOI] [PubMed] [Google Scholar]
- Jin H, Fu M, Duan Z, Duan S, Li M, Dong X, Liu B, Feng D, Wang J, Peng L, et al. (2018) LOW PHOTOSYNTHETIC EFFICIENCY 1 is required for light-regulated photosystem II biogenesis in Arabidopsis. Proc Natl Acad Sci USA 115: E6075–E6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J, Sobotka R, Tichý M, Yu J, Konik P, Halada P, Nixon PJ, Komenda J (2014) Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26: 1200–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Reisinger V, Müller BC, Dobáková M, Granvogl B, Eichacker LA (2004) Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J Biol Chem 279: 48620–48629 [DOI] [PubMed] [Google Scholar]
- Komenda J, Nickelsen J, Tichý M, Prásil O, Eichacker LA, Nixon PJ (2008) The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ (2012) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W, Wang DN, Fujiyoshi Y (1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature 367: 614–621 [DOI] [PubMed] [Google Scholar]
- Link S, Engelmann K, Meierhoff K, Westhoff P (2012) The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol 160: 2202–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Lu Y. (2016) Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front Plant Sci 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Eichacker LA (1999) Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell 11: 2365–2377 [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Takahashi K, Tanaka R, Nagata N, Kiss AZ, Funk C, Nomura Y, Nakagami H, Jansson S, Shinozaki K (2018) Stable accumulation of photosystem II requires ONE-HELIX PROTEIN1 (OHP1) of the light harvesting-like family. Plant Physiol 176: 2277–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N, Junge W (2015) Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu Rev Biochem 84: 659–683 [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: From cyanobacteria to plants. Annu Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliano C, Saracco G, Barber J (2013) Structural, functional and auxiliary proteins of photosystem II. Photosynth Res 116: 167–188 [DOI] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plöchinger M, Schwenkert S, von Sydow L, Schröder WP, Meurer J (2016) Functional update of the auxiliary proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in maintenance and assembly of PSII. Front Plant Sci 7: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Rokka A, Suorsa M, Saleem A, Battchikova N, Aro EM (2005) Synthesis and assembly of thylakoid protein complexes: Multiple assembly steps of photosystem II. Biochem J 388: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Pakrasi HB (2004) Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem 279: 45417–45422 [DOI] [PubMed] [Google Scholar]
- Schägger H. (2006) Tricine-SDS-PAGE. Nat Protoc 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P (2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JR. (2015) The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol 66: 23–48 [DOI] [PubMed] [Google Scholar]
- Shestakov SV, Anbudurai PR, Stanbekova GE, Gadzhiev A, Lind LK, Pakrasi HB (1994) Molecular cloning and characterization of the ctpA gene encoding a carboxyl-terminal processing protease: Analysis of a spontaneous photosystem II-deficient mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 269: 19354–19359 [PubMed] [Google Scholar]
- Shi LX, Hall M, Funk C, Schröder WP (2012) Photosystem II, a growing complex: Updates on newly discovered components and low molecular mass proteins. Biochim Biophys Acta 1817: 13–25 [DOI] [PubMed] [Google Scholar]
- Su X, Ma J, Wei X, Cao P, Zhu D, Chang W, Liu Z, Zhang X, Li M (2017) Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 357: 815–820 [DOI] [PubMed] [Google Scholar]
- Torabi S, Umate P, Manavski N, Plöchinger M, Kleinknecht L, Bogireddi H, Herrmann RG, Wanner G, Schröder WP, Meurer J (2014) PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 26: 1183–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (2000) Biogenesis of the chloroplast-encoded D1 protein: Regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Duan Z, Zhang J, Peng L (2016) Biogenesis factor required for ATP synthase 3 facilitates assembly of the chloroplast ATP synthase complex. Plant Physiol 171: 1291–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pu H, Duan Z, Li Y, Liu B, Zhang Q, Li W, Rochaix JD, Liu L, Peng L (2018) Nucleus-encoded protein BFA1 promotes efficient assembly of the chloroplast ATP synthase coupling factor 1. Plant Cell 30: 1770–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]