Genetic analyses reveal the relationship among the three key molecular components involved in regulating Fe-dependent root developmental responses to phosphate deficiency in Arabidopsis.
Abstract
The inhibition of primary root (PR) growth is a major developmental response of Arabidopsis (Arabidopsis thaliana) to phosphate (Pi) deficiency. Previous studies have independently uncovered key roles of the LOW PHOSPHATE RESPONSE1 (LPR1) ferroxidase, the tonoplast-localized ALUMINUM SENSITIVE3 (ALS3)/SENSITIVE TO ALUMINUM RHIZOTOXICITY1 (STAR1) transporter complex, and the SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1; a transcription factor)-ALUMINUM-ACTIVATED MALATE TRANSPORTER1 (ALMT1; a malate transporter) regulatory module in mediating this response by controlling iron (Fe) homeostasis in roots, but how these three components interact to regulate PR growth under Pi deficiency remains unknown. Here, we dissected genetic relationships among these three key components and found that (1) STOP1, ALMT1, and LPR1 act downstream of ALS3/STAR1 in controlling PR growth under Pi deficiency; (2) ALS3/STAR1 inhibits the STOP1-ALMT1 pathway by repressing STOP1 protein accumulation in the nucleus; and (3) STOP1-ALMT1 and LPR1 control PR growth under Pi deficiency in an interdependent manner involving the promotion of malate-dependent Fe accumulation in roots. Furthermore, this malate-mediated Fe accumulation depends on external Pi availability. We also performed a detailed analysis of the dynamic changes in the tissue-specific Fe accumulation patterns in the root tips of plants exposed to Pi deficiency. The results indicate that the degree of inhibition of PR growth induced by Pi deficiency is not linked to the level of Fe accumulated in the root apical meristem or the elongation zone. Our work provides insights into the molecular mechanism that regulates the root developmental response to Pi deficiency.
Plant growth and development and agricultural production depend on the availability of nutrients in soil. The deficiency of inorganic phosphate (Pi) is a common abiotic stress and has become one of the most limiting factors for agricultural production (Raghothama, 2000; López-Arredondo et al., 2014). The remodeling of root architecture (i.e. the inhibition of primary root [PR] growth and the promotion of lateral root formation) is a major developmental response of several plant species to Pi deficiency (Vance et al., 2003). Rather than a consequence of the metabolic damage caused by nutrient shortage, this response is believed to result from an active developmental reprogramming that helps plants more efficiently forage for Pi in topsoil (Abel, 2017). One piece of genetic evidence supporting this notion is that a mutation in a single gene can result in the insensitivity of PR growth to Pi deficiency (Reymond et al., 2006; Sánchez-Calderón et al., 2006; Svistoonoff et al., 2007).
The molecular mechanism that controls the Pi deficiency-induced inhibition of PR growth has been studied extensively in the model plant Arabidopsis (Arabidopsis thaliana; Desnos, 2008; Péret et al., 2011, 2014; Abel, 2017). Pi deficiency-induced inhibition of PR growth begins with a reduction in cell elongation followed by the progressive exhaustion of meristematic cells. At later stages, cell division ceases, and cells in the former elongation and meristematic regions of the PR undergo premature differentiation (Sánchez-Calderón et al., 2005). In this process, the root tip has a critical role in sensing the decrease in local, external Pi levels (Ticconi et al., 2004; Svistoonoff et al., 2007; Ward et al., 2008; Thibaud et al., 2010). In addition, Pi-deficient (−Pi) plants accumulate a high level of iron (Fe) in roots, and the presence of Fe in the growth medium is essential for the inhibition of PR growth induced by Pi deficiency (Misson et al., 2005; Hirsch et al., 2006; Svistoonoff et al., 2007; Ward et al., 2008; Zheng et al., 2009).
Over the last 20 years, many Arabidopsis mutants with altered sensitivity of PR growth to Pi deficiency have been identified. Most of them, however, also exhibited developmental abnormality under Pi sufficiency (Ma et al., 2003; Miura et al., 2005; Nacry et al., 2005; Jiang et al., 2007; Mayzlish-Gati et al., 2012; Singh et al., 2014). This indicates that these mutations, rather than specifically disturbing the Pi-sensing and -signaling pathways, have pleotropic effects. The first Pi-specific mutant identified is the Arabidopsis low phosphate response1 (lpr1) mutant that is insensitive to Pi deficiency-induced inhibition of PR growth but has a normal growth phenotype under Pi sufficiency (Svistoonoff et al., 2007). LPR1 and its homolog LPR2 belong to a multicopper oxidase family, and their mutations together have an additive effect on the PR response to Pi deficiency. Müller et al. (2015) found that LPR1 is localized to both the endoplasmic reticulum and cell walls and possesses a ferroxidase activity that converts Fe2+ to Fe3+. Pi deficiency also increases the accumulation of Fe3+ in the root apoplast (Müller et al., 2015), but in lpr1 and lpr1 lpr2, the level of Fe3+ in the root apoplast is greatly reduced. In contrast, the Arabidopsis mutant phosphate deficiency response2, which is hypersensitive to Pi deficiency-induced inhibition of PR growth, accumulates more Fe3+ in the root apoplast than the wild type (Ticconi et al., 2004; Müller et al., 2015). Müller et al. (2015) proposed that the overaccumulated Fe3+ in the apoplast of −Pi roots generates a high level of reactive oxygen species (ROS), resulting in an increased deposition of callose in cell walls and plasmodesmata. The increased callose deposition in plasmodesmata then interferes with the intercellular movement of SHR protein, a key transcription factor involved in the maintenance of the root stem cell niche in the root apical meristem (RAM), and thus impairs PR growth. Therefore, the degree of inhibition of PR growth is linked to the level of Fe3+ in the RAM. A recent study, however, indicated that callose deposition in the RAM is not required for the meristem exhaustion induced by Pi deficiency (Gutiérrez-Alanís et al., 2017).
Belal et al. (2015) and our group (Dong et al., 2017) recently identified another Pi-specific Arabidopsis mutant, hypersensitive to Pi starvation10 (hps10), whose PR growth is hypersensitive to Pi deficiency. HPS10 encodes the previously reported ALUMINUM SENSITIVE3 (ALS3), which is involved in plant tolerance to aluminum toxicity (Larsen et al., 2005). ALS3 and its interacting protein SENSITIVE TO ALUMINUM RHIZOTOXICITY1 (STAR1) form a putative ATP-binding cassette transporter complex in tonoplasts (Dong et al., 2017), but its exact transport substrate remains unknown. In contrast to lpr1 and lpr1 lpr2, hps10 (als3) and star1 overaccumulate Fe3+ in roots. Two other research groups have independently reported two other Pi-specific Arabidopsis mutants, sensitive to proton rhizotoxicity1 (stop1) and aluminum-activated malate transporter1 (almt1), whose PR growth is insensitive to Pi deficiency (Balzergue et al., 2017; Mora-Macías et al., 2017). Like als3, both stop1 and almt1 are hypersensitive to aluminum toxicity (Hoekenga et al., 2006; Iuchi et al., 2007). STOP1 is a zinc finger-type transcription factor, and ALMT1 is a malate transporter localized on the plasma membranes of root cells. STOP1 binds directly to the promoter of ALMT1 and up-regulates its transcription, resulting in the excretion of malate from root cells and an increase in plant tolerance to aluminum (Tokizawa et al., 2015). In both stop1 and almt1 mutants, the accumulation of Fe in the root apex is greatly reduced (Balzergue et al., 2017; Mora-Macías et al., 2017). Balzergue et al. (2017) proposed that the rapid inhibition of PR growth upon plant exposure to Pi deficiency results from the high accumulation of Fe in the root elongation zone (EZ), which is mediated by the STOP1-ALMT1 pathway. Following the rapid inhibition of cell elongation is a slow inhibition of cell proliferation in the RAM and meristematic cell exhaustion, in which the STOP1-ALMT1 pathway does not play a role. In contrast to Balzergue et al. (2017), Mora-Macías et al. (2017) hypothesized that the STOP1-ALMT1 pathway mediates Fe accumulation in the stem cell niche, which is responsible for the Pi deficiency-induced exhaustion of the cells in the RAM. In both studies, the authors proposed that it is the formation of a malate-Fe3+ complex that leads to the production of ROS through Fe redox cycling. Although the independent studies on the action mechanisms of LPR1, ALS3/STAR1, and STOP1-ALMT1 all point to a role of Fe in mediating PR growth under Pi deficiency, the underlying genetic relationships among these three components remain elusive. Also, additional data are needed to test the hypothesis that the degree of inhibition of PR growth occurred with the level of Fe in the EZ or in the stem cell niche.
In this work, we determined the genetic relationship among ALS3/STAR1, STOP1-ALMT1, and LPR1 and how these components interact to regulate PR growth under Pi deficiency. Our results also provide insights into the link between the inhibition of PR growth and the dynamic changes in Fe accumulation in roots during plant exposure to Pi deficiency.
RESULTS
STOP1, ALMT1, and LPR1 Act Downstream of ALS3 to Mediate Pi Deficiency-Induced Inhibition of PR Growth
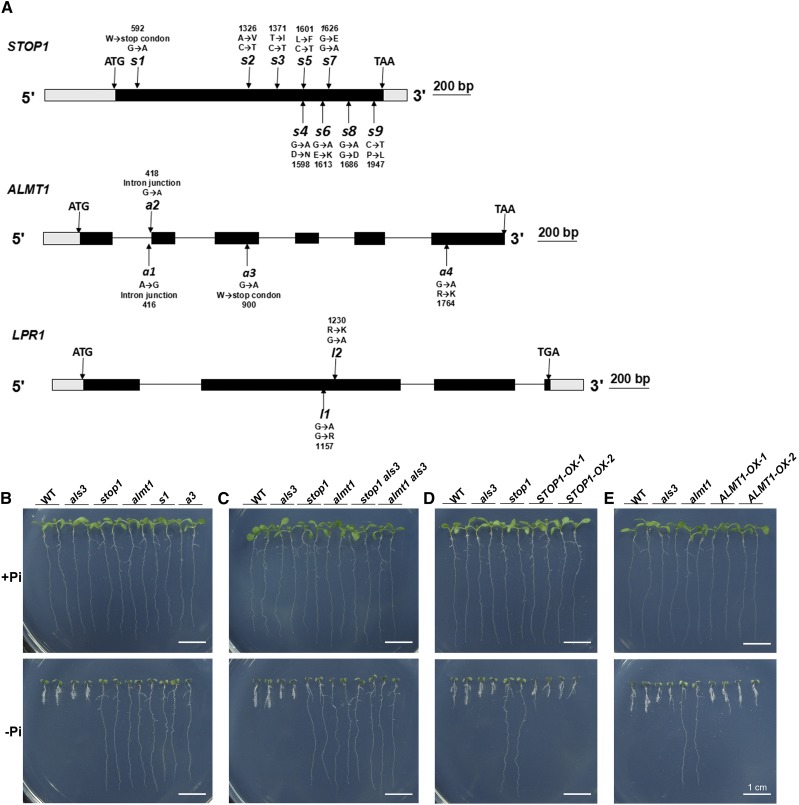
To identify the molecular components that interact with ALS3 in the regulation of PR growth under −Pi conditions, we performed a large-scale screen for the genetic suppressors of als3-3 (SALK_004094); for convenience, we hereafter refer to this line as als3. We mutagenized als3 seeds using ethylmethane sulfonate (EMS) and directly germinated M2 seeds on −Pi medium for the analysis of root growth phenotypes. At 6 d after germination (DAG), we compared the PR lengths of M2 versus wild-type seedlings. We isolated 130 suppressors of als3 whose PR growth in response to Pi deficiency was either similar to that of the wild type (Supplemental Fig. S1A) or whose PR growth was insensitive to Pi deficiency (Supplemental Fig. S1B). We then selected 43 suppressors whose PR growth was insensitive to the inhibition caused by Pi deficiency for further studies. The mutations in STOP1, ALMT1, or LPR1 were reported previously to cause PR growth to be insensitive to Pi deficiency (Svistoonoff et al., 2007; Balzergue et al., 2017; Mora-Macías et al., 2017). Therefore, we sequenced these three genes in 43 Pi deficiency-insensitive suppressors and identified nine suppressors (designated s1–s9) with a mutation in STOP1, four suppressors (designated a1–a4) with a mutation in ALMT1, and two suppressors (designated l1 and l2) with a mutation in LPR1. These mutations generated premature stop codons, caused amino acid conversions, or affected RNA splicing (Fig. 1A). In this work, we focused on these 15 suppressors that carried the mutations in STOP1, ALMT1, or LPR1. When grown on a Pi-sufficient (+Pi) medium, these suppressors did not differ morphologically from the wild type (Fig. 1B; Supplemental Fig. S2A).
Figure 1.
Molecular lesions and growth phenotypes of the genetic suppressors of als3. A, Diagrams showing the structure of the STOP1, ALMT1, and LPR1 genes and the positions of the mutations (arrows) in als3 suppressors. Black boxes, gray boxes, and horizontal lines represent the coding regions, untranslated regions, and introns, respectively. B to E, Morphologies of 6-d-old seedlings of the wild type (WT), various mutants, and als3 suppressors grown on +Pi and −Pi media. B, The wild type, als3, stop1, almt1, s1, and a3. C, The wild type, als3, stop1 als3, and almt1 als3. D, The wild type, als3, stop1, and two STOP1-OX lines. E, The wild type, als3, almt1, and two ALMT1-OX lines. Bars = 1 cm.
Because all of these suppressors had similar PR growth phenotypes under both Pi sufficiency and deficiency, s1, a3, and l1 were chosen as representative of each category for further study. We grew these three suppressors along with T-DNA insertion mutants of STOP1, ALMT1, and LPR1 on −Pi medium. As reported previously (Svistoonoff et al., 2007; Balzergue et al., 2017; Mora-Macías et al., 2017), the PR growth of stop1 (SALK_114108), almt1 (SALK_009629c), and lpr1 (SALK_016297) was insensitive to Pi deficiency (Fig. 1B; Supplemental Fig. S2A). The PR growth phenotypes of s1, a3, and l1 were similar to those of stop1, almt1, and lpr1 (Fig. 1B; Supplemental Figs. S2A and S3), suggesting that the mutations in STOP1, ALMT1, or LPR1 were responsible for the suppression of the als3 phenotype.
To confirm that the mutations in STOP1, ALMT1, and LPR1 suppress the als3 phenotypes, we crossed stop1 and almt1 with als3. Like the stop1 and almt1 single mutants, the PR growth of both the stop1 als3 and almt1 als3 double mutants also was not inhibited by Pi deficiency (Fig. 1C; Supplemental Fig. S3). Previously, we also found that the lpr1 als3 double mutant had similar PR growth to the lpr1 single mutant under Pi deficiency (Dong et al., 2017). In this work, we further generated the STOP1- and LPR1-overexpressing lines (STOP1-OX and LPR1-OX) using the Cauliflower mosaic virus (CaMV) 35S promoter (Supplemental Fig. S4, A and B). Balzergue et al. (2017) described a transgenic line that expressed an ALMT1-GFP fusion gene driven by the Arabidopsis UBQ10 promoter and could complement the almt1 mutant phenotype. We found that this UBQ10::ALMT1-GFP line (hereafter referred to as ALMT1-OX) also overexpressed the ALMT1 gene (Supplemental Fig. S4C). When these three types of overexpressing lines were grown on −Pi medium, their PRs were shorter than those of the wild type (Fig. 1, D and E; Supplemental Figs. S2 and S3). On +Pi medium, the PR growth of STOP1- and ALMT1-OX lines did not differ from that of the wild type (Fig. 1, D and E), but the PR length of the LPR1-OX line was only approximately two-thirds of that of the wild type (Supplemental Figs. S2B and S3). Taken together, these results indicate that STOP1, ALMT1, and LPR1 act downstream of ALS3 to mediate the Pi deficiency-induced inhibition of PR growth.
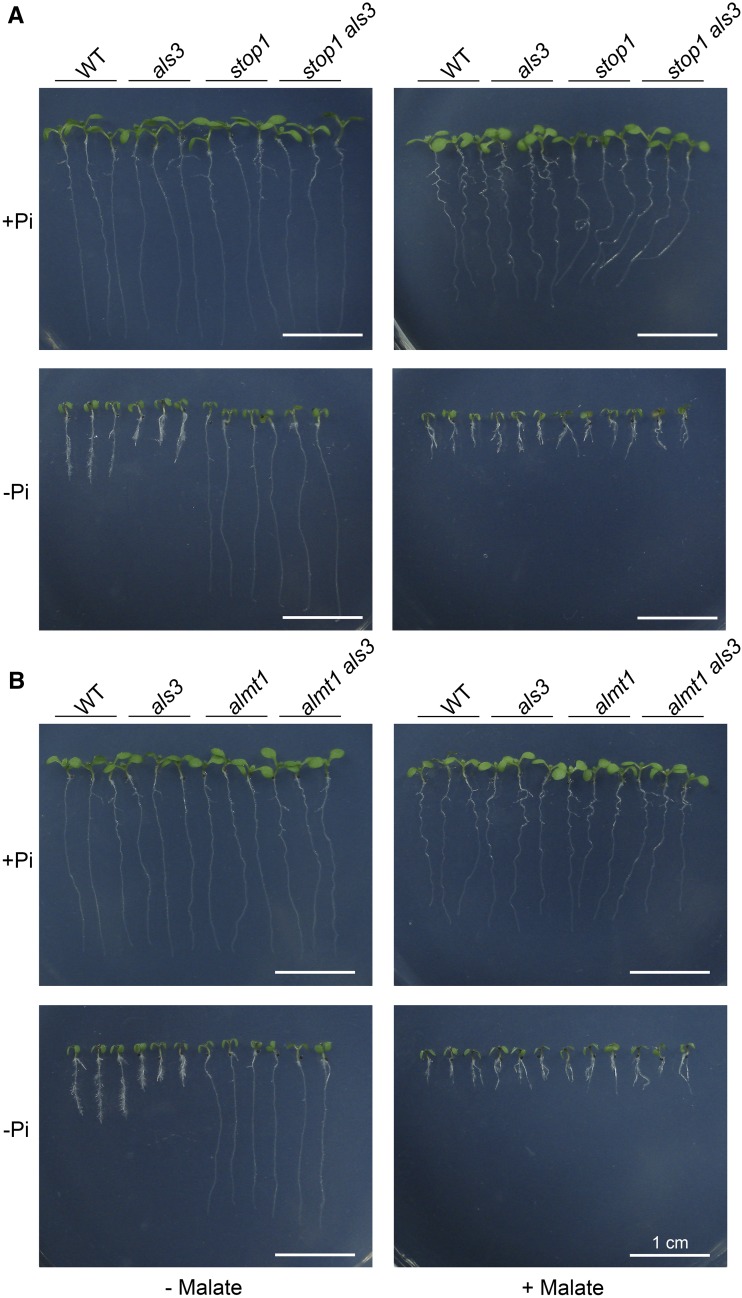
Finally, we grew the wild type and various mutants on −Pi medium supplemented with 1 mm malate. On this medium, the PR growth phenotypes of the stop1 and almt1 single mutants and of the stop1 als3 and almt1 als3 double mutants resembled those of the wild type (Fig. 2; Supplemental Fig. S5). The PR of all these lines on −Pi/malate medium was even shorter than that on −Pi medium without the addition of malate. These results further demonstrated that the suppression of the als3 hypersensitive root phenotype in the double mutants resulted from their reduced ability to excrete malate into the rhizosphere, which is consistent with the function of ALMT1 as a malate transporter.
Figure 2.
Effects of malate on the root growth of seedlings. Morphologies are shown for 6-d-old seedlings of the wild type (WT), als3, stop1, and stop1 als3 (A) and of the wild type, als3, almt1, and almt1 als3 (B) grown on +Pi and −Pi media with or without 1 mm malate. Bars = 1 cm.
Dynamic Changes in Fe Accumulation in Roots of Plants Exposed to Pi Deficiency
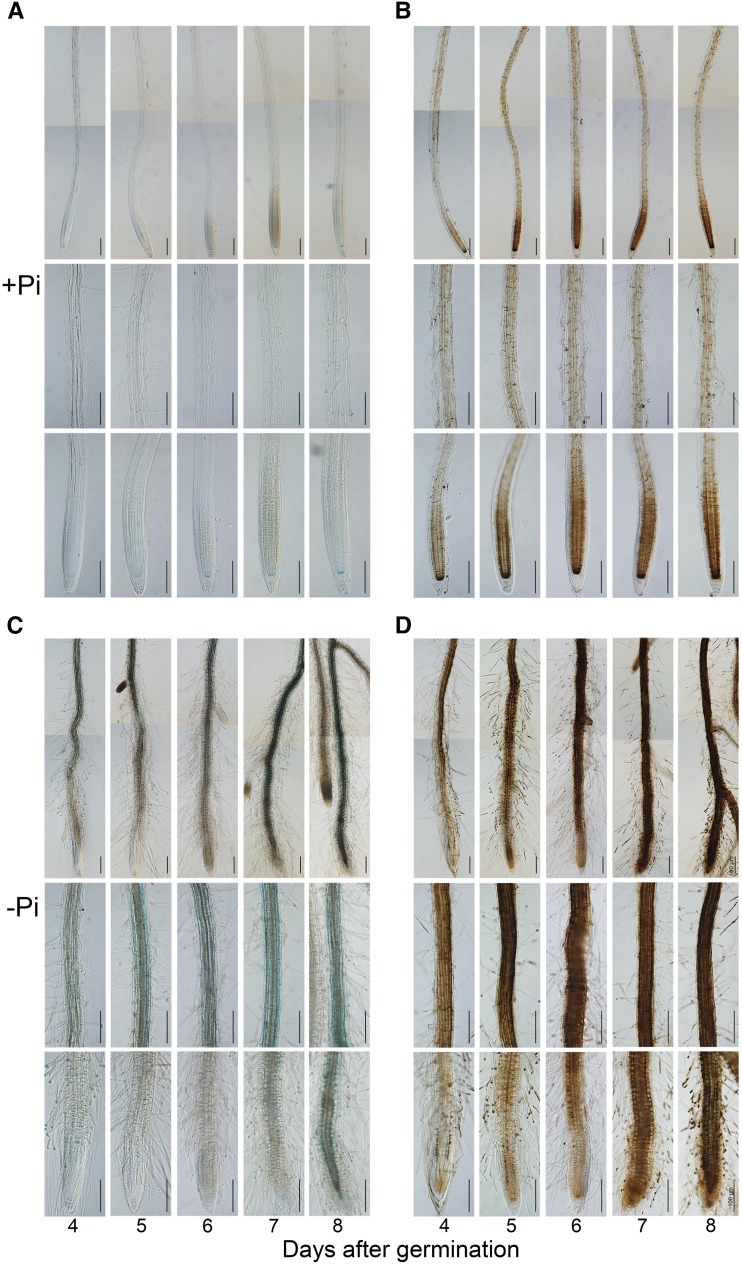
We and other researchers reported previously that the Pi deficiency-induced inhibition of PR growth depends on the accumulation of Fe in roots (Ward et al., 2008; Müller et al., 2015; Balzergue et al., 2017; Dong et al., 2017; Mora-Macías et al., 2017). To determine whether the degree of the inhibition of PR growth is linked directly to the level of Fe accumulated in a specific region of the root, we conducted a time-course analysis of Fe accumulation patterns in roots of +Pi and −Pi seedlings. We directly germinated wild-type seeds on +Pi or −Pi medium and assessed PR length and Fe accumulation at 3 to 8 DAG. PR lengths of +Pi and −Pi seedlings were similar at 3 DAG but differed thereafter (i.e. PR length was less on −Pi medium than on +Pi medium; Supplemental Fig. S6A). We used the Perls and Perls/diaminobenzidine (DAB) staining methods to assess Fe accumulation in the roots. The Perls method mainly stains Fe3+, and the Perls/DAB method stains both Fe2+ and Fe3+. The two methods revealed similar staining patterns. In the +Pi seedlings, the overall Fe staining in the RAM increased from 4 to 6 DAG and then remained unchanged from 6 to 8 DAG (Fig. 3, A and B), although the PR continued to grow from 6 to 8 DAG (Supplemental Fig. S6A). For all +Pi seedlings, Fe accumulation was substantial in the stem cell niche, which included the quiescent center and its surrounding initials. The other tissue layers of the RAM, except the epidermis, also were well stained. No Fe staining was evident in the root caps. At 4, 5, and 6 DAG, Fe accumulation in the root apex was much lower for −Pi seedlings than for +Pi seedlings (Fig. 3, C and D). During the same period, a strong Fe staining was evident in the maturation zone (MZ) of −Pi seedlings. At 4 DAG, the EZ of the −Pi seedlings had undergone premature differentiation to form root hairs, and the RAM was much smaller for the −Pi seedlings than for the +Pi seedlings. At 7 and 8 DAG, the RAM was completely exhausted in the −Pi seedlings, and the strong Fe staining along the root axis extended to the tip of the PR, which had stopped growing (Supplemental Fig. S6A). We speculated that the reduction of Fe staining in the root apex of the −Pi seedlings at 4, 5, and 6 DAG was due to the loss of viability of the cells in the RAM and the EZ.
Figure 3.
Fe accumulation patterns in roots of wild-type seedlings grown on +Pi and −Pi media. Fe in roots is shown at 4 to 8 DAG, as indicated by Perls staining (A and C) and Perls/DAB staining (B and D). The top, middle, and bottom rows in each section are photographs of the whole root, a part of the MZ, and the root tip, respectively. The root images of the top rows are composed of two tiled images. Roots of representative seedlings are shown. Bars = 100 μm.
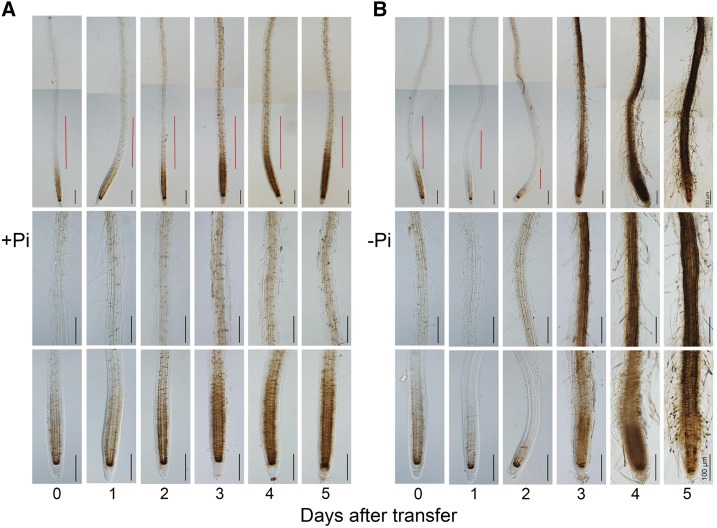
At 1 and 2 DAG, the root was too small for Fe staining, so the above direct germination method on −Pi medium was not proper to analyze the early changes in Fe accumulation when plants were exposed to Pi deficiency. Therefore, we stained seedling roots at different times after they were transferred from +Pi to −Pi medium. We first germinated wild-type seeds on +Pi medium and transferred the seedlings to +Pi or −Pi medium 3 DAG. We then recorded PR length at 0 to 5 d after transfer (DAT) and performed Fe staining using the Perls/DAB method. At 0 DAT (right before transfer), strong Fe staining was evident in the RAM of all seedlings (Fig. 4A). After the seedlings were transferred to +Pi medium, Fe staining in roots increased gradually from 1 to 3 DAT but remained unchanged from 3 to 5 DAT (Fig. 4A). These results were consistent with those of the +Pi seedlings in the nontransfer experiment (Fig. 3A). On −Pi medium at 1 DAT, Fe staining along the root axis was much less than at 0 DAT (Fig. 4B). On −Pi medium at 2 DAT, Fe staining in the RAM and EZ was still much weaker than at 0 DAT, but the Fe accumulation in the MZ began to increase; at the same time, the RAM had become much smaller (Fig. 4B), and PR growth was greatly inhibited (Supplemental Fig. S6B). This reduction in Fe accumulation also was evident in seedlings that were germinated and grown on +Pi medium for 8 d and then transferred to −Pi medium for 2 d (Supplemental Fig. S7). This result indicated that Pi deficiency rapidly reduced Fe accumulation during the early exposure time regardless of the ages of the seedlings when they were transferred to −Pi medium. For seedlings grown for either 3 or 8 d on +Pi medium before transfer to −Pi medium, Fe staining intensity in the stem cell niche was similar at 0 DAT (before transfer) versus 2 DAT to −Pi medium (Fig. 4B; Supplemental Fig. S7). From 3 to 5 DAT to −Pi medium, Fe staining along the whole root axis became stronger, and the cells in the RAM and EZ underwent premature differentiation, which caused the MZ to move close to the tip of the PR (Fig. 4B).
Figure 4.
Fe accumulation patterns in roots of wild-type seedlings at 0 to 5 DAT from +Pi medium to +Pi or −Pi medium. Fe accumulation was assessed by Perls/DAB staining. Wild-type seeds were first germinated on +Pi medium for 3 d and then transferred to +Pi medium (A) or −Pi medium (B). The top, middle, and bottom rows in each section are photographs of the whole root (the red lines indicate the positions of the EZ), a part of the MZ, and the root tip, respectively. The root images of the top rows are composed of two tiled images. Roots of representative seedlings are shown. Bars = 100 μm.
Suppression of the als3 Phenotype by the Mutation in STOP1 and ALMT1 Is Associated with the Suppression of Fe Accumulation in the Root Maturation Zone
We have shown previously that suppression of the als3 hypersensitive root phenotype by the mutation in LPR1 occurs with the suppression of Fe overaccumulation in roots of als3 (Dong et al., 2017). Therefore, we wondered whether the suppression of the als3 phenotype by the mutations in STOP1 and ALMT1 also was linked to the suppression of Fe accumulation in roots. We directly germinated seeds of the wild type, als3, stop1, almt1, stop1 als3, almt1 als3, and the STOP1- and ALMT1-OX lines on +Pi medium. At 3 DAG, we transferred the seedlings to +Pi or −Pi medium. The Fe staining patterns in roots were similar at 1 versus 2 DAT, as revealed by the Perls/DAB method (Fig. 5; Supplemental Fig. S8). At 2 DAT to +Pi or –Pi medium, Fe staining patterns in the roots were similar for all genotypes (Fig. 5). In addition, for the seedlings transferred to −Pi medium, Fe staining intensity in the stem cell niche did not obviously differ among all genotypes. These results indicated that the suppression of the als3 hypersensitive PR phenotype was not linked to the suppression of Fe accumulation in the RAM and EZ during the early exposure of plants to Pi deficiency.
Figure 5.
Fe accumulation patterns in the roots of the wild type (WT), various mutants and overexpressing lines, and the stop1 als3 and almt1 als3 double mutants as indicated by Perls/DAB staining. Seeds of the different genotypes were germinated on +Pi medium and grown for 3 d before they were transferred to +Pi medium (A) or −Pi medium (B). At 2 DAT, the roots of the seedlings were stained for Fe. The top, middle, and bottom rows in each section are photographs of the whole root, a part of the MZ, and the root tip, respectively. The root images of the top rows are composed of two tiled images. Roots of representative seedlings are shown. Bars = 100 μm.
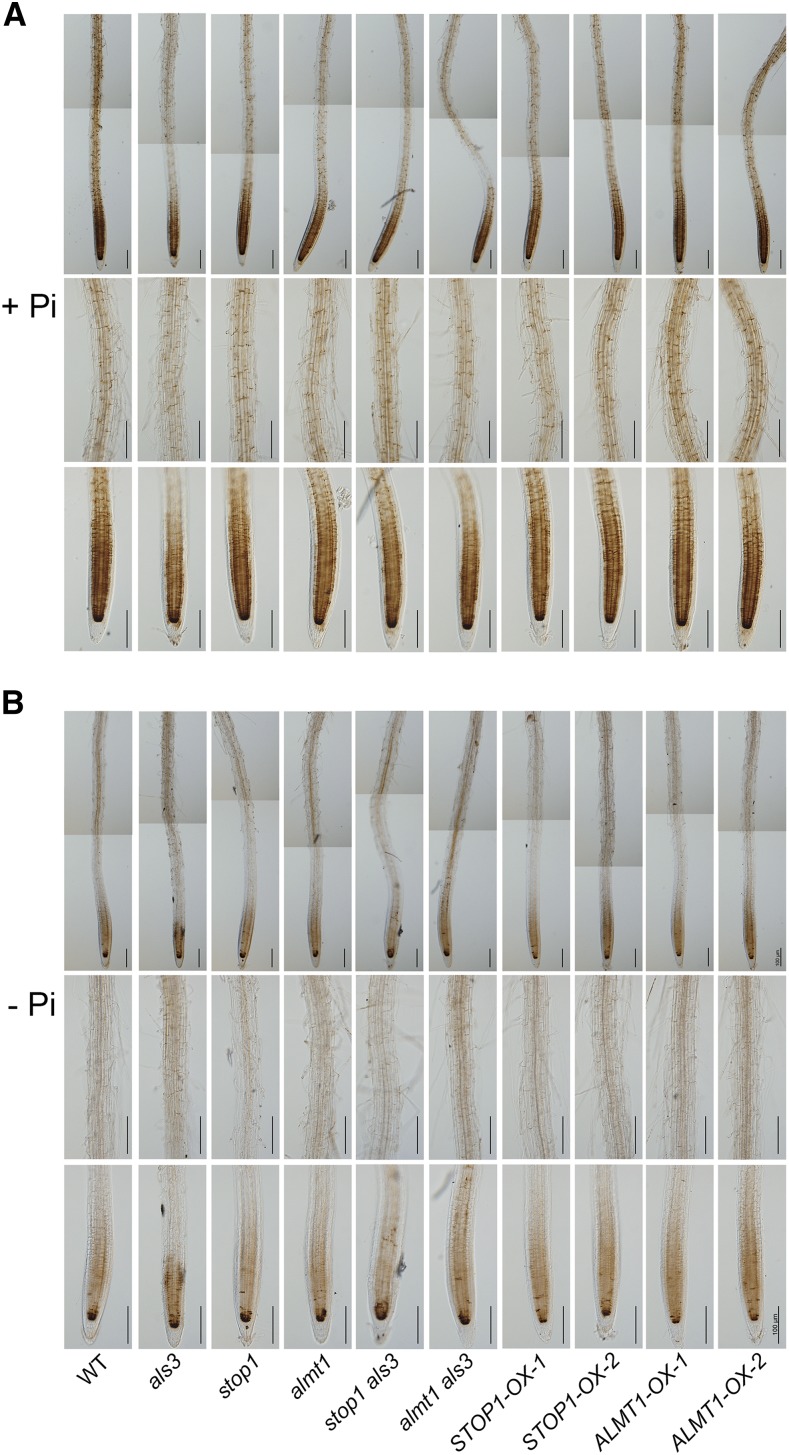
Next, we compared the levels of Fe in the MZ of the roots of −Pi seedlings among the different genotypes. We directly germinated the seeds on +Pi and −Pi media and subjected the roots to Fe staining at 6 DAG. At this stage on +Pi medium, all genotypes showed a similar Fe accumulation pattern to the wild type (Fig. 6A). On −Pi medium, however, the EZ had become invisible, and the RAM was almost completely exhausted in the wild type, als3, and STOP1- and ALMT1-OX lines (Fig. 6B). The als3, STOP1-OX, and ALMT1-OX lines, which had a hypersensitive PR phenotype, had stronger Fe staining than the wild type. In contrast, stop1, almt1, stop1 als3, and almt1 als3, which produced long PRs on −Pi medium, had weaker Fe staining than the wild type. These results indicated that the suppression of the als3 phenotype by the mutation of STOP1 and ALMT1 occurred with the suppression of Fe accumulation in the MZ of roots.
Figure 6.
Fe accumulation patterns in the roots of 6-d-old seedlings of the wild type (WT), various mutants and overexpressing lines, and the stop1 als3 and almt1 als3 double mutants grown on +Pi and −Pi media. Fe in roots grown on +Pi medium (A) or –Pi medium (B) was determined by Perls/DAB staining. The top and bottom rows in each section are photographs of the MZ and the root apex, respectively. Roots of representative seedlings are shown. Bars = 100 μm.
The fact that knockout of STOP1 or ALMT1 and overexpression of STOP1 or ALMT1 affected Fe accumulation patterns under Pi deficiency but not under Pi sufficiency (Fig. 6) also indicated that the malate-mediated accumulation of Fe in roots depends on external Pi availability.
ALS3/STAR1 Suppresses the Transcription of ALMT1 by Repressing the Accumulation of STOP1 Protein in the Nucleus
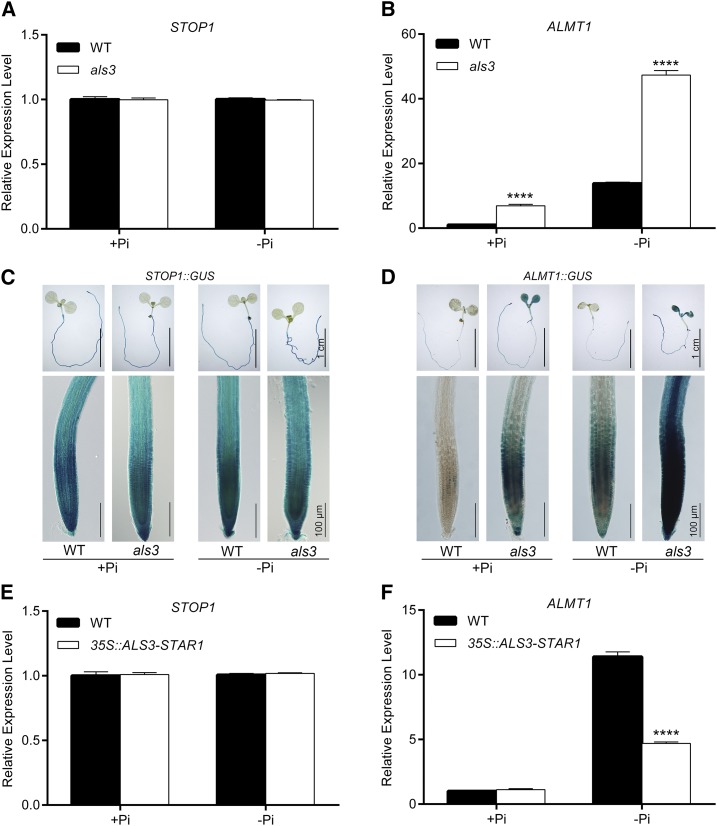
To understand how ALS3 and its partner protein STAR1 interact with STOP1 and ALMT1 to mediate PR growth under Pi deficiency, we examined the effects of the mutations in ALS3 or STAR1 on the transcription of STOP1 and ALMT1 and the effects of the mutations in STOP1 and ALMT1 on the transcription of ALS3 and STAR1. Reverse transcription quantitative PCR (RT-qPCR) analyses indicated that the mRNA levels of STOP1 in the roots of the wild type, als3, and star1 (Arabidopsis Biological Resource Center stock no. CS384144) were similar under both Pi sufficiency and deficiency (Fig. 7A; Supplemental Fig. S9A). The mRNA levels of ALMT1 in the wild type were induced by Pi deficiency, but this induction was much higher in als3 and star1 than in the wild type (Fig. 7B; Supplemental Fig. S9B). In contrast, the mRNA levels of ALS3 were unchanged in stop1 and almt1 compared with the wild type (Supplemental Fig. S9, C and D). We also analyzed the expression patterns of the GUS gene driven by the STOP1 (STOP1::GUS) or ALMT1 (ALMT1::GUS) promoter in the wild-type and als3 backgrounds. The expression level of STOP1::GUS was not altered in als3 (Fig. 7C), but the expression of ALMT1::GUS was much greater in als3 than in the wild type (Fig. 7D). Together, these results demonstrated that ALS3/STAR1 suppresses the transcription of ALMT1 but not that of STOP1.
Figure 7.
Transcriptional regulation of STOP1 and ALMT1 by ALS3. A and B, RT-qPCR analyses of the expression of STOP1 (A) and ALMT1 (B) in the roots of 6-d-old seedlings of the wild type (WT) and als3. C and D, Expression patterns of STOP1::GUS (C) and ALMT1::GUS (D) transgenes in the roots of 6-d-old seedlings in the wild-type and als3 backgrounds. The top row in each section shows whole seedlings, and the bottom row shows root tips. Bars = 1 cm (top rows) and 100 μm (bottom rows). E and F, RT-qPCR analyses of the expression of STOP1 (E) and ALMT1 (F) in 6-d-old wild type and 35S::ALS3-STAR1 seedlings. In A, B, E, and F, ACTIN2 was used as an internal control. The experiments were repeated three times, and representative results are shown. The values are means + sd of three samples each with three technical replicates. Asterisks indicate significant differences from the wild type (Student’s t test, ****, P < 0.0001).
Next, we examined the expression of STOP1 and ALMT1 in transgenic plants that overexpressed an ALS3-STAR1 fusion gene under the direction of the CaMV 35S promoter (Supplemental Fig. S10). The PR growth of this line was less sensitive to Pi deficiency than that of the wild type (Fig. 8C; Supplemental Fig. S11); that is, its phenotype was similar to that of the plants overexpressing both ALS3 and STAR1 (Dong et al., 2017). This indicated that the ALS3-STAR1 fusion protein is functionally equivalent to the ALS3/STAR1 protein complex. The mRNA level of STOP1 in the 35S::ALS3-STAR1 plant was similar to that in the wild type under both +Pi and −Pi conditions (Fig. 7E); however, the induction of ALMT1 expression was largely suppressed in this line (Fig. 7F). These results provided further evidence that ALS3/STAR1 suppresses the transcription of ALMT1 but not that of STOP1.
Figure 8.
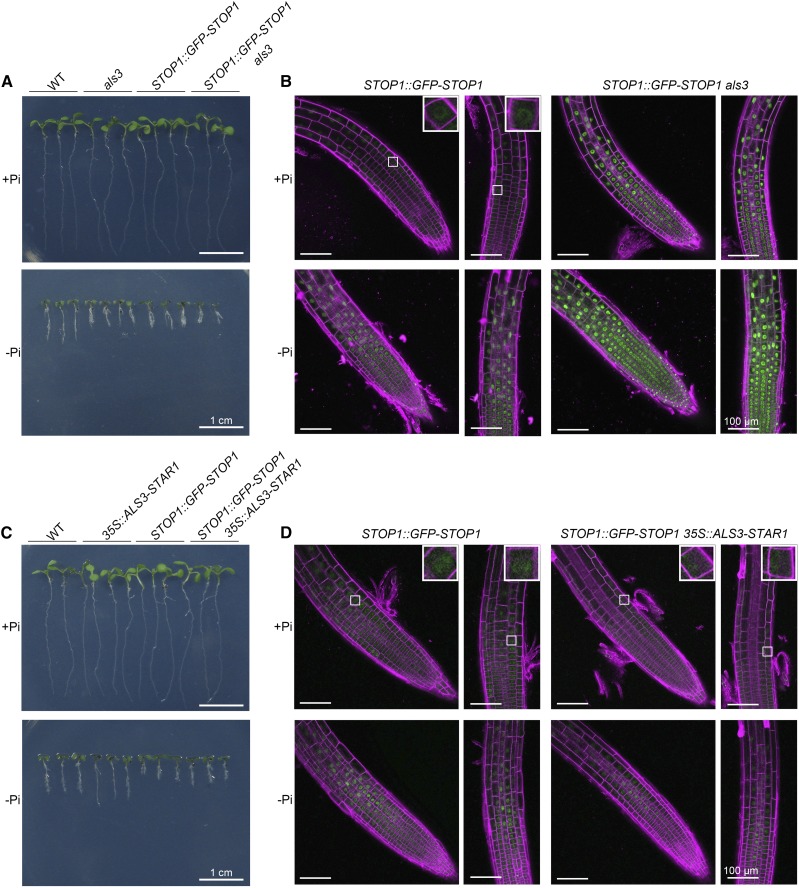
Accumulation of STOP1 protein in the nucleus of root cells of different genotypes. A and C, Morphologies of 6-d-old seedlings of the various genotypes grown on +Pi and −Pi media. B, GFP signals of STOP1::GFP-STOP1 in roots of wild-type (WT) and als3 seedlings grown on +Pi and −Pi media. D, GFP signals of STOP1::GFP-STOP1 in roots of wild-type and 35S::ALS3-STAR1 seedlings. For the observation of GFP signals, seeds with different genotypes were germinated on +Pi medium and grown for 3 d. The seedlings then were transferred to +Pi or −Pi medium for 24 h before they were photographed. The cell walls were stained with propidium iodide (shown in magenta). The insets in B and D show closeup views of representative root cells. Bars = 1 cm (A and C) and 100 μm (B and D).
STOP1 promotes the expression of ALMT1 by binding directly to its promoter (Tokizawa et al., 2015; Balzergue et al., 2017). Because ALS3/STAR1 does not affect the transcription of STOP1, we wondered whether ALS3/STAR1 suppresses the transcription of ALMT1 by modulating the accumulation of STOP1 protein in the nucleus. To test this hypothesis, we crossed a STOP1::GFP-STOP1 line (Balzergue et al., 2017) with als3 and star1. The expression of GFP-STOP1 mRNA was 8 times higher in the STOP1::GFP-STOP1 seedlings than in the wild type under both Pi sufficiency and deficiency (Supplemental Fig. S12A), and PRs were shorter for STOP1::GFP-STOP1 seedlings than for the wild type on −Pi medium (Fig. 8A; Supplemental Fig. S11). On −Pi medium, the PRs of STOP1::GFP-STOP1/als3 and STOP1::GFP-STOP1/star1 seedlings were as short as that of als3 (Fig. 8A; Supplemental Fig. S11) and star1 (Supplemental Figs. S11 and S13A). On both +Pi and −Pi media, the mRNA levels of STOP1 were similar in als3, star1, and the wild-type background (Supplemental Fig. S12B). In the STOP1::GFP-STOP1 plant, a weak GFP signal was observed in the nucleus on +Pi medium, and this signal was largely increased under −Pi conditions (Fig. 8, B and D; Supplemental Fig. S13B), as reported previously by Balzergue et al. (2017). The GFP signals in the nucleus were much stronger in the als3 and star1 plants than in the wild type under both +Pi and −Pi conditions (Fig. 8B; Supplemental Fig. S13B). When the STOP1::GFP-STOP1 construct was introduced into the 35S::ALS3-STAR1 plant, there was no obvious change in the mRNA level of STOP1 (Supplemental Fig. S12B), but the GFP fluorescence was lower than in the STOP1::STOP1-GFP plant on −Pi medium (Fig. 8D). The quantification of the GFP-STOP1 signals in different lines under both +Pi and −Pi conditions is shown in Supplemental Figure S14. Finally, we analyzed the mRNA levels of ALMT1 in the stop1 als3 double mutant by RT-qPCR. High expression of ALMT1 in als3 was suppressed completely in stop1 als3 (Supplemental Fig. S15).
Taken together, these results demonstrated that ALS3/STAR1 suppresses the transcription of ALMT1 by repressing the accumulation of STOP1 protein in the nucleus.
ALS3 and LPR1 Do Not Affect Each Other’s Transcription
That the mutation of LPR1 suppressed the als3 hypersensitive root phenotype indicated that LPR1 and ALS3 function in the same pathway and that LPR1 acts downstream of ALS3. To determine whether ALS3 and LPR1 affect the transcription of each other, we first compared the mRNA levels of LPR1 in the wild type and als3. RT-qPCR analysis indicated that, in the wild type, LPR1 was constitutively expressed under +Pi and −Pi conditions and that its expression in als3 also was similar to that of the wild type under both +Pi and −Pi conditions (Supplemental Fig. S16A). We further confirmed this result by analyzing LPR1::GUS expression in the wild type and als3. In the LPR1::GUS line (a kind gift from Dr. Steffen Abel), GUS staining was evident in the stem cell niche and in all the tissue layers in the RAM and EZs except the epidermis under both +Pi and −Pi conditions (Supplemental Fig. S16B). This expression pattern was unchanged in the als3 background. Similarly, the mRNA level of ALS3 was similar in lpr1 and the wild type under both +Pi and −Pi conditions (Supplemental Fig. S16C). These results indicated that LPR1 and ALS3 do not affect each other’s transcription.
STOP1/ALMT1 and LPR1 Mediate PR Growth under Pi Deficiency in an Interdependent Manner
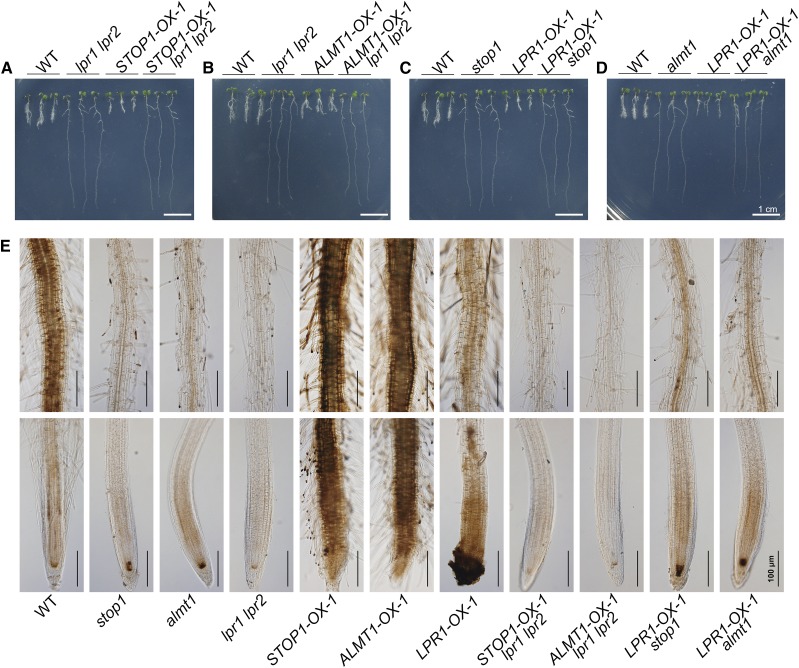
Because stop1, almt1, and lpr1 mutants had long PR phenotypes and because the mutation of STOP1, ALMT1, or LPR1 suppressed the als3 short-PR phenotype under Pi deficiency, we wanted to determine the genetic relationship between STOP1/ALMT1 and LPR1. The STOP1-OX-1 and ALMT1-OX-1 lines were crossed with lpr1 lpr2, and the LPR1-OX-1 line was crossed with stop1 or almt1. All of the lines generated through genetic crosses had growth phenotypes that resembled the phenotype of the mutants under −Pi conditions (Fig. 9, A–D). These results indicate that STOP1/ALMT1 and LPR1 mediate PR growth under Pi deficiency in an interdependent manner.
Figure 9.
Morphologies and Fe staining patterns of the roots of 6-d-old seedlings of various genotypes. A to D, Morphologies of 6-d-old seedlings of the wild type (WT), various mutants and overexpressing lines, and the lines derived from various genetic crosses grown on +Pi and −Pi media. E, Fe accumulation as indicated by Perls/DAB staining in the roots of 6-d-old seedlings of the various genotypes grown on −Pi medium. The top and bottom rows are photographs of the root MZ and the root apex, respectively. Roots of representative seedlings are shown. Bars = 1 cm (A–D) and 100 μm (E).
Previously, Balzergue et al. (2017) found that the expression of LPR1::GUS was not altered in stop1 and the expression of ALMT1::GUS was not altered in lpr1. Our RT-qPCR analyses also indicated that STOP1/ALMT1 and LPR1 did not affect each other’s transcription (Supplemental Fig. S16, A–C). Furthermore, we noticed that, in the l1 suppressor, the hyperaccumulation of ALMT1 transcripts was reduced only slightly by the lpr1 mutation (Supplemental Fig. S17), although the als3 short-root phenotype was converted to a long-root phenotype. This result provided further evidence that the mutation of LPR1 did not affect the transcription of ALMT1. Therefore, our experimental data conflicted with the report by Mora-Macías et al. (2017), who showed that the expression of LPR1 was down-regulated in stop1 and almt1 mutants. Based on our results, the interdependence of STOP1/ALMT1 and LPR1 functions apparently cannot be explained by their effects on each other’s transcription.
The Interdependence of STOP1/ALMT1 and LPR1 Functions under Pi Deficiency Involves the Promotion of Malate-Dependent Fe Accumulation in Roots
To determine the nature of the interdependence of STOP1/ALMT1 and LPR1 functions in regulating PR growth under Pi deficiency, we examined the levels of Fe accumulation in roots in various mutants and overexpressing lines by directly germinating seeds on −Pi medium. At 6 DAG, the Fe staining in the root apex among the wild type, stop1, and almt1 was similar. lpr1 lpr2 had weaker staining, and STOP1-OX-1 and ALMT1-OX-1 had much stronger staining, than the wild type (Fig. 9E). Interestingly, LPR1-OX-1 also had strong staining in the RAM, but its staining included the epidermis and root cap, which was not observed for STOP1-OX-1 and ALMT1-OX-1. The Fe accumulation in the MZ of STOP1-OX-1 lpr1 lpr2 and ALMT1-OX-1 lpr1 lpr2 was similar to that in lpr1 lpr2 (Fig. 9E), which occurred with the reversion of their hypersensitive PR growth phenotype to the insensitive PR growth phenotype (Fig. 9, A and B). The Fe staining pattern in LPR1-OX-1 stop1 and LPR1-OX-1 almt1 was similar to that in stop1 and almt1 (Fig. 9E), which occurred with the reversion of the hypersensitive PR growth phenotype of LPR1-OX-1 to the insensitive PR growth phenotypes of stop1 and almt1 (Fig. 9, C and D). Together, these results indicated that the interdependence of LPR1 and STOP1/ALMT1 in regulating PR growth under Pi deficiency involves the promotion of malate-dependent Fe accumulation in roots. Because the Fe accumulation pattern in LPR1-OX was different from that in STOP1-OX-1 and ALMT1-OX-1 (Fig. 9E), we also concluded that LPR1 and STOP1/ALMT1 regulate Fe accumulation in roots via different mechanisms.
DISCUSSION
The inhibition of PR growth is a major developmental response of Arabidopsis to Pi deficiency. Although several key components (i.e. the LPR1 ferroxidases, the ALS3/STAR1 transporter complex, and the STOP1-ALMT1 regulatory module) have been identified to regulate PR growth under Pi deficiency (Svistoonoff et al., 2007; Müller et al., 2015; Balzergue et al., 2017; Dong et al., 2017), how these components interact with each other to exert their effects is unclear.
To search for the molecular components that interact with ALS3/STAR1, we performed a genetic screen for the suppressors of als3. One group of suppressors that displayed insensitive PR growth phenotypes under Pi deficiency contained a mutation in the STOP1, ALMT1, or LPR1 gene (Fig. 1; Supplemental Fig. S2). In contrast, transgenic plants overexpressing any of these three genes exhibited opposite phenotypes. These results further confirmed the roles of STOP1, ALMT1, and LPR1 in regulating PR growth under Pi deficiency. Using genetic and molecular approaches, we had three main findings. First, STOP1, ALMT1, and LPR1 act downstream of ALS3/STAR1 in mediating PR growth under Pi deficiency (Fig. 1; Supplemental Fig. S2). Second, ALS3/STAR1 suppresses the transcription of ALMT1 by repressing the accumulation of STOP1 protein in the nucleus, but the transcription of ALS3 and STAR1 is not affected by STOP1 or ALMT1 (Figs. 7 and 8; Supplemental Figs. S9 and S13). Third, the reciprocal suppression of the hypersensitive PR growth phenotypes and Fe overaccumulation of STOP1/ALMT1-OX and LPR1-OX lines by LPR1 and STOP1/ALMT1 mutations under Pi deficiency demonstrated that the functions of these two components are interdependent (Fig. 9). This functional interdependence is not due to their effects on each other’s transcription, which differs from the report of Mora-Macías et al. (2017). Based on these three results, we propose a working model of how these molecular components mediate PR growth under Pi deficiency, as illustrated in Figure 10 and discussed in the following two paragraphs.
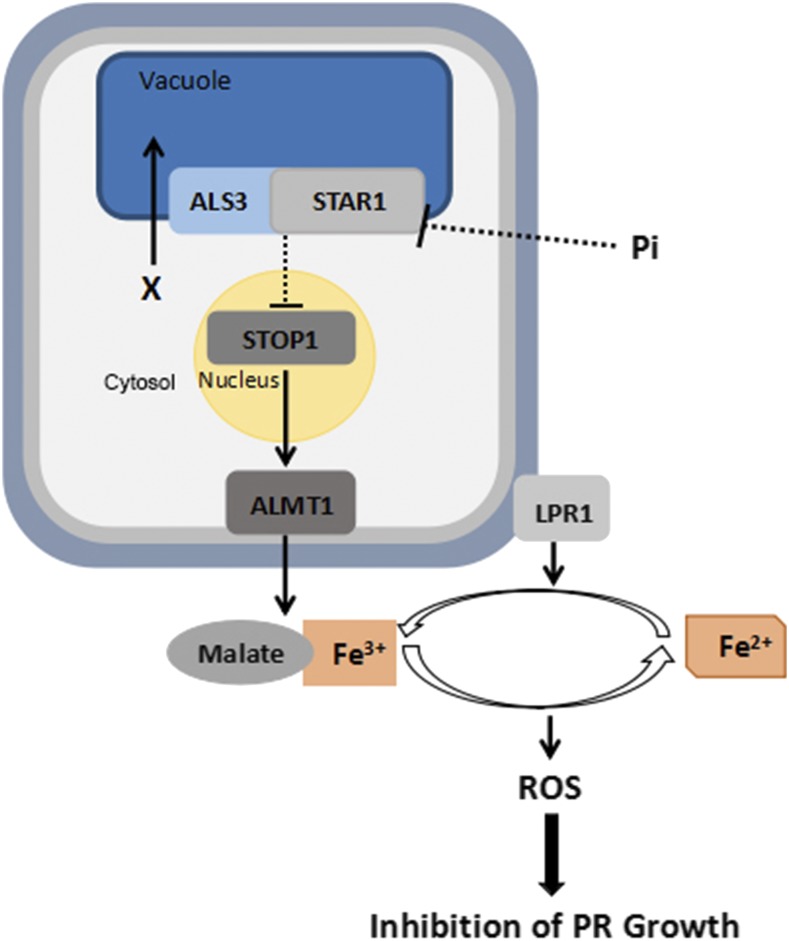
Figure 10.
Working model showing ALS3/STAR1, STOP1, ALMT1, and LPR1 interactions to regulate PR growth under Pi deficiency. Arrows indicate promotion, and perpendicular lines indicate suppression. Dotted lines indicate indirect interactions. X, An unidentified metabolite or ion; gray line surrounding the cell, the plasma membrane; light blue line surrounding the cell, the cell wall.
Under Pi sufficiency, the active ALS3/STAR1 transporter complex transports an unidentified metabolite or ion into vacuoles, which reduces the concentration of this unknown substrate in the cytosol. The low concentration of this metabolite or ion in the cytosol directly or indirectly reduces the accumulation of STOP1 protein in the nucleus, which, in turn, suppresses the transcription of ALMT1. The suppression of ALMT1 transcription results in a decreased excretion of malate into the apoplast of the root apex. The low level of malate in the apoplast, therefore, is unable to form enough malate-Fe3+ complex to allow Fe3+ to enter redox cycling to generate ROS, which could cross-link the cell wall components to inhibit cell elongation or to interfere with cell division in the RAM. The level of malate in the apoplast, however, might not be the only factor that determines the final amount of the malate-Fe3+ complex in the apoplast under Pi sufficiency. This is because the STOP1- and ALMT1-OX lines, which presumably accumulate high levels of malate in the apoplast of the root apex, still have similar levels of Fe accumulation and root growth phenotypes to the wild type. More likely, the microenvironment of the root apoplast under Pi sufficiency does not favor the binding of malate to Fe3+ to form the malate-Fe3+ complex; therefore, PR growth is not inhibited in the STOP1- and ALMT1-OX lines under Pi sufficiency.
Under Pi deficiency, an unknown mechanism reduces the protein level or the activity of the ALS3/STAR1 transporter, which results in the overaccumulation of that unidentified metabolite or ion in the cytosol. The increased accumulation of the metabolite or ion promotes the accumulation of STOP1 protein in the nucleus. The elevated abundance of STOP1 protein in the nucleus, in turn, increases the transcription of ALMT1, resulting in a high efflux of malate into the apoplast. At the same time, the microenvironment of the root apoplast becomes more favorable for the formation of the malate-Fe3+ complex. In addition, the expression of Fe acquisition genes, such as IRON REGULATED TRANSPORTER1 and FERRIC REDUCTION OXIDASE2, is suppressed dramatically in Pi-deficient roots (Misson et al., 2005; Thibaud et al., 2010; Lei et al., 2011; Li and Lan, 2015; Hoehenwarter et al., 2016) The suppressed expression of FERRIC REDUCTION OXIDASE2, which functions in reducing Fe3+ to Fe2+, also helps maintain a high level of Fe3+ in the root apoplast. Together, these changes accelerate the formation of the malate-Fe3+ complex in an LPR1-dependent manner. The formation of malate-Fe3+ complexes channels Fe3+ into redox cycling to generate ROS, which then inhibit PR growth. The requirement of the simultaneous presence of malate and Fe3+ to inhibit PR growth can well explain the nature of the interdependence of the functions of STOP1-ALMT1 and LPR1. That exogenous application of malate to −Pi medium can restore the PR growth of stop1 and almt1 single mutants and stop1 als3 and almt1 als3 double mutants to that of the wild type further supports this notion (Fig. 2; Balzergue et al., 2017; Mora-Macías et al., 2017). Our model also explains why the mutation of LPR1 could still suppress the hypersensitive PR growth phenotype of als3, although ALS3 and LPR1 do not affect each other’s transcription (Supplemental Fig. S16). This is because in the l1 suppressor, the root apoplast cannot maintain a basal level of Fe3+ to form the malate-Fe3+ complex, although the l1 suppressor has a superinduction of ALMT1 transcription (Supplemental Fig. S17D). Similarly, the STOP1-OX lpr1 lpr2 and ALMT1-OX lpr1 lpr2 lines have an insensitive PR growth phenotype, because these lines lack enough Fe3+ to form the malate-Fe3+ complex even though they contain high levels of malate in the root apoplast.
Although our work has elucidated the genetic relationship among ALS3/STAR1, STOP1-ALMT1, and LPR1 and has further confirmed the role of Fe in mediating Pi deficiency-induced inhibition of PR growth, how the inhibition of PR growth is linked to changes in Fe accumulation in roots is still an open question. Müller et al. (2015) reported that when wild-type Arabidopsis seedlings were transferred from +Pi to −Pi medium, within 20 h, Fe accumulation was increased greatly along the whole root axis, including the EZ and the stem cell niche. By comparing the Fe staining patterns in the wild type and the mutants with opposite PR growth phenotypes, Müller et al. (2015) hypothesized that the degree of inhibition of PR growth by Pi deficiency is simply linked to the levels of Fe accumulation in the stem cell niche and the EZ (i.e. the more Fe accumulated in the stem cell niche and the EZ, the more severe the inhibition of PR growth by Pi deficiency). Mora-Macías et al. (2017) found that the mutation in STOP1 and ALMT1 reduced Fe accumulation in the stem cell niche in seedlings that had been exposed to Pi deficiency for 5 d and that exogenous application of malate to −Pi medium restored Fe accumulation in the stem cell niche. Therefore, they proposed that Fe accumulation in the stem cell niche was responsible for the inhibition of PR growth by Pi deficiency. Balzergue et al. (2017), however, noticed that at 2 d after seedlings were transferred to −Pi medium, the level of Fe in the stem cell niche of stop1 and almt1 did not obviously differ from that in the wild type. Instead, they found that the Fe staining in the EZ was stronger in the wild type than in stop1, almt1, or lpr1. Therefore, they proposed that the rapid inhibition of PR growth during plant early exposure to Pi deficiency was caused by the increased accumulation of Fe in the EZ but not in the stem cell niche. Because all three hypotheses or inferences (Müller et al., 2015; Balzergue et al., 2017; Mora-Macías et al., 2017) were based on samples taken at only one or two time points after plants were exposed to Pi deficiency, or due to the different experimental conditions (e.g. different recipes were used for −Pi medium by the different research groups), the results from these studies still might not be sufficient to provide a definite answer about the relationship between the degree of inhibition of PR growth and the levels of Fe accumulation in the stem cell niche and the EZ.
To further investigate the relationship between Fe accumulation and the Pi deficiency-induced inhibition of PR growth in more detail, we carried out a time-course analysis of the Fe accumulation patterns in roots. During the first 2 d after seedlings were transferred from +Pi to −Pi medium, the overall Fe staining in the RAM and the EZ was largely decreased (Fig. 4; Supplemental Fig. S7). The levels of Fe in the stem cell niche, however, did not obviously change, which was consistent with the results reported by Balzergue et al. (2017). Furthermore, we found that, at 2 DAT, the Fe staining intensity in the stem cell niche of the wild type and various mutants and overexpressing lines was similar even though the inhibition of PR growth had already occurred (Balzergue et al., 2017; Supplemental Fig. S6B). Therefore, these results demonstrated that the rapid inhibition of PR growth by exposure to Pi deficiency was not linked to the level of Fe in the stem cell niche. We also noticed that 2 d after seedlings were transferred from +Pi medium to −Pi medium, the Fe staining intensity in the EZ and the RAM did not obviously differ among the wild type, als3, stop1, and almt1 (Fig. 5). This indicated that the degree of the inhibition of PR growth by Pi deficiency also is not simply linked to the level of Fe in the EZ. Therefore, the great reduction of Fe accumulation in the root apex in the first 2 DAT to −Pi medium might be regarded as an early stress response that prevents the damage that would result from the rapid and high production of ROS. Instead, we found that the sensitivity of PR growth of various genotypes occurred with the level of Fe in the MZ at 6 DAG. At this stage, almost the entire RAM and EZ underwent premature differentiation (Fig. 6). Because the cells in the MZ no longer elongate, the level of Fe in the MZ cannot be used to explain the differences in the sensitivity of PR growth under Pi deficiency (i.e. the level of Fe in the MZ is probably a consequence rather than a cause of the sensitivity of PR growth to Pi deficiency). In other words, the level of Fe in the MZ, therefore, might be used as an indicator of the sensitivity of PR growth to Pi deficiency. The increased accumulation of Fe in the MZ of −Pi roots might be due to the increased activity of LPR1 and ALMT1, which increased the formation of the Fe-malate complex in the root apoplast. Alternatively, the increased accumulation of Fe in the MZ of −Pi roots might be due to the increased uptake of Fe by roots because of the increased formation of root hairs induced by P deficiency. Determining the exact cause of the overaccumulation of Fe in the MZ of −Pi roots will require further investigation.
In summary, this study has elucidated the genetic relationship among the three key components in the regulatory pathway that controls the Pi deficiency-induced inhibition of PR growth. Our detailed analyses of the dynamic changes in Fe accumulation during plant exposure to Pi deficiency also indicate that the degree of the inhibition of PR growth is not simply linked to the level of Fe accumulated in the RAM or EZ. The next challenge will be to understand the molecular mechanism of how the mobilization of Fe accumulation in roots affects PR growth under Pi deficiency.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Columbia-0 background. The T-DNA insertion lines SALK_004094 (als3-3), SALK_114108 (stop1), SALK_009629C (almt1), SALK_016297 (lpr1), and CS384144 (star1) were obtained from the Arabidopsis Biological Resource Center. The lpr1 lpr2 mutant and the STOP1::GUS, ALMT1::GUS, and STOP1::GFP-STOP1 lines were generated as reported previously (Svistoonoff et al., 2007; Balzergue et al., 2017). The LPR1::GUS line was a kind gift from Dr. Steffen Abel. All double or triple mutants were generated by genetic crosses between these lines. The presence of the T-DNA in the genomic DNA of the SALK lines was confirmed by PCR analysis using primers specific for each T-DNA insertion. The primers used to confirm the T-DNA insertion are listed in Supplemental Table S1.
Arabidopsis seeds were surface sterilized in 20% (v/v) bleach for 10 min and then washed three times with sterile-distilled water. After being stratified at 4°C for 2 d, the seeds were sown on petri plates containing +Pi medium or −Pi medium. The standard +Pi medium was one-half-strength Murashige and Skoog medium with 1% (w/v) Suc, 0.1% (w/v) MES, and 0.8% (w/v) agarose (Biowest Regular Agarose G-10) or 1.2% (w/v) agar (Sigma-Aldrich). For the −Pi medium, KH2PO4 in the +Pi medium was replaced with K2SO4. The pH was adjusted to 5.8 for both +Pi and −Pi media. All experiments used agarose-containing medium, except that agar-containing medium was used for the experiments concerning the analysis of the expression of the promoter::GUS transgene. The plates with seeds were placed vertically in a growth room with a photoperiod of 16 h of light and 8 h of dark at 22°C to 24°C. The light intensity was 100 µmol m–2 s–1.
Mutagenesis and Suppressor Screening
The als3 mutant was used for suppressor screening. About 100,000 als3 seeds (M1 seeds) were incubated with 0.6% (v/v) EMS (Sigma-Aldrich; catalog no. M0880) for 10 h with gentle agitation. EMS was inactivated with 100 mm sodium thiosulfate for 10 min. The seeds were washed 10 times with sterile-distilled water and sown on +Pi medium. At 6 DAG, the seedlings were transferred to soil and grown to maturity. The M2 seeds were harvested in 1,200 pools with each pool containing seeds from 150 M1 plants. For each pool, 50 M2 seeds were grown on −Pi medium for 6 d. The seedlings with an insensitive PR growth phenotype were selected and grown in soil to produce seeds. The insensitive PR growth phenotype of each putative suppressor line was confirmed in the next generation.
Identification of stop1, almt1, and lpr1 Mutations
Genomic DNAs were extracted from the leaves of all suppressor lines. For each genomic DNA, the STOP1, ALMT1, and LPR1 genes were sequenced using primers specific for these three genes. The sequences of the primers used for sequencing are listed in Supplemental Table S1.
Quantification of PR Length
For the quantification of PR length, the seedlings were photographed and the images of roots were analyzed by the software ImageJ. For each genotype, 15 roots were used for analysis. The experiments were repeated three times, and representative results are shown.
Vector Construction and Plant Transformation
For plant overexpression vector construction, the wild-type STOP1 and LPR1 genes were PCR amplified from genomic DNA extracted from Columbia-0 plants and were cloned into the site between the CaMV 35S promoter and the NOS terminator on the pZH01 vector using the Gibson assembly cloning method (Gibson et al., 2009). The resulting constructs 35S::STOP1 and 35S::LPR1 carried a hygromycin-resistant gene as the selectable marker for plant transformation.
For the construction of the plant vector expressing an ALS3-STAR1 fusion gene, the ALS3 coding sequence (CDS) was PCR amplified from the Columbia-0 cDNA using ALS3 fusion LP/RP primers and cloned into the site between the CaMV 35S promoter and the NOS terminator on the pCAMBIA1300 vector using the Gibson assembly cloning method, which resulted in a 35S::ALS3 vector. Then, the STAR1 CDS was PCR amplified from the Columbia-0 cDNA using STAR1 fusion LP/RP primers and cloned into the site between the ALS3 CDS and the NOS terminator on the 35S::ALS3 vector using the Gibson assembly cloning method, which resulted in a 35S::ALS3-STAR1 vector. Finally, two oligonucleotides were annealed to form a double-stranded DNA fragment that encodes a 15-amino acid linker sequence. This DNA fragment was inserted into the site between the ALS3 CDS and the STAR1 CDS on the 35S::ALS3-STAR1 vector, which resulted in the final 35S::ALS3-STAR1 plant transformation vector. This vector also carried a hygromycin-resistant gene as the selectable marker for plant transformation.
These three constructs were mobilized into the Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis plants using the floral dip method (Clough and Bent, 1998). The stable transgenic lines were selected on hygromycin-containing medium. The primers used for the construction of the vectors are listed in Supplemental Table S2.
RT-qPCR
Total RNAs in roots of 6-d-old seedlings were extracted using the Highpure Total RNA Mini kit (Magen). A 2-μg quantity of RNA was reverse transcribed to cDNAs using Moloney murine leukemia virus reverse transcriptase (Takara). RT-qPCR was performed as described by Song et al. (2016). SYRB Fast qPCR Master Mix (KAPA) was used for RT-qPCR analyses on a Bio-Rad CFX96 real-time PCR system. ACTIN2 was used as an internal control. Each experiment was repeated three times with three technical replicates, and similar results were obtained. Representative results are shown. The primers used for RT-qPCR are listed in Supplemental Table S3.
Perls and Perls/DAB Staining Assays
Perls staining was performed as described previously with minor modifications (Roschzttardtz et al., 2009). In brief, the roots were excised from the seedlings and submerged directly into the staining solution of a Perls stain kit (Solarbio). After 30 min in the staining solution, the samples were rinsed with sterile-distilled water two times.
Perls/DAB staining was performed according to Balzergue et al. (2017) with minor modifications. The final concentration of DAB solution used for staining was 0.025%. The roots stained by both Perls and Perls/DAB methods were stored in 0.1 m Na-Pi buffer (pH 7.4) before being photographed. All stained samples were cleared on glass slides with an 80% (v/v) chloral hydrate glycerol clearing solution (diluted with 0.1 m Na-Pi buffer, pH 7.4). The Fe staining patterns in roots were examined using a 20× objective with a differential interference contrast microscope (Olympus BX51) equipped with a camera (Olympus DP71). For each genotype, 15 roots were subjected to Fe staining. The experiments were repeated three times, and representative results are shown.
GUS Histochemical Staining Assays
The histochemical staining and analyses of GUS activity were carried out as described by Jefferson et al. (1987). To avoid overstaining, 6-d-old STOP1::GUS, ALMT1::GUS, and LPR1::GUS seedlings in all genetic backgrounds were stained for 50 min, 3 h, and overnight, respectively.
Confocal Microscopy
Roots excised from 6-old-seedlings were dipped in 30 mm propidium iodide for 10 s to stain cell walls. The stained samples were washed twice in sterile-distilled water before the fluorescence signals were observed with an LSM710 confocal microscope (Zeiss). The excitation and emission wavelengths were 488 nm and 491 to 535 nm for GFP and 561 nm and 600 to 675 nm for propidium iodide staining, respectively. The captured fluorescence images were processed and analyzed with Zen Black and Zen Blue software. GFP signals shown in the same figure were collected using the same parameters. The intensity of GFP fluorescence signals was measured using Zen Blue software. For each genotype, five roots with 10 cells per root were used to measure the intensity of GFP fluorescence. The experiments were repeated three times.
Accession Numbers
Sequence data from this article can be found in the TAIR database under the following accession numbers: ALS3 (AT2G37330), STAR1 (AT1G67940), LPR1 (AT1G23010), LPR2 (AT1G71040), STOP1 (AT1G34370), and ALMT1 (AT1G08430).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Morphologies of M2 seedlings of EMS-mutagenized als3 grown on −Pi medium.
Supplemental Figure S2. Morphologies of wild-type, als3, lpr1, l1, and LPR1-OX seedlings grown on +Pi and −Pi media.
Supplemental Figure S3. PR length of 6-d-old seedlings of the wild type, various mutants and overexpressing lines, and the als3 suppressors grown on +Pi and −Pi media.
Supplemental Figure S4. Relative expression levels of STOP1, LPR1, and ALMT1 in roots of wild-type and overexpressing line seedlings.
Supplemental Figure S5. PR lengths of 6-d-old seedlings of the wild type and various mutants grown on +Pi and −Pi media with or without malate.
Supplemental Figure S6. Change in PR length over time of wild-type seedlings grown on +Pi and −Pi media.
Supplemental Figure S7. Fe accumulation patterns in roots of wild-type seedlings exposed to Pi deficiency.
Supplemental Figure S8. Fe accumulation patterns in roots of the wild type, various mutants and overexpressing lines, and als3 suppressors.
Supplemental Figure S9. Expression levels of STOP1 and ALMT1 in star1 seedlings and of ALS3 in stop1 and almt1 seedlings.
Supplemental Figure S10. Expression levels of ALS3 and STAR1 in wild-type and 35S::ALS3-STAR1 seedlings.
Supplemental Figure S11. PR lengths of the wild type, als3, star1, and various transgenic lines grown on +Pi and −Pi media.
Supplemental Figure S12. Expression levels of STOP1 in wild-type and STOP1::GFP-STOP1 seedlings and GFP-STOP1 in seedlings of various genetic backgrounds.
Supplemental Figure S13. Accumulation of STOP1 protein in the nucleus of root cells of the wild type and star1.
Supplemental Figure S14. Intensity of GFP fluorescence signals in seedlings of lines with the pSTOP1::GFP-STOP1 transgene in different genetic backgrounds.
Supplemental Figure S15. Expression levels of ALMT1 in wild-type, als3, and stop1 als3 seedlings.
Supplemental Figure S16. Expression levels of LPR1 in wild-type and als3 seedlings and of ALS3 in wild-type and lpr1 seedlings.
Supplemental Figure S17. Effects of mutations in STOP1/ALMT1 and LPR1 on each other’s transcription.
Supplemental Table S1. Primers used to verify all T-DNA insertion lines.
Supplemental Table S2. Primers used for the construction of plant transformation vectors.
Supplemental Table S3. Primers for RT-qPCR.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the seed stocks of the mutant lines and Dr. Steffen Abel of Leibniz Institute of Plant Biochemistry for providing the LPR1::GUS line.
Footnotes
This work was supported by the Ministry of Science and Technology of China (grant no. 2016YFD0100700) and the National Natural Science Foundation of China (grant nos. 31670256 and 31700217).
References
- Abel S. (2017) Phosphate scouting by root tips. Curr Opin Plant Biol 39: 168–177 [DOI] [PubMed] [Google Scholar]
- Balzergue C, Dartevelle T, Godon C, Laugier E, Meisrimler C, Teulon JM, Creff A, Bissler M, Brouchoud C, Hagège A, et al. (2017) Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat Commun 8: 15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belal R, Tang R, Li Y, Mabrouk Y, Badr E, Luan S (2015) An ABC transporter complex encoded by Aluminum Sensitive 3 and NAP3 is required for phosphate deficiency responses in Arabidopsis. Biochem Biophys Res Commun 463: 18–23 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Desnos T. (2008) Root branching responses to phosphate and nitrate. Curr Opin Plant Biol 11: 82–87 [DOI] [PubMed] [Google Scholar]
- Dong J, Piñeros MA, Li X, Yang H, Liu Y, Murphy AS, Kochian LV, Liu D (2017) An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol Plant 10: 244–259 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Alanís D, Yong-Villalobos L, Jiménez-Sandoval P, Alatorre-Cobos F, Oropeza-Aburto A, Mora-Macías J, Sánchez-Rodríguez F, Cruz-Ramírez A, Herrera-Estrella L (2017) Phosphate starvation-dependent iron mobilization induces CLE14 expression to trigger root meristem differentiation through CLV2/PEPR2 signaling. Dev Cell 41: 555–570.e3 [DOI] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Hoehenwarter W, Mönchgesang S, Neumann S, Majovsky P, Abel S, Müller J (2016) Comparative expression profiling reveals a role of the root apoplast in local phosphate response. BMC Plant Biol 16: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA, Cançado GM, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, et al. (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA 103: 9738–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M (2007) Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 104: 9900–9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X (2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Geisler MJ, Jones CA, Williams KM, Cancel JD (2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J 41: 353–363 [DOI] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D (2011) Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol 156: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lan P (2015) Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopsis roots. BMC Res Notes 8: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L (2014) Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65: 95–123 [DOI] [PubMed] [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP (2003) Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayzlish-Gati E, De-Cuyper C, Goormachtig S, Beeckman T, Vuylsteke M, Brewer PB, Beveridge CA, Yermiyahu U, Kaplan Y, Enzer Y, et al. (2012) Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol 160: 1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Macías J, Ojeda-Rivera JO, Gutiérrez-Alanís D, Yong-Villalobos L, Oropeza-Aburto A, Raya-González J, Jiménez-Domínguez G, Chávez-Calvillo G, Rellán-Álvarez R, Herrera-Estrella L (2017) Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc Natl Acad Sci USA 114: E3563–E3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S (2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell 33: 216–230 [DOI] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P (2005) A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T (2011) Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Péret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L (2014) Root architecture responses: In search of phosphate. Plant Physiol 166: 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Roschzttardtz H, Conejero G, Curie C, Mari S (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 51: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L (2006) Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Fridman Y, Friedlander-Shani L, Tarkowska D, Strnad M, Savaldi-Goldstein S (2014) Activity of the brassinosteroid transcription factors BRASSINAZOLE RESISTANT1 and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE RESISTANT2 blocks developmental reprogramming in response to low phosphate availability. Plant Physiol 166: 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yu H, Dong J, Che X, Jiao Y, Liu D (2016) The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet 12: e1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37: 801–814 [DOI] [PubMed] [Google Scholar]
- Tokizawa M, Kobayashi Y, Saito T, Kobayashi M, Satoshi I, Nomoto M, Tada Y, Yamamoto YY, Koyama H (2015) SENSITIVE TO PROTON RHIZOTOXICITY1, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and other transcription factors are involved in ALUMINUM-ACTIVATED MALATE TRANSPORTER1 expression. Plant Physiol 167: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Uhde-Stone C, Allan D (2003) Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, et al. (2009) Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol 151: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]