Four epidermal patterning factor-like genes expressed at the periphery of the meristem restrict shoot apical meristem size and promote leaf initiation when sensed by ERECTA family receptors.
Abstract
The shoot apical meristem (SAM) enables the formation of new organs throughout the life of a plant. ERECTA family (ERf) receptors restrict SAM size and promote initiation of leaves while simultaneously supporting establishment of correct phyllotaxy. In the epidermis and during organ elongation ERf activity is regulated by a family of Epidermal Patterning Factor-Like (EPFL) secreted Cys-rich small proteins. Here we show that ERfs play a critical role in communication between the SAM leaf boundary and the central zone in Arabidopsis (Arabidopsis thaliana). Ectopic expression of ERECTA in the central zone using the CLAVATA3 promoter is sufficient to restrict meristem size and promote leaf initiation. Genetic analysis demonstrated that four putative ligands: EPFL1, EPFL2, EPFL4, and EPFL6 function redundantly in the SAM. These genes are expressed at the SAM-leaf boundary and in the peripheral zone. Previously EPFL4 and EPFL6 have been linked with elongation of aboveground organs. Here we demonstrate that EPFL1 and EPFL2 promote organ elongation as well. In addition, we show that expression of ERECTA in the central zone of the SAM has a strong impact on elongation of internodes and pedicels and growth of leaves. These results suggest that ERfs can stimulate organ growth cell nonautonomously.
Cell-to-cell communications coordinate numerous processes during plant development. Plant cells use both small organic molecules and peptides as message carriers. Plasma membrane-localized receptor-like kinases sense the majority of peptides and some organic molecules and then activate appropriate developmental programs. The ability of a receptor to sense multiple signals and the variety of responses a signal may trigger enable the complexity and plasticity of developmental programs.
The ERECTA family (ERf) signaling pathway was initially linked to aboveground organ elongation (Torii et al., 1996). Since then it has become clear that ERf receptors also regulate numerous other developmental processes such as stomata formation, leaf initiation, shoot apical meristem (SAM) structure, and flower differentiation (Shpak, 2013). In Arabidopsis (Arabidopsis thaliana), the family consists of three genes: ERECTA, ERECTA-LIKE1 (ERL1), and ERL2 (Shpak et al., 2004). The contribution of an individual receptor to the regulation of a particular developmental response varies. For example, ERECTA is the primary receptor regulating organ elongation whereas ERL1 plays a leading role in the regulation of stomata spacing. In the SAM these receptors function redundantly with single and double mutants exhibiting extremely weak or no phenotypes (Chen et al., 2013). The activity of ERf receptors is regulated by a family of 11 secreted Cys-rich small proteins from the Epidermal Patterning Factor/EPF-like (EPF/EPFL) family (Shimada et al., 2011). Three proteins—EPF1, EPF2, and STOMAGEN (EPFL9)—regulate stomata development (Hara et al., 2007, 2009; Hunt and Gray, 2009; Hunt et al., 2010; Sugano et al., 2010). Based on the phenotypes of mutants and on the fact that EPF2 is able to induce phosphorylation of downstream signaling components, EPF1 and EPF2 are thought to activate the receptors (Hara et al., 2007, 2009; Hunt and Gray, 2009; Lee et al., 2015). STOMAGEN competes with EPF1 and EPF2 for binding to ERfs but is unable to activate the downstream cascade, and thus functions as an antagonist (Ohki et al., 2011; Lee et al., 2015). Two ligands, EPFL4 and EPFL6, stimulate aboveground organ elongation (Abrash et al., 2011; Uchida et al., 2012). Another ligand, EPFL2, has been shown to regulate the shape of leaf margins (Tameshige et al., 2016). The rice (Oryza sativa) ortholog of EPFL1 induces awn elongation (Bessho-Uehara et al., 2016). No function for the remaining four potential ligands has been established. Selection of which ligands can bind to ERf receptors on a surface of an individual cell depends on the presence of the coreceptor TOO MANY MOUTHS, which promotes binding of EPF1, EPF2, and STOMAGEN and inhibits binding of EPFL4 and EPFL6 (Lin et al., 2017). The binding of ligands to ERfs or to ERf/TOO MANY MOUTHS complexes does not cause significant conformational changes or induce homodimerization of ERfs (Lin et al., 2017). ERfs might function in a complex with receptor-like kinases of the SERK family, which could potentially assist ERfs in activation of downstream targets (Meng et al., 2015). A MAP kinase cascade consisting of YODA, MKK4/5/7/9, and MPK3/6 transmits the signal downstream of ERfs (Bergmann et al., 2004; Wang et al., 2007; Lampard et al., 2009, 2014; Meng et al., 2012). The cascade is regulated by MAP KINASE PHOSPHATASE1 (Tamnanloo et al., 2018). How the signal is transmitted from the receptors to the cascade is not known.
Here we focus on ERf signaling in the SAM, a small but complex structure that must tightly control the proliferation and differentiation of its constituent cells. The SAM contains three different regions: the central zone with a pool of undifferentiated, slowly dividing cells; the peripheral zone where leaf and flower primordia are initiated; and the underlying rib zone, which provides cells for internodes. As cells are continually transitioned from the central zone into the other two, cell-to-cell communications are essential to maintain a relatively constant number of stem cells. These communications are achieved through a negative feedback loop consisting of the receptor/ligand pair CLAVATA1 (CLV1)/CLAVATA3 (CLV3) and the transcription factor WUSCHEL (WUS) (Clark, 2001). Presumably, the rate of cell proliferation and differentiation in the peripheral zone and the rib zone is also tightly controlled to ensure a consistent rate of organ initiation and uniformity of size; however, how this is achieved is not known. In addition, leaves and flowers develop in a specific geometric pattern. In Arabidopsis the SAM forms leaves and flowers at 137.5° angles to each other, producing a spiral pattern of these organs around the stem. The formation of auxin maxima determines the position of organ primordia (Sluis and Hake, 2015). ERfs play a critical role in these processes—the vegetative SAM of er erl1 erl2 is dramatically wider and has a much broader central zone exhibiting increased expression of WUS (Chen et al., 2013; Uchida et al., 2013), and leaf primordia are initiated at a significantly reduced rate with almost random divergence angles (Chen et al., 2013). The changes in leaf initiation in er erl1 erl2 correlate with abnormal auxin distribution as determined by a DR5rev:GFP marker and decreased PIN1 expression in the vasculature (Chen et al., 2013).
To gain insight into the function of ERfs in the SAM, we explored their pattern of expression and searched for ligands that are perceived by ERfs. While ERfs are endogenously expressed throughout the SAM, their expression in the central region by the CLV3 promoter is most efficient in rescuing the meristematic defects of er erl1 erl2, compared to expression in the peripheral zone by the KANADI (KAN) promoter. Interestingly, ERECTA expression under the CLV3 promotor is also able to rescue leaf size and stem elongation phenotypes, suggesting that those parameters might be controlled by ERfs indirectly from distant tissues. Based on the phenotype of the quadruple mutant, ERfs can sense four ligands in the SAM: EPFL1, EPFL2, EPFL4, and EPFL6. Two of these ligands (EPFL1 and EPFL2) are expressed in the boundary region in the embryo and in the vegetative SAM. EPFL4 and EPFL6 are expressed at the periphery of the vegetative SAM. EPFL expression on the periphery of the meristem is critical as the epfl1 epfl2 epfl4 epfl6 mutant can be rescued by EPFL1 expressed under the KAN promoter but not CLV3. Our data suggest that ERfs coordinate development of the central zone and the peripheral regions of the SAM.
RESULTS
Expression of ERECTA in the Central Zone is Most Efficient in Regulating the SAM Size
Based on an in situ analysis and a reporter gene assay, ERfs are expressed broadly in the vegetative SAM and throughout forming leaf primordia (Yokoyama et al., 1998; Shpak et al., 2005; Uchida et al., 2013). A gene expression profile of the inflorescence SAM suggests similar expression of ERfs in the central zone, the peripheral zone, and in the organizing center with only ERL1 being upregulated in the central zone (Yadav et al., 2009). In this experiment the zones were defined by CLV3, FILAMENTOUS FLOWER, and WUS expression, respectively. We were interested in how the meristematic expression of ERfs affects plant morphology and whether ERECTA expression in a specific zone is sufficient to rescue defects observed in the er erl1 erl2 mutant. With this goal in mind, five different promoters were chosen. The SHOOTMERISTEMLESS (STM) promoter was used to express the gene throughout the SAM (Long et al., 1996). The CLV3 and WUS promoters were used to drive ERECTA expression in the central zone and in the organizing center, respectively (Mayer et al., 1998; Fletcher et al., 1999). The AINTEGUMENTA (ANT) promoter was used to induce ERECTA expression in the peripheral zone and broadly in the forming leaf primordia (Elliott et al., 1996). We expected the KAN promoter to express ERECTA at the outer edges of the peripheral zone and on the abaxial side of leaf primordia (Kerstetter et al., 2001; Yadav et al., 2014). The ERECTA sequence placed behind the described promoters contained all 26 introns. Previous work suggested that endogenous ERECTA cis-regulatory elements are localized in the promoter (Yokoyama et al., 1998), with introns being essential for mRNA stability; mRNA produced by intronless ERECTA is degraded at the 3′ end (Karve et al., 2011). Thus it is unlikely that introns contributed to the pattern of expression in the generated transgenic lines.
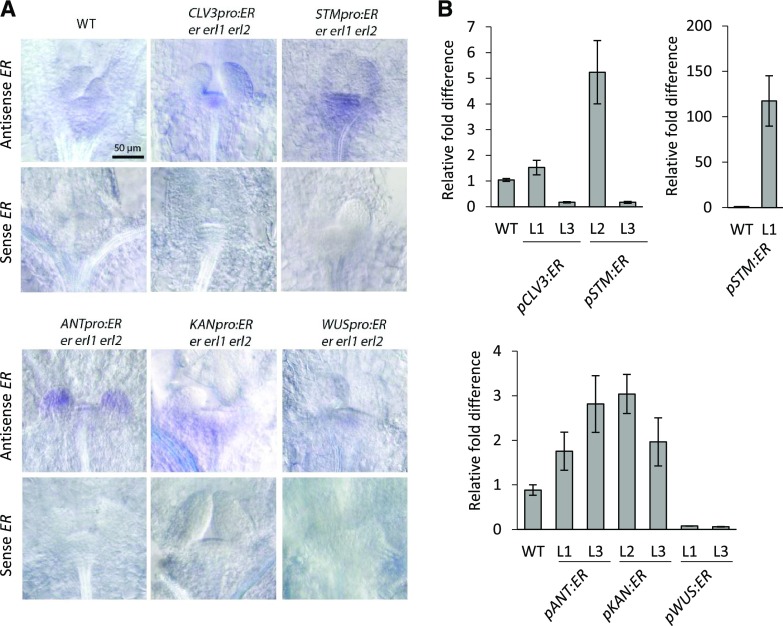
To examine ERECTA expression in the generated transgenic lines, we performed in situ hybridization on 3-d-old seedlings (Fig. 1A). In the wild type, ERECTA was detected throughout the SAM and in leaf primordia, although the signal was very weak. In the pCLV3:ER and pWUS:ER transgenic plants, ERECTA was expressed as expected in the central zone and the organizing center, respectively. Based on both in situ and RT-qPCR, ERECTA expression was considerably lower in the pWUS:ER lines compared to all other transgenic lines (Fig. 1). Most importantly, in neither pCLV3:ER nor in pWUS:ER transgenic lines was ERECTA detected outside of the SAM. In pSTM:ER transgenic lines, a signal was observed throughout the SAM and sometimes on the abaxial side of leaf primordia. The strength of the in situ signal and its appearance outside of the meristem varied greatly, consistent with variable expression of ERECTA in those lines as determined by RT-qPCR (Fig. 1B). In the pANT:ER transgenic plants, in situ analysis detected ERECTA in the outer L1 layer of the SAM and throughout young organ primordia. A similar pattern, including expression in the L1 layer of the SAM, was observed previously when a 6.5-kb ANT promoter was used to drive GUS expression (An et al., 2004). In pKAN:ER transgenic plants, the majority of the signal was detected in the peripheral zone with very low expression in leaf primordia. Thus, the CLV3 and WUS promoters drove expression of ER as expected, with the STM, ANT, and KAN promoters expressing ERECTA in slightly different patterns, suggesting that expression of genes under exogenous promoters should always be coupled with analysis of their expression.
Figure 1.
Ectopic expression of ERECTA (ER) in the SAM using heterologous promoters. A, Representative DIC images of in situ hybridization with a sense and an antisense probe for ER using 3-DPG T3 or T4 transgenic seedlings. B, RT-qPCR analysis of ER in 5-DPG seedlings of wild-type (WT) and transgenic plants. The average of three biological replicates is presented. Error bars represent se. All images are under the same magnification.
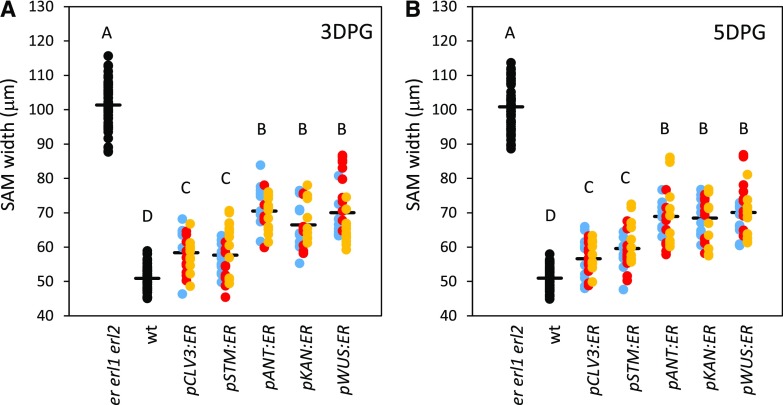
To understand how zone-specific expression of ERECTA affects the SAM size, we analyzed transgenic seedlings 3 d post germination (DPG) and 5 DPG (Fig. 2). For each of the constructs, we have combined the data from three independent transgenic lines. Because we are interested in the differences between constructs and not between lines within a construct, the combined data improves the estimate of the mean parameter and reduces the sd of the mean, better resolving the differences between the constructs and controls. Because natural variation is expected to be present between lines, the net effect of combining the data is to broaden the overall distribution instead of narrowing it. Even with the additional broadening, the effect of the various constructs can be clearly seen in the raw data in Figure 2. The size of the meristem in lines expressing ERECTA throughout the meristem (under the STM promoter) or in the central zone (under the CLV3 promoter) was rescued more efficiently compared to the other transgenic lines in both 3-DPG and 5- DPG samples (Fig. 2). The expression of ERECTA in the organizing center under the WUS promoter, in leaf primordia and the L1 layer of the SAM under the ANT promoter, and in the peripheral zone under the KAN promoter, were also able to rescue the SAM size but did so less efficiently (Fig. 2). This result cannot be attributed to the low expression of ERECTA in pANT:ER and pKAN:ER lines as determined by both RT-qPCR and in situ (Fig. 1, A and B). The reduced ability of pWUS:ER to rescue SAM size is probably only partially due to the low expression of ERECTA in those lines as low expression of ERECTA in pCLV3:ER line #3 and pSTM:ER lines #3 is sufficient to rescue the SAM size (Figs. 1B and 2). Thus, ERECTA can affect the meristem size when expressed in a variety of tissues, but it is most efficient when expressed in the central zone.
Figure 2.
Expression of ERECTA in the central zone (pCLV3:ER) or broadly in the meristem (pSTM:ER) rescues SAM size defects the most effectively. SAM width measurements were performed by DIC microscopy using 3-DPG (A) and 5-DPG (B) seedlings. The width of SAM was measured as indicated by an arrow in Figure 8C. Three genetically independent transgenic lines were analyzed (line #1 is blue; line #2 is red; line #3 is yellow; n for each transgenic line = 7 to 11). The mean is indicated as a thick horizontal line. In all cases presented, the sd of the mean was less than 1.5 mm and can be considered insignificant. Different letters indicate significant difference at P < 0.05, as determined by one-way ANOVA with Tukey’s post test.
Expression of ERECTA in the Central Zone of the SAM is Most Efficient in Regulating Leaf Initiation
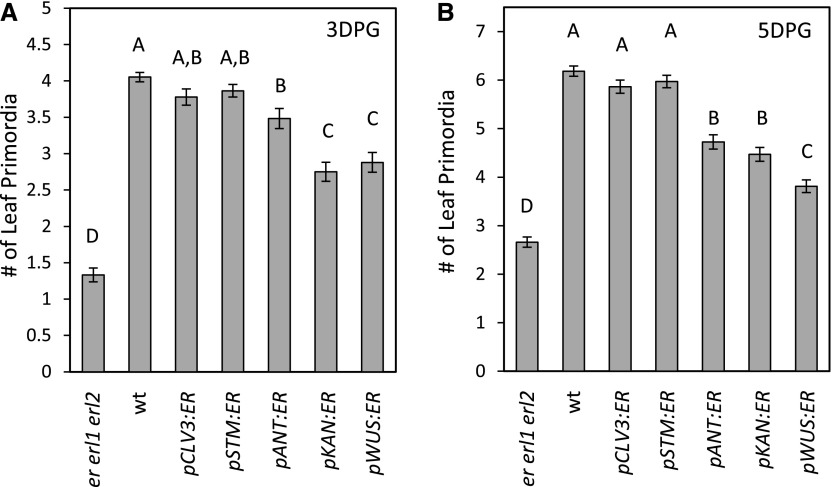
ERfs promote leaf initiation (Chen et al., 2013). At 3 DPG and 5 DPG, the er erl1 erl2 mutant forms on average 0.33 ± 0.03 and 0.42 ± 0.03 times as many leaves compared to the wild type. Out of the five promoters used, CLV3 and STM were the most efficient in rescuing leaf initiation defects (Fig. 3). Plants expressing the pCLV3:ER and pSTM:ER constructs in the er erl1 erl2 background formed very similar numbers of leaves at 3 DPG and an indistinguishable number of leaves at 5 DPG compared to the wild type. The pANT:ER transgenic plants had on average 0.85 ± 0.05 and 0.80 ± 0.06 times as many leaves compared to the wild type at 3 d and 5 d, respectively. It is not clear whether expression in the leaf primordia or in the L1 layer of the meristem is responsible for this phenotype. Expression of ERECTA in the peripheral zone using the KAN promoter or in the organizing center using the WUS promoter were the least efficient in enhancement of leaf initiation (Fig. 3). It is interesting to note that expression of ERECTA in the organizing center has an effect at all on leaf initiation in the peripheral zone, suggesting that at least to some extent ERfs regulate leaf initiation indirectly. Based on the phenotypes of pCLV3:ER, pKAN:ER, and pWUS:ER transgenic plants, we conclude that ERfs can regulate leaf initiation indirectly and they do so the most efficiently when expressed in the central zone of the meristem.
Figure 3.
Expression of ERECTA in the central zone (pCLV3:ER) or broadly in the meristem (pSTM:ER) rescues leaf initiation most efficiently. The number of leaf primordia formed was measured by DIC microscopy using 3-DPG (A) and 5-DPG (B) seedlings. Three genetically independent transgenic lines were analyzed for each construct and the data were combined to determine the mean (n for each transgenic line = 7 to 11; total n for each construct = 27 to 65). Error bars represent se. Different letters indicate significant difference at P < 0.05, as determined by one-way ANOVA with Tukey’s post test.
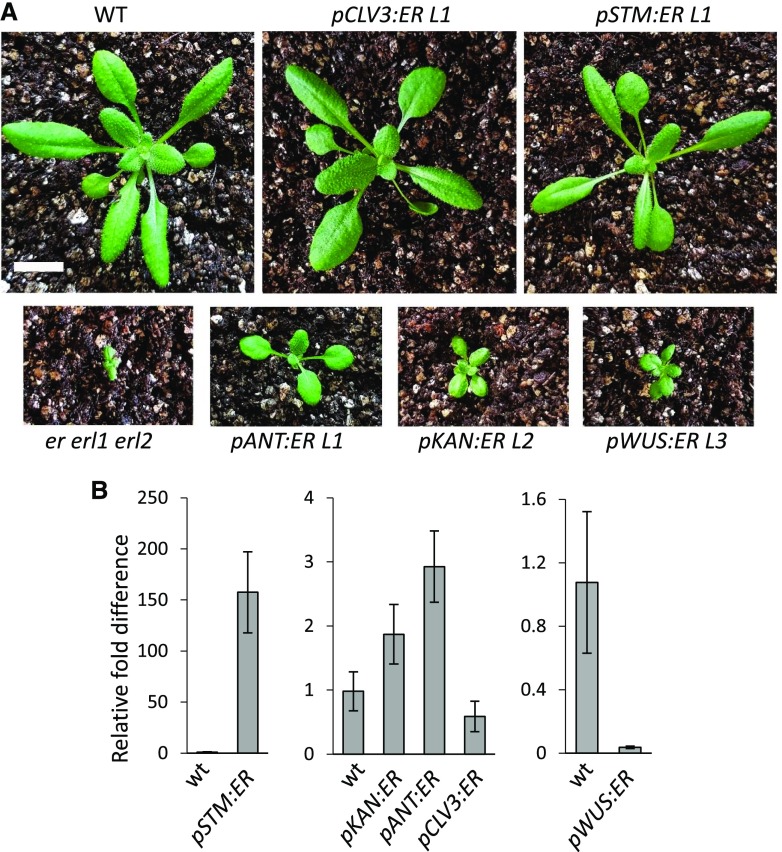
ERECTA Expression in the SAM Can Alter Leaf Expansion and Stem Elongation
In addition to SAM size and leaf initiation, the expression of ERECTA under the utilized promoters altered other aspects of plant development. Two out of the five constructs, pSTM:ER and pANT:ER, were able to rescue infertility of er erl1 erl2 (Supplemental Fig. S1), consistent with their expression during both early and late stages of flower development (Elliott et al., 1996; Long et al., 1996). CLV3 and WUS are expressed only during early stages of flower development (Mayer et al., 1998; Fletcher et al., 1999), and thus it is not surprising that ERECTA expressed under promoters of those genes cannot rescue fertility defects. Although the KAN promoter is active on the abaxial side of initiating floral organs and in the tissue that gives rise to ovules (Kerstetter et al., 2001), that expression was not sufficient to rescue infertility of er erl1 erl2 (Supplemental Fig. S1A).
The ERf genes are not only important for leaf initiation but also for leaf expansion (Shpak et al., 2004). In the three independent transgenic lines analyzed, the STM promoter led to very different levels of ERECTA transcription from approximately 100 to 150 times more than the wild type in line #1 to approximately five times less in line #3 (Fig. 1B). The expression was observed both in young primordia (Fig. 1A) and in mature leaves (Fig. 4B). The different levels of ERECTA expression were reflected in the size and shape of leaves, with fully rescued leaf expansion in line #1 and a minor increase in leaf expansion in line #3 (Supplemental Fig. S1B). Two pCLV3:ER lines also varied in the levels of ERECTA expression. The pCLV3:ER line #1 expressed approximately 1.5 more ERECTA compared to the wild type, and line #3 about six times less (Fig. 1B). This difference of expression again was reflected in different leaf sizes (Supplemental Fig. S1B). Comparison of line #3 pSTM:ER and line #3 pCLV3:ER, which on the level of the whole seedling express similar amounts of ERECTA, suggests that expression under the CLV3 promoter is more efficient in promoting leaf expansion (Fig. 1B and Supplemental Fig. S1B). Interestingly, ERECTA expression directly in leaves using the KAN and ANT promoters only weakly altered leaf size (Fig. 4). The leaf size in pKAN:ER line #2, which had twice as much ERECTA in mature leaves compared to the wild type, was very similar to the size of leaves in pWUS:ER line #3 where ERECTA was barely detectable if even present. The most revealing line is pANT:ER line #1, which expressed relatively high levels of ERECTA throughout both young primordia and older leaves but only partially rescued leaf size. Taken together, these data suggest that ERfs can regulate leaf size indirectly from the SAM.
Figure 4.
Expression of ERECTA using the CLV3 or STM promoters most efficiently rescues leaf shape defects of the er erl1 erl2 mutant. A, 20-DPG plants, bar = 1 cm. B, RT-qPCR analysis of ER in leaves of wild-type (wt) and T3 to T6 transgenic plants. The average of three biological replicates is presented. Error bars represent se. All images are under the same magnification.
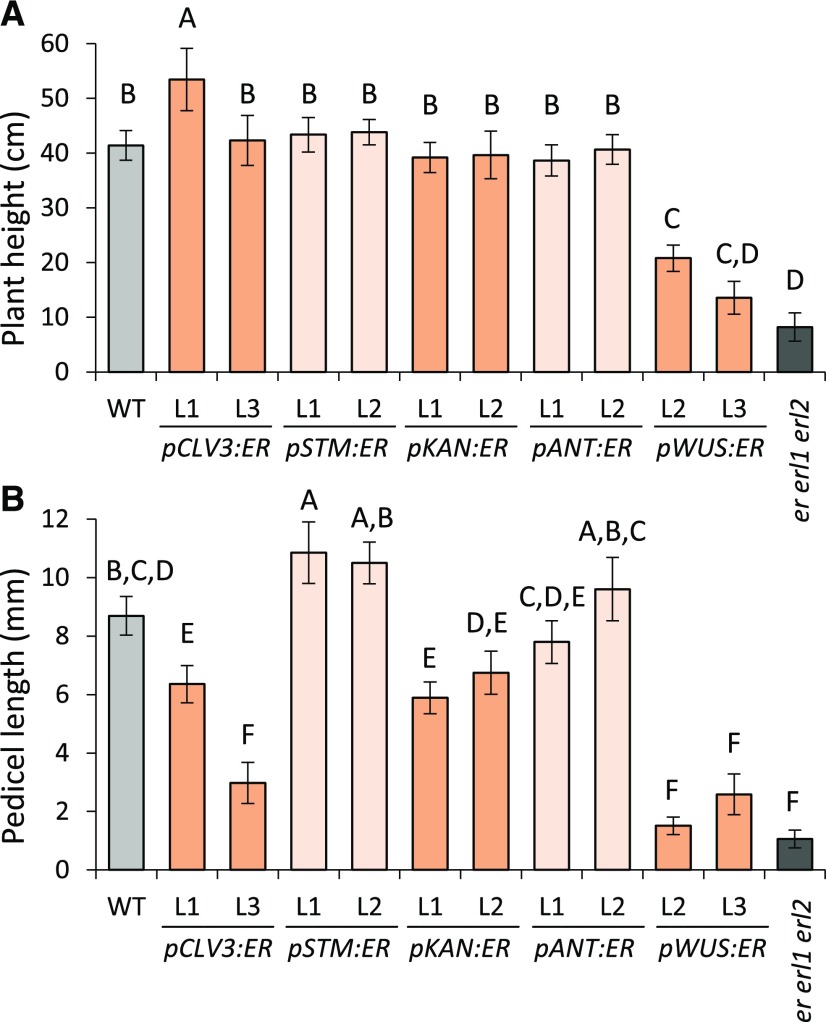
To further explore how ectopic expression of ERECTA affects plant growth, we analyzed plant height and pedicel lengths. Previously it was shown that ERECTA expression in the phloem using the SUC2 promoter was able to rescue height and pedicel length in the erecta mutant (Uchida et al., 2012). Here we show that expression in a variety of tissues rescues elongation defects of er erl1 erl2 (Fig. 5). ERECTA most efficiently stimulated stem growth when expressed under CLV3 and STM promoters, which is most noticeable when one observes younger plants (Supplemental Fig. S1C). Given enough time, ERECTA under the control of four promoters, CLV3, STM, ANT and KAN, fully rescued final plant height (Fig. 5A). ERECTA under the same four promoters also stimulated pedicel elongation in er erl1 erl2 (Fig. 5B), with pCLV3:ER and pKAN:ER being the least efficient. This may be at least partially related to their inability to rescue fertility, because we previously demonstrated that pedicels attached to unfertilized siliques are approximately 2 mm shorter compared to those attached to fertilized siliques (Bundy et al., 2012). Unexpectedly, even low expression of ERECTA in the organizing center of the SAM using the WUS promoter had a small but statistically significant effect on both stem and pedicel elongation (Fig. 5; Supplemental Fig. S1C). These results suggest that expression of ERECTA in the phloem is not obligatory for regulation of organ elongation and ERECTA expressed in other tissues can promote organ elongation as well.
Figure 5.
Expression of ERECTA under a variety of promoters can fully or partially rescue elongation of stem and pedicels in the er erl1 erl2 mutant. Plant height (A) and pedicel length (B) were measured in mature 2-month-old plants. Two independent transgenic lines were analyzed; n = 10 to 30 for heights and n = 64 for pedicel length. Error bars represent sd. Different letters indicate significant difference at P < 0.01, as determined by one-way ANOVA with Tukey’s post test.
Expression Pattern of EPFL1, EPFL2, EPFL4, and EPFL6 Near the SAM
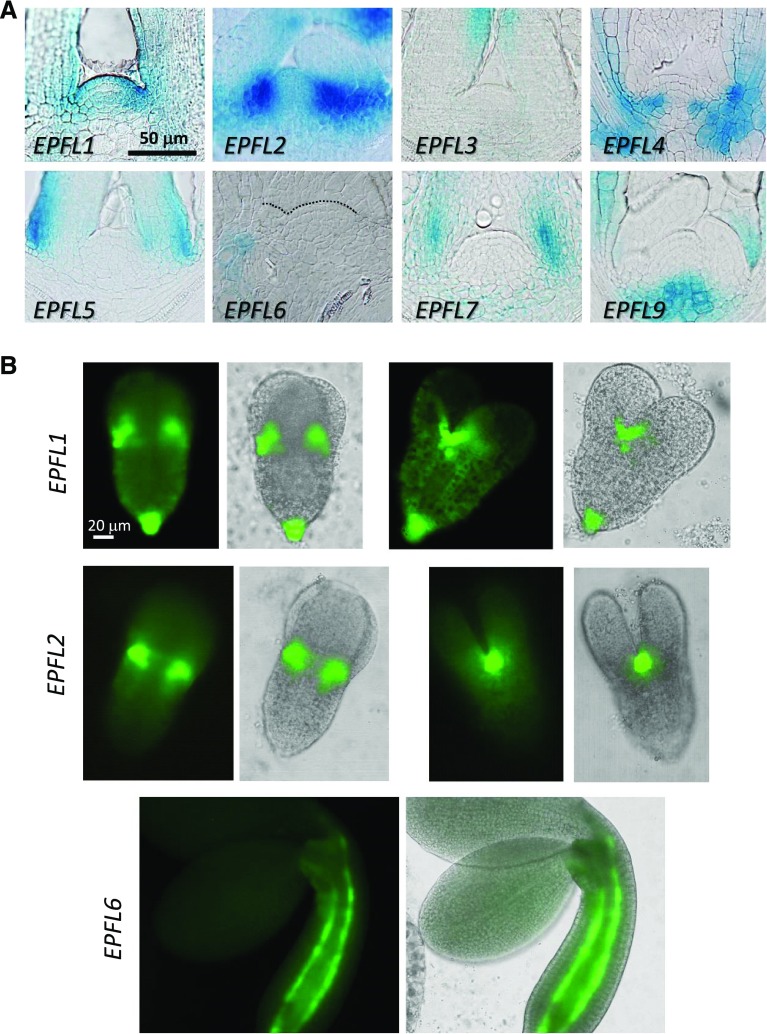
The activity of ERf receptors is regulated by a group of secreted small proteins from the EPF/EPFL family (Shimada et al., 2011). To narrow down the group of ligands that might be perceived by ERfs in the SAM, we investigated EPF/EPFL expression patterns using the GUS and GFP transcriptional reporter assays. Analysis of whole mount seedlings using the GUS assay suggested that EPF1 and EPF2 are expressed in epidermis, and specifically in developing stomata, but not in the SAM (Supplemental Fig. S2). We were unable to detect expression of EPFL8 at that developmental stage. The expression of other genes near the meristematic region was further examined by sectioning (Fig. 6A). Three genes, EPFL3, EPFL5/CHALLAH-LIKE1, and EPFL7, were expressed in different regions of leaf primordia: EPFL3 on the adaxial side of leaves at some distance from the SAM; EPFL5 at the base of leaf primordia, especially on the abaxial size; and EPFL7 in the internal tissues at the base of leaf primordia. Four genes were clearly expressed near the meristematic region: EPFL1, EPFL2, EPFL4/CLL2, and EPFL9/STOMAGEN. We observed expression of EPFL1 at the boundary and in L1 of the SAM and on the adaxial side of forming leaf primordia. EPFL2 exhibited extremely strong expression at the boundary. EPFL4 was expressed in the L3 layer of the peripheral zone. EPFL9 was expressed in the rib zone of the meristem. Because EPFL9 is an antagonist of ERfs (Lee et al., 2015) and currently mutants in that gene are unavailable, we did not investigate it any further. EPFL6/CHAL was not expressed directly in the SAM but its expression was detected in the inner tissues underneath leaf primordia peripheral to the SAM. Next, we used epifluorescence microscopy to analyze EPFL expression during embryogenesis. Out of 11 genes, EPFL1, EPFL2, and EPFL6 were expressed in the developing embryos. EPFL1 and EPFL2 were expressed very highly in the peripheral regions of the embryonic SAM where margins of cotyledons meet (Fig. 6B). EPFL1 was also expressed in the epidermis of the hypocotyl and in the root apical meristem. We detected expression of EPFL6 in hypocotyl only during late embryogenesis. It was expressed in two cell files that are presumably endodermis.
Figure 6.
A reporter gene assay of the EPF/EPFL gene family in the SAM demonstrates distinct patterns of expression. A, Longitudinal sections of shoot apices of T2 7-DPG or 10-DPG wild-type seedlings expressing indicated pEPFL:EGFP-GUS constructs. The dotted line in the EPFL6 insert emphasizes the L1 layer of the SAM. B, Epi-fluorescence microscopy of plants expressing pEPFL1:EGFP-GUS and pEPFL2:EGFP-GUS in torpedo embryos and pEPFL6:EGFP-GUS in bend cotyledons embryos. For the first two constructs, the same embryo is represented from two different perspectives. All images are under the same magnification in (A) and in (B).
EPFL1, EPFL2, EPFL4, and EPFL6 Partially Redundantly Regulate Elongation of Plant Organs
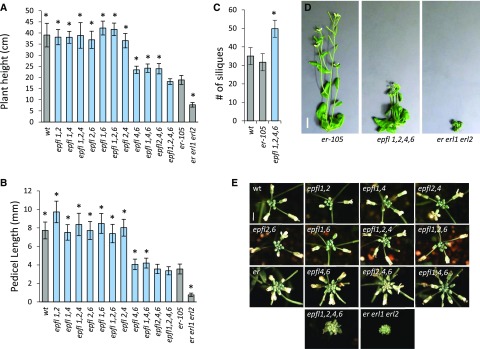
Due to their expression near the meristematic region, we investigated the function of EPFL1, EPFL2, EPFL4, and EPFL6 in plant development. The epfl4/cll2-1 and epfl6/chal-2 single mutants are null alleles carrying T-DNA insertions with no visible phenotype (Abrash and Bergmann, 2010; Abrash et al., 2011; Uchida et al., 2012). The epfl2-1 allele is a null allele carrying a transposon insertion and the mutant exhibits diminished leaf tooth growth (Tameshige et al., 2016). The epfl1-1 allele is a null allele carrying a transposon insertion and the mutant has no visible phenotype (Supplemental Figs. S3 and S4). To understand the function of these genes we created all possible combinations of double and triple mutants. The epfl4 epfl6 plants are shorter in stature compared to the wild type but are slightly taller compared to er-105 (Fig. 7, A and B; Abrash et al., 2011; Uchida et al., 2012). None of the other double mutants displayed a significant reduction in elongation of stems or pedicels (Fig. 7, A and B). Addition of the epfl1 mutation to epfl4 epfl6 did not change stem and pedicel elongation, whereas the presence of epfl2 in the epfl4 epfl6 background slightly reduced elongation of pedicels, leading to formation of more compact inflorescence (Fig. 7, B and E). The epfl1 epfl2 epfl4 epfl6 mutant reached a final height comparable to that of the erecta single mutant; however, it grew drastically slower and took an additional four weeks to achieve maturity compared to erecta (Fig. 7, A and D). In this respect epfl1 epfl2 epfl4 epfl6 is similar to er erl1 erl2, which is also characterized by an extended period of growth and a longer lifespan (Kosentka et al., 2017). We observed that the extended life span of epfl1 epfl2 epfl4 epfl6 leads to increased number of siliques formed on the main stem (Fig. 7C). Taken together, these data suggest that although EPFL4 and EPFL6 play the primary role in stimulation of stem and pedicel elongation, EPFL1 and EPFL2 also contribute to this process.
Figure 7.
EPFL1, EPFL2, EPFL4, and EPFL6 synergistically regulate stem and pedicel elongation with EPFL4 and EPFL6 playing the key role. A, Height of fully grown plants (n = 27 to 46 except er erl1 erl2, n = 12). B, Lengths of mature pedicels on the main stem (n = 100 to 120). C, Number of siliques on the main stem (n = 10). A to C, Bars represent the average; error bars represent sd. Values significantly different from er-105 are indicated by asterisks (based on Student t test; P < 0.001). D, 6-week-old plants of er-105, epfl1-1 epfl2-1 epfl4 epfl6, and er-105 erl1-2 erl2-1. Scale bar: 1 cm. E, Influorescence apices from the wild type, er, er erl1 erl2, and various combinations of epfl mutants. Bar = 25 mm. All images are under the same magnification in (D) and in (E). wt, wild type.
In addition to changes in elongation of aboveground organs we also observed changes in silique growth, fertility, and apical dominance (Supplemental Fig. S4.) Of all double mutants, epfl1 epfl6 formed the shortest siliques, suggesting the primary role for these two genes is in fruit development (Supplemental Fig. S4B). Fertility was reduced in the epfl1 epfl2 epfl6 and epfl1 epfl4 epfl6 mutants, and epfl1 epfl2 epfl4 epfl6 plants are infertile (Supplemental Fig. S4A). In addition, all four genes contribute partially redundantly to establishment of apical dominance (Supplemental Fig. S4C). No obvious changes in the formation of stomata were observed (Supplemental Fig. S5).
EPFL1, EPFL2, EPFL4, and EPFL6 Redundantly Regulate SAM Size and Leaf Initiation
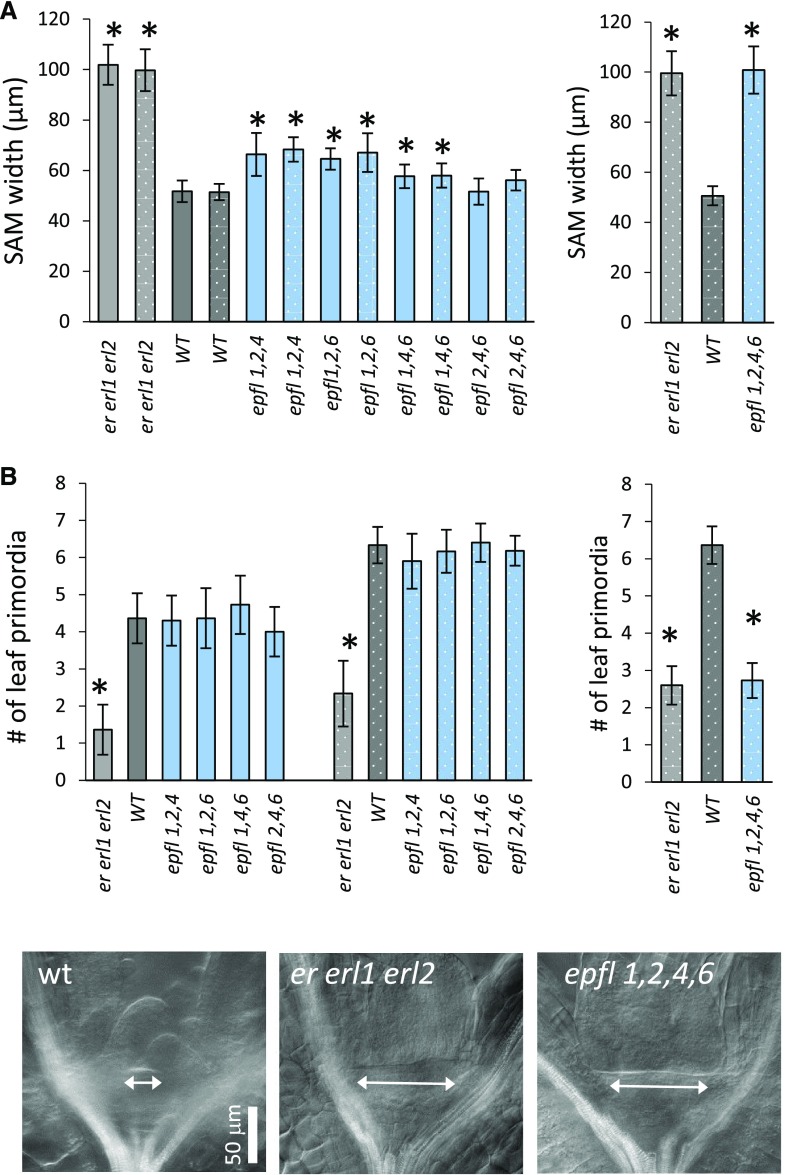
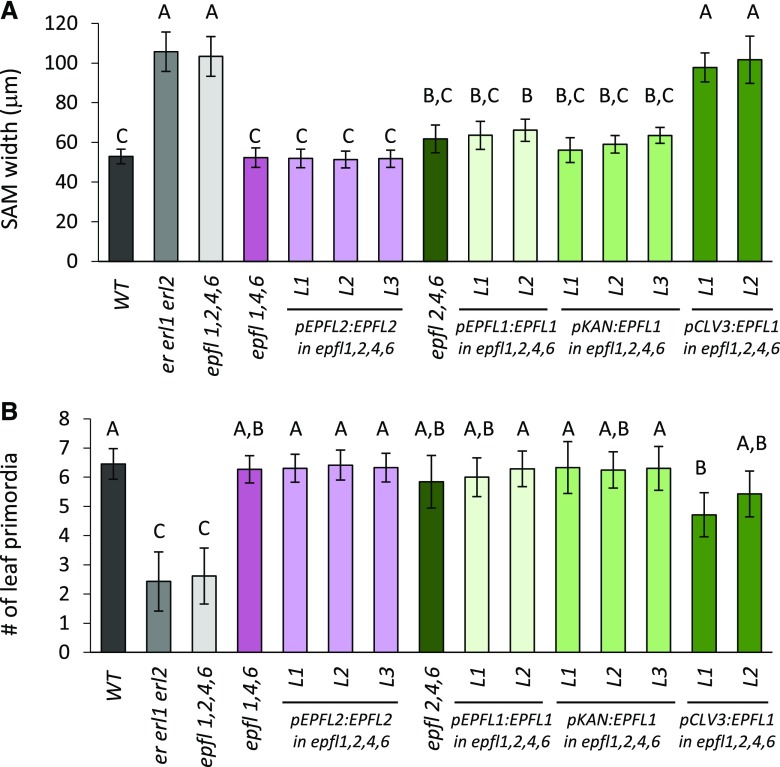
Analysis of triple epfl mutants demonstrated a slight but statistically significant increase of meristem size in triple mutant combinations: epfl1 epfl2 epfl4; epfl1 epfl2 epfl6; and epfl1 epfl4 epfl6 (Fig. 8A). There were no significant changes in the rate of leaf initiation (Fig. 8B). Because the epfl1 epfl2 epfl4 epfl6 mutant is infertile and the epidermal phenotype cannot be used to identify it in the progeny of erfl1/+ epfl2 epfl4 epfl6 plants, 30 seedlings with slightly shorter petioles of cotyledons were genotyped for epfl1-1 before fixation for differential interference contrast (DIC) microscopy. This allowed us to identify 10 epfl1 epfl2 epfl4 epfl6 mutants. The following analysis demonstrated that in terms of meristem size and leaf initiation rate epfl1 epfl2 epfl4 epfl6 is indistinguishable from er erl1 erl2 (Fig. 8), suggesting that the four genes EPFL1, EPFL2, EPFL4, and EPFL6 regulate meristem function. To confirm that the phenotype is due to mutations in the EPFL genes and not to some other overlooked mutations, we independently expressed EPFL1 and EPFL2 under their endogenous promoters in epfl1 epfl2 epfl4 epfl6. Both constructs rescued meristematic defects (Fig. 9), promoted stem and pedicel elongation (Supplemental Fig. S6), and rescued fertility defects in multiple independent transgenic lines. To test whether ligands have to be coexpressed with ERfs in the central zone or if they function from the peripheral zone, EPFL1 was expressed in epfl1 epfl2 epfl4 epfl6 under CLV3 and KAN promoters. The expression under KAN fully rescued both meristem size and leaf initiation, whereas expression under CLV3 had no effect on the meristem size and only partially rescued leaf initiation (Fig. 9). In addition, expression under KAN but not the CLV3 promoter rescued organ elongation defects (Supplemental Fig. S6). Taken together, these data suggest that four EPFL genes, EPFL1, EPFL2, EPFL4, and EPFL6, redundantly regulate maintenance of meristem size and promote leaf initiation with expression in the peripheral zone being sufficient for their function.
Figure 8.
EPFL1, EPFL2, EPFL4, and EPFL6 redundantly regulate the size of the SAM and the rate of leaf initiation. Comparison of the SAM width (A) and the number of formed leaf primordia (B) in the wild type, er erl1 erl2, and epfl family mutants determined by DIC microscopy at 3 DPG (solid bars) and 5 DPG (dotted bars). Bars represent the average; error bars represent sd. n = 10 to 11. Values significantly different from the wild type are indicated by asterisks (based on Student t test; P < 0.006). C, DIC images of meristematic regions in the wild type, er erl1 erl2, and epfl1,2,4,6 at 3 DPG. The meristem width is displayed with an arrow. All images are under the same magnification in (C). wt, wild type.
Figure 9.
The meristematic phenotype of epfl1,2,4,6 can be fully rescued by expression of EPFL1 or EPFL2 under endogenous promoters or by expression of EPFL1 under KAN promoter but not CLV3. Comparison of the SAM width (A) and the number of formed leaf primordia (B) in the wild type (WT), selected mutants as indicated and in independent transgenic lines expressing indicated constructs in epfl1,2,4,6 background as determined by DIC microscopy in 5-DPG seedlings. Bars represent the average; error bars represent sd. n = 7 to 14. Different letters indicate significant difference at P < 0.01, as determined by one-way ANOVA with Tukey’s post test.
DISCUSSION
The ERf signaling pathway first appeared in early land plants and has evolved to regulate multiple aspects of plant development (Villagarcia et al., 2012; Shpak, 2013; Takata et al., 2013). Whereas a species typically contains only two to three ERf receptors, the EPF/EPFL family of putative ligands is relatively large, with 10 or more genes being typical (Takata et al., 2013; Zhang et al., 2018). Each individual EPF/EPFL is expressed in a unique spatio-temporal pattern and often controls a specific developmental process. For example, in Arabidopsis, rice and the moss Physcomitrella patens EPF1 orthologs control stomata development (Hara et al., 2007, 2009; Hunt and Gray, 2009; Caine et al., 2016, 2018).
The first indication that ERECTA signaling might contribute to regulation of SAM structure came from the analysis of higher order mutants. It was observed that the er mutation enhances meristematic defects of CLV pathway mutants and suppresses those of the uni-1D/+ mutant (Diévart et al., 2003; Durbak and Tax, 2011; Uchida et al., 2011). Later, analysis of the er erl1 erl2 mutant demonstrated that ERfs synergistically inhibit expansion of the vegetative meristem and promote leaf initiation (Chen et al., 2013; Uchida et al., 2013). While the CLAVATA pathway regulates meristem height, ERECTA signaling restricts the meristem width and functions independently of CLAVATA (Mandel et al., 2014, 2016). Understanding a signaling pathway depends on knowing the identity of cells involved in sending and receiving the signal. ERf receptors are expressed throughout the SAM and in forming leaf primordia (Yokoyama et al., 1998; Uchida et al., 2013), but that does not mean that their expression in all those areas is necessary for regulation of meristem expansion and/or leaf initiation. To uncover the regions where ERfs are critical for meristem maintenance and organ initiation, we expressed ERECTA under a range of promoters in the er erl1 erl2 mutant. Unexpectedly, expression of ERECTA under all five promoters, STM, CLV3, KAN, ANT, and WUS, in different and in some cases nonoverlapping areas of the meristem reduced meristem size and promoted leaf initiation, suggesting that ERfs can have an impact on meristem function when expressed in a variety of locations. Simultaneously, expression of ERECTA throughout the meristem under the STM promoter or in the central zone under the CLV3 promoter had the strongest impact on the meristem width and organ initiation, implying that the function of ERfs in the central zone is paramount. It is interesting to note that expression of ERECTA under the WUS promoter elements is insufficient to fully rescue meristematic defects of er erl1 erl2 while expression of CLV1 under the same promoter elements fully rescues the clv1 mutant (Nimchuk et al., 2015), which reinforces the distinctiveness of these two signaling pathways.
The next question is: What signals are perceived by ERfs in the SAM? There are 11 EPF/EPFLs in Arabidopsis. Analysis of mutants suggests that four genes, EPFL1, EPFL2, EPFL4, and EPFL6, contribute to meristem size establishment and promotion of leaf initiation. These genes function redundantly with triple mutants exhibiting no or very weak meristematic phenotypes. EPFL1, EPFL2 and EPFL4, EPFL6 belong to two closely related clades with the stomata-regulating EPF1, EPF2, and EPFL9 genes being more distantly related (Takata et al., 2013). Both clades have one additional gene, EPFL3 and EPFL8, respectively, which are neither expressed near the meristematic region nor seem to be essential for SAM regulation. EPFL4 and EPFL6 are verified ERf ligands as they bind directly to ERfs (Lee et al., 2012; Lin et al., 2017). EPFL2 has been shown to bind to Erf-containing complexes, which suggests that genes belonging to that clade are likely to encode ERf ligands (Tameshige et al., 2016). Because all four genes have the potential to suppress stomata development when expressed in the epidermal tissue layer, they are likely to be agonists of ERf receptors (Abrash et al., 2011). EPFL1 and EPFL2 are expressed during embryogenesis in the boundary region between two cotyledons at the periphery of the SAM. After germination, they are expressed in the analogous region at the border of the meristem and previously formed leaf primordia. Expression of EPFL1 in the border zone is consistent with gene expression profiling of the inflorescence SAM, which indicated upregulated EPFL1 expression at the periphery of the SAM (Yadav et al., 2009). EPFL2 was classified as a boundary-enriched gene in a TRAP-seq experiment that was done using 7-d-old seedlings (Tian et al., 2014). The boundary zone has a low rate of cell divisions, low auxin accumulation, and high expression of CUC genes, which is similar to another location where EPFL2 is expressed—the sinus of leaf teeth (Tameshige et al., 2016; Wang et al., 2016). EPFL4 and EPFL6 are expressed near the SAM after germination with EPFL4 in the internal layers of the peripheral zone and EPFL6 in the border region at some distance from SAM.
ERf expression in the central zone and the expression of EPFLs at the periphery of the meristem or at the bases of leaf primordia suggest that the ERf signaling pathway enables communications between the border region and the central zone. This conclusion is also supported by the ability of EPFL1 to rescue the quadruple mutant phenotype when expressed under the KAN but not the CLV3 promoter. Taken together, our data suggest that overlap in the expression of EPFLs and ERfs expression at the outer boundary of the central zone of the meristem restricts SAM width and promotes leaf initiation.
Recently it has been proposed that ERfs function in the L1 layer of the meristem where they sense signals coming from internal layers of the SAM (Kimura et al., 2018). Our data are inconsistent with this conclusion. Kimura et al. utilized only two promoters to interrogate the function of ERfs and did not measure the meristematic parameters at multiple developmental points using numerous samples to obtain statistically significant data. Because the expression of ERECTA in many different regions of the meristem alters behavior of meristematic cells, it is important to obtain quantitative measurements for precise comparisons. Moreover, it is necessary to take into account the differences in the expression levels of ERECTA. For example, while ERECTA expressed under the ANT, KAN, and WUS promoters rescues meristem defects in a similar manner, the first two promoters drive ERECTA expression at much higher levels, suggesting that the SAM is much more sensitive to ERECTA that is localized in the organizing center. In addition, our data suggest that ligands are expressed endogenously at the boundary of the meristem and in the peripheral zone and not in the internal layers. EPFL expression in the internal layers driven by the CLV3 promoter cannot efficiently rescue the meristematic defects of the epfl1 epfl2 epfl4 epfl6 mutant. Although Kimura and colleagues state that EPFLs are secreted in the internal layers of the SAM, data supporting that conclusion is not provided in their paper.
Expression of a gene under an exogenous promoter is a popular approach to interrogate gene function in a specific tissue. This approach has been effective in revealing the function of ERfs. Here, we would like to emphasize some issues associated with this approach. First, to prove that a gene controls a particular process from a specific tissue, it is necessary to use a sizable range of exogenous promoters. Because expression of ERECTA in a variety of nonoverlapping tissues has an effect on meristematic processes and elongation of organs, we believe that the use of a limited number of promoters has been misleading (Uchida et al., 2012; Kimura et al., 2018). Second, the expression pattern of a gene under an exogenous promoter can differ from what is expected, and it is essential to evaluate the actual expression pattern. For example, while in situ data suggest that ANT is expressed in leaf and flower primordia (Elliott et al., 1996; Long and Barton, 2000), the commonly used 6.5-kb promoter of that gene also drives expression in the L1 layer of the meristem (An et al., 2004). Finally, some promoters can lead to a variety of expression levels and expression patterns. An example is the STM promoter. In situ data indicate that STM is expressed throughout the SAM and is downregulated in the forming organ primordia (Long et al., 1996; Long and Barton, 2000). However, in transcriptional reporter assays the STM promoter induced diverse expression patterns that differed from the endogenous; The reporters were expressed underneath the SAM in the cells of the hypocotyl, in the vascular cells of the leaf primordia, preferentially at the boundary of SAM, or in the peripheral region but not the central region of the SAM (Kim et al., 2003; Verkest et al., 2005; Landrein et al., 2015). Similarly, in our experiments we observed STM expression underneath the SAM and in leaf primordia. Moreover, expression levels between created transgenic lines varied more than 500-fold. Hypothetically, these differences in expression could be due to inconsistent epigenetic regulation of the STM promoter in new locations (Katz et al., 2004).
By expressing ERECTA in different regions of the SAM, we anticipated the rescue of meristematic phenotypes. What we did not expect was to rescue the elongation of aboveground organs. ERf genes promote elongation of internodes, pedicels, petioles, siliques, leaves, and flower organs (Torii et al., 1996; Shpak et al., 2004). Previously it has been proposed that ERfs promote internode and pedicel growth by enabling cell-to-cell communication between the endodermis and phloem (Uchida et al., 2012). Our data suggest that ERECTA can promote organ elongation when expressed in a variety of locations including the central zone of the SAM. Most significantly, our data suggest that expression in the phloem is not essential for ERfs to promote elongation of organs. How can ERfs regulate organ elongation from the SAM? We can envision several mechanisms. Internodes are initially formed through activity of the peripheral zone that generates progenitor cells for epidermis, cortex, and of the rib zone that supplies cells for the central cylinder. As observed above, the activity of ERECTA in the central zone promotes initiation of leaves in the peripheral zone. Thus, it is not a big stretch to imagine that ERfs promote proliferation of cells surrounding forming leaf primordia. Alternatively, ERfs might regulate growth of internodes indirectly, for example through controlling homeostasis of hormones such as auxin or gibberellin. This latter possibility can account for the ERf’s ability to regulate organ growth when expressed in a variety of tissues, including from the phloem and the SAM. Our data indicate that the understanding of ERf’s role in organ elongation is incomplete, and requires further investigation.
The phenotype of the epfl1 epfl2 epfl4 epfl6 mutant suggests that, in addition to regulation of meristem structure, all four genes promote elongation of internodes and pedicels, with EPFL4 and EPFL6 playing the major role in this process. The expression pattern of EPFL1 and EPFL2 in internodes and pedicels and their precise role in organ elongation is yet to be established. Although the quadruple mutant grows much more slowly than epfl4 epfl6, its final size is only slightly below that of the er mutant and is considerably bigger compared to er erl1 erl2. This result suggests that either other ligands contribute to regulation of organ elongation or perhaps ERf regulates organ elongation by two mechanisms: in response to ligand binding and independently of ligand binding. Previously we demonstrated that the kinase dead ERECTA partially rescues organ elongation when expressed in er erl1 erl2 (Kosentka et al., 2017). If the main outcome of EPF/EPFL binding is the activation of the ERf kinase domain, then it would be expected that the phenotype of epfl1 epfl2 epfl4 epfl6 would resemble that of the kinase dead receptor, favoring the second hypothesis of two different mechanisms of ERf function in organ elongation.
MATERIALS AND METHODS
Generation of Transgenic Plants
Four different promoters (STM, ANT, KAN, and WUS) were independently cloned into pPZP222 vectors that carried the genomic ERECTA sequence and the endogenous 1.9 kb ERECTA terminator. The endogenous terminator does not have any regulatory sequences as all regulatory elements are localized in the ERECTA promoter (Yokoyama et al., 1998). In the constructs, the 35S promoter drives expression of the selective marker gentamycin. To prevent this promoter from influencing expression of the transgenes, ERECTA and the selective marker were cloned in the head-to-tail orientation. Using the longer ERECTA terminator (1.9 kb) instead of the 35S terminator (∼200 bp) served the purpose of introducing spacer DNA between the two promoters to further reduce interactions between the transgene and the 35S promoter. pSTM:ER (pPZK 311) was generated by amplifying a 4.62-kb region upstream of the STM start site. A similar 4.5-kb STM promoter region has been used and was analyzed in Verkest et al. (2005). pWUS:ER (pPZK 310) was created by amplifying a 4.5-kb region upstream of the WUS start site. This promoter region has been used previously by Yadav et al. (2009). pANT:ER (pPZK 315) was created by amplifying a 4.3-kb region upstream of the ANT start site as in Grandjean et al. (2004). pKAN:ER (pPZK 312) was generated by amplifying a 3.6-kb region upstream of the KAN start site as in Wu et al. (2008). The fifth construct pCLV3:ER (pPZK317) was generated slightly differently due to the presence of an enhancer in the terminator of CLV3 (Brand et al., 2002). The genomic ERECTA sequence was inserted into pPZP222 between the 1.5-kb sequence upstream of the CLV3 start site and the 1.2-kb sequence downstream of the CLV3 stop codon. All promoter/ERECTA/terminator sequences were cloned into pPZP222 between BamHI and XbaI restriction sites. All created constructs were examined by the restriction analysis and sequencing of amplified regions.
The described plasmids were transformed into an Agrobacterium tumefaciens strain GV3101/pMP90 by electroporation and introduced into er erl1/+ erl2 plants by the floral dip method. The er-105 erl1-2 erl2-1 mutant has been described in Shpak et al. (2004). The T1 transgenic plants were selected based on gentamicin resistance. Kanamycin resistance was used to identify erl1-/+ or erl1−/− lines in the T2 generation. In the T3 or T4 generation, we selected lines that are homozygous for the transgene based on gentamicin resistance.
To generate pEPFL1:EGFP-GUS, a 1.5-kb fragment upstream of the EPFL1 start site was PCR-amplified and inserted into p-ENTR/topo (Invitrogen) and recombined using LR recombinase (Invitrogen) into pKGWFS7 (Karimi et al., 2005). To clone the promoters of EPF1 (2.7 kb), EPF2 (2.7 kb), EPFL2 (3 kb), EPFL3 (2.9 kb), EPFL4 (2.8 kb), EPFL6 (2.9 kb), EPFL7 (1.5 kb), EPFL8 (2.4 kb), and EPFL9 (2 kb) in front of EGFP-GUS a modified version of the Rapid one-step recombinational cloning method was used (Fu et al., 2008). The promoter regions were amplified by PCR using gene-specific primers that also contained shortened AttL1 or AttL2 sequences. Each fragment was extended using attL1-T2.1 and attL2-T2.1 primers to produce complete AttL sequences on both sides of each fragment. The generated fragments were recombined into pKGWFS7 using LR recombinase (Invitrogen). Primer sequences can be found in Supplemental Table S1.
The generated pEPFL:EGFP-GUS plasmids were introduced into wild-type plants as described above. The transgenic plants were selected based on kanamycin resistance. pEPFL5:GUS transgenic plants were described in Abrash and Bergmann (2010) and Abrash et al. (2011).
To generate pEPFL1:EPFL1 a 3.3-kb fragment encompassing a 2-kb region upstream of the EPFL1 start site and 0.8-kb region downstream of the stop codon was amplified and cloned into pPZP222. pEPFL2:EPFL2 was generated by amplifying a 4.2-kb fragment including 2.5 kb upstream of the EPFL2 start codon and 1 kb downstream of the stop codon. In pKAN:EPFL1 and pCLV3:EPFL1 constructs, we used the same promoter regions as in pKAN:ER and pCLV3:ER and the EPFL1 sequence that included introns. The pKAN:EPFL1 construct contains the endogenous 0.8-kb EPFL1 terminator whereas pCLV3:EPFL1 contains the 1.2-kb sequence downstream of the CLV3 stop codon.
In Situ Analysis
In situ hybridization was performed as described in Hejátko et al. (2006) using 3-DPG T3 and T4 transgenic or wild-type seedlings. One-kb cDNA region of ERECTA between the SacI and XhoI restriction sites was cloned into pBluescript II and used as the template for in vitro transcription with T3 (Promega) and T7 (Invitrogen) RNA polymerases to make the sense and antisense probes, respectively. To generate EPFL probes, their full-length coding DNA sequences were amplified using wild-type cDNA and primers that contained the T7 promoter sequence near either the start or the stop codons. All probes were hydrolyzed to produce fragments with an average length of about 0.3 Kb. Representative images were taken using DIC microscopy.
Analysis of Mutant Phenotypes
For measurements of leaf number and SAM size by DIC microscopy, seedlings were grown on plates containing modified Murashige and Skoog medium supplemented with vitamins (Research Products International) and 1% (w/v) Suc. Selected 3-DPG and 5-DPG seedlings were incubated in a solution of 9:1 ethanol/acetic acid overnight, rehydrated using an ethanol series (90%, 80%, 70%, and 50%) and cleared in a chloral hydrate solution (chloral hydrate/water/glycerol 8:1:1). The pSTM:ER transgenic lines were analyzed in the T4 or T5 generations, as they were homozygous for the transgene and the erl1 mutation. The pWUS:ER, pKAN:ER, pANT:ER, and pCLV3:ERECTA transgenic lines were analyzed in the T3 generation. These were homozygous for the transgene but were segregated for erl1. The er erl1 erl2 plants used for analyses were identified based on the presence of stomata clusters in cotyledons. Microscopic observations were done using an Eclipse 80i microscope (Nikon) with DIC optics and NIS-Elements BR imaging software (Nikon) was used for measurements. For measurement of plant height and pedicel length and to observe leaf growth, plants were grown as described in Kosentka et al. (2017).
Generation of the epfl Double, Triple, and Quadruple Mutants
The epfl1-1 (CS104435) transposon-insertion mutant (Columbia background) was obtained from the Arabidopsis Biological Resource Center. The Epfl2-1 (CSHL ET5721) transposon-insertion mutant (Landsberg erecta background) was received from Cold Spring Harbor Laboratory and outcrossed three times to epfl1-1 to obtain epfl1-1 epfl2-1 in the Columbia background. The absence of the er-1 mutation in epfl1-1 epfl2-1 was confirmed by sequencing. The epfl1 epfl2 double mutant was crossed with epfl4 epfl6 /cll2-1 chal-2 (Abrash et al., 2011) to obtain new combinations of mutations. Epfl double, triple, and quadruple mutants were identified by genotyping with epfl1-1 and epfl2-1 primers from Supplemental Table S2 and with epfl4/cll2-1, epfl6/chal-2 specific primers described in Abrash et al. (2011). We used a three-primer PCR for genotyping of epfl1-1 and epfl2-1. During genotyping of epfl1-1, the primers epfl1.436.rev and 3dspm were used to amplify an approximately 200-bp fragment and the primers epfl1.436.rev and epfl1.74 were used to amplify a 387-bp fragment. During genotyping of epfl2-1, the primers epfl2.1 and gus.43.rc were used to amplify an approximately 700-bp fragment and the primers epfl2.1 and epfl2.540.rev were used to amplify a 575-bp fragment. Because the epfl1 epfl2 epfl4 epfl6 mutant is infertile, for the morphological analysis we obtained it from the progeny of erfl1/+ epfl2 epfl4 epfl6 plants.
The GUS Reporter Gene and Assay and Microscopy
GUS staining was performed as described in Sessions et al. (1999) using 5-DPG T2 or T3 transgenic seedlings. Multiple independent transgenic lines were analyzed for each construct to find a consistent pattern of expression. Depending on the level of the signal, the concentration of ferricyanide and ferrocyanide in the staining buffer varied between 2 and 6 mM. After staining, the samples were dehydrated with a graded ethanol series up to 50% ethanol, fixed in FAA solution (3.7% formaldehyde/5% acetic acid/50% ethanol) for 30 min, dehydrated with a graded series of ethanol to 100% ethanol, infiltrated with polymethacryl resin Technovit 7100, and then embedded and polymerized in Technovit 7100 (Heraeus Kulzer). Eight-micrometer sections were prepared using a Leica RM-2255 microtome. Pictures were obtained using an Eclipse 80i microscope (Nikon) and a 12-megapixel cooled color DXM-1200c (Nikon) camera. A C-FL B-2A (Nikon) filter cube was used to observe the GFP signal.
Reverse Transcription PCR
Total RNA was isolated from five DPG seedlings and from fully expanded leaves using a Spectrum Plant Total RNA Kit (Sigma-Aldrich). RNA was treated with RQ1 DNase (Promega) and first-strand cDNA was synthesized using 150 ng of RNA with a ProtoScript II RT-PCR Kit (New England Biolabs) according to the manufacturer’s instructions. Quantitative PCR was performed using the CFX96 Real Time System (BioRad) with Sso Evagreen Supermix reagent (BioRad). Each experiment used three technical replicates and three biological replicates to calculate relative fold difference of ERECTA to ACTIN-2 expression. Bio-Rad CFX Manager was used to calculate cycle threshold values and the fold difference in gene expression was calculated using the delta-delta-Ct algorithm (2−ΔΔCt). Primers and annealing temperatures are listed in Supplemental Table S3.
Accession Numbers
Arabidopsis Genome Initiative numbers for the genes discussed here are as follows: ER (At2g26330), ERL1 (At5g62230), ERL2 (At5g07180), EPFL1 (At5g10310), EPFL2 (At4g37810), EPFL4 (At4g14723), and EPFL6 (At2g30370).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The effect of ERECTA expression under different promoters on plant morphology.
Supplemental Figure S2. The GUS reporter gene assay of the EPF/EPFL gene family.
Supplemental Figure S3. Epfl1-1 is a null mutant with a transposon insertion in the second exon.
Supplemental Figure S4. EPFL1, EPFL2, EPFL4, and EPFL6 partially redundantly regulate flower development and apical dominance.
Supplemental Figure S5. The epfl1,2,4,6 mutant does not exhibit obvious stomata patterning defects.
Supplemental Figure S6. Expression of EPFL1 under the endogenous and KAN promoters and EPFL2 under endogenous promoter rescues elongation of stem and pedicels in the epf 1,2,4,6.
Supplemental Table S1. Primers used for cloning.
Supplemental Table S2. Primers used for genotyping epfl1-1 and epfl2-1.
Supplemental Table S3. Primers used for RT-PCR.
Acknowledgments
We thank Dominique Bergmann for sharing with us seeds of the epfl4 epfl6 mutant and pEPFL5:GUS.
Footnotes
This work was supported by the National Science Foundation (IOS-0843340 to E.D.S.) and by Hunsicker Research Incentive Award (to E.D.S.).
Articles can be viewed without a subscription.
References
- Abrash EB, Bergmann DC (2010) Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 [DOI] [PubMed] [Google Scholar]
- Abrash EB, Davies KA, Bergmann DC (2011) Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bessho-Uehara K, Wang DR, Furuta T, Minami A, Nagai K, Gamuyao R, Asano K, Angeles-Shim RB, Shimizu Y, Ayano M, et al. (2016) Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc Natl Acad Sci USA 113: 8969–8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Grünewald M, Hobe M, Simon R (2002) Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol 129: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy MGR, Thompson OA, Sieger MT, Shpak ED (2012) Patterns of cell division, cell differentiation and cell elongation in epidermis and cortex of Arabidopsis pedicels in the wild type and in ERECTA. Plos ONE 7: e46262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Chater CC, Kamisugi Y, Cuming AC, Beerling DJ, Gray JE, Fleming AJ (2016) An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 143: 3306–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Yin X, Sloan J, Harrison EL, Mohammed U, Fulton T, Biswal AK, Dionora J, Chater CC, Coe RA, et al. (2018) Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MK, Wilson RL, Palme K, Ditengou FA, Shpak ED (2013) ERECTA family genes regulate auxin transport in the shoot apical meristem and forming leaf primordia. Plant Physiol 162: 1978–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE. (2001) Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol 2: 276–284 [DOI] [PubMed] [Google Scholar]
- Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, Clark SE (2003) CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbak AR, Tax FE (2011) CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 189: 177–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Fu C, Wehr DR, Edwards J, Hauge B (2008) Rapid one-step recombinational cloning. Nucleic Acids Res 36: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean O, Vernoux T, Laufs P, Belcram K, Mizukami Y, Traas J (2004) In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16: 74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T (2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hejátko J, Blilou I, Brewer PB, Friml J, Scheres B, Benková E (2006) In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Hunt L, Gray JE (2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Hunt L, Bailey KJ, Gray JE (2010) The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol 186: 609–614 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karve R, Liu W, Willet SG, Torii KU, Shpak ED (2011) The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA 17: 1907–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Oliva M, Mosquna A, Hakim O, Ohad N (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D (2003) Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Tasaka M, Torii KU, Uchida N (2018) ERECTA-family genes coordinate stem cell functions between the epidermal and internal layers of the shoot apical meristem. Development 145: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosentka PZ, Zhang L, Simon YA, Satpathy B, Maradiaga R, Mitoubsi O, Shpak ED (2017) Identification of critical functional residues of receptor-like kinase ERECTA. J Exp Bot 68: 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Lukowitz W, Ellis BE, Bergmann DC (2009) Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Wengier DL, Bergmann DC (2014) Manipulation of mitogen-activated protein kinase kinase signaling in the Arabidopsis stomatal lineage reveals motifs that contribute to protein localization and signaling specificity. Plant Cell 26: 3358–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, Laufs P, Takeda S, Aida M, Traas J, et al. (2015) Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. eLife 4: e07811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YCL, Putarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Zhang L, Han Z, Yang X, Liu W, Li E, Chang J, Qi Y, Shpak ED, Chai J (2017) A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev 31: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218: 341–353 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mandel T, Moreau F, Kutsher Y, Fletcher JC, Carles CC, Eshed Williams L (2014) The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development 141: 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel T, Candela H, Landau U, Asis L, Zelinger E, Carles CC, Williams LE (2016) Differential regulation of meristem size, morphology and organization by the ERECTA, CLAVATA and class III HD-ZIP pathways. Development 143: 1612–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S (2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L (2015) Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM (2015) Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki S, Takeuchi M, Mori M (2011) The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat Commun 2: 512. [DOI] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky MF (1999) The Arabidopsis thaliana MERISTEM LAYER1 promoter specifies epidermal expression in meristems and young primordia. Plant J 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Shimada T, Sugano SS, Hara-Nishimura I (2011) Positive and negative peptide signals control stomatal density. Cell Mol Life Sci 68: 2081–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED. (2013) Diverse roles of ERECTA family genes in plant development. J Integr Plant Biol 55: 1238–1250 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Sluis A, Hake S (2015) Organogenesis in plants: Initiation and elaboration of leaves. Trends Genet 31: 300–306 [DOI] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I (2010) Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244 [DOI] [PubMed] [Google Scholar]
- Takata N, Yokota K, Ohki S, Mori M, Taniguchi T, Kurita M (2013) Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One 8: e65183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameshige T, Okamoto S, Lee JS, Aida M, Tasaka M, Torii KU, Uchida N (2016) A secreted peptide and its receptors shape the auxin response pattern and leaf margin morphogenesis. Curr Biol 26: 2478–2485 [DOI] [PubMed] [Google Scholar]
- Tamnanloo F, Damen H, Jangra R, Lee JS (2018) MAP KINASE PHOSPHATASE1 controls cell fate transition during stomatal development. Plant Physiol 178: 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C, et al. (2014) An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Syst Biol 10: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Igari K, Bogenschutz NL, Torii KU, Tasaka M (2011) Arabidopsis ERECTA-family receptor kinases mediate morphological alterations stimulated by activation of NB-LRR-type UNI proteins. Plant Cell Physiol 52: 804–814 [DOI] [PubMed] [Google Scholar]
- Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, Tasaka M, Torii KU (2012) Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc Natl Acad Sci USA 109: 6337–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Shimada M, Tasaka M (2013) ERECTA-family receptor kinases regulate stem cell homeostasis via buffering its cytokinin responsiveness in the shoot apical meristem. Plant Cell Physiol 54: 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes CLd, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L (2005) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagarcia H, Morin AC, Shpak ED, Khodakovskaya MV (2012) Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana. J Exp Bot 63: 6493–6504 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hasson A, Rossmann S, Theres K (2016) Divide et impera: Boundaries shape the plant body and initiate new meristems. New Phytol 209: 485–498 [DOI] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA (2008) KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc National Acad Sci USA 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV (2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Xie M, Girke T, Reddy GV (2014) A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development 141: 2735–2744 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Kato A, Torii KU, Komeda Y (1998) The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J 15: 301–310 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li S, Xue S, Yang S, Huang J, Wang L (2018) Phylogenetic and CRISPR/Cas9 studies in deciphering the evolutionary trajectory and phenotypic impacts of rice ERECTA genes. Front Plant Sci 9: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]