SUMO conjugation activity causes the formation of SUMO nuclear bodies, which strongly overlap with COP1-containing nuclear bodies due to a VP motif in the E3 ligase SIZ1 that acts as bridge protein.
Abstract
Attachment of the small ubiquitin-like modifier (SUMO) to substrate proteins modulates their turnover, activity, or interaction partners. However, how this SUMO conjugation activity concentrates the proteins involved and the substrates into uncharacterized nuclear bodies (NBs) remains poorly understood. Here, we characterized the requirements for SUMO NB formation and for their subsequent colocalization with the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a master regulator of plant growth. COP1 activity results in degradation of transcription factors, which primes the transcriptional response that underlies elongation growth induced by darkness and high ambient temperatures (skoto- and thermomorphogenesis, respectively). SUMO conjugation activity alone was sufficient to target the SUMO machinery into NBs. Colocalization of these bodies with COP1 required, in addition to SUMO conjugation activity, a SUMO acceptor site in COP1 and the SUMO E3 ligase SAP and Miz 1 (SIZ1). We found that SIZ1 docks in the substrate-binding pocket of COP1 via two valine-proline peptide motifs, which represent a known interaction motif of COP1 substrates. The data reveal that SIZ1 physically connects COP1 and SUMO conjugation activity in the same NBs that can also contain the blue-light receptors CRYPTOCHROME 1 and CRYPTOCHROME 2. Our findings thus suggest that sumoylation stimulates COP1 activity within NBs. Moreover, the presence of SIZ1 and SUMO in these NBs explains how both the timing and amplitude of the high-temperature growth response is controlled. The strong colocalization of COP1 and SUMO in these NBs might also explain why many COP1 substrates are sumoylated.
SUMO (small ubiquitin-like modifier) is an essential protein modification in Arabidopsis (Arabidopsis thaliana) that is associated with >1,000 targets (Saracco et al., 2007; Miller et al., 2010; Rytz et al., 2018). Its attachment (sumoylation) is catalyzed in two steps by the SUMO E1 ACTIVATING ENZYME (SAE1/SAE2 heterodimer) and the SUMO E2 CONJUGATING ENZYME (SCE1; Colby et al., 2006; Saracco et al., 2007). SCE1 recognizes directly and modifies a short consensus motif ΨKxE in substrates (where Ψ denotes a bulky hydrophobic residue, K the acceptor Lys, x any residue, and E is Glu; Bernier-Villamor et al., 2002; Yunus and Lima, 2006). Despite this motif, 25%–50% of the SUMO substrates are modified at other sites in humans (Hendriks et al., 2014; Lamoliatte et al., 2017). Modification of these nonconsensus sites involves at least two mechanisms. First, certain E3 ligases orient SCE1, which then allows SUMO transfer to nonconsensus sites (Yunus and Lima, 2009; Gareau and Lima, 2010; Streich and Lima, 2016). Second, the presence of SUMO-interaction motifs (SIMs) in substrates promotes sumoylation at nonconsensus sites (Zhu et al., 2008; Flotho and Melchior, 2013). These SIMs are typified by a stretch of three to four aliphatic residues (Val, Ile, Leu) flanked by a series of acidic and phosphorylated residues.
SUMO is mostly attached as a monomeric adduct, but SUMO chains are also formed (Colby et al., 2006). SUMO chains are recognized by SUMO-targeted Ubiquitin E3 ligases (StUbls) that mark chain-modified proteins for degradation (Perry et al., 2008; Guo et al., 2014). Distant homologs of StUbls have been identified in Arabidopsis (Elrouby et al., 2013), but their function remains to be elucidated for plants. SUMO chain formation involves binding of a second SUMO to a charged SCE1 ∼ SUMO donor complex (Bencsath et al., 2002; Knipscheer et al., 2007; Streich and Lima, 2016). This second SUMO interacts noncovalently via a SIM-like interaction with SCE1 at a site distant from its catalytic pocket.
Interestingly, SUMO and its sumoylation machinery often resides in nuclear bodies (NBs) of various shapes and sizes in eukaryotes, including plants, as is the case in Arabidopsis for the SUMO ligase SAP and Miz 1 (SIZ1; Miura et al., 2005; Cheong et al., 2009; Kim et al., 2016), the SUMO protease OVERLY TOLERANT TO SALT 2 (Conti et al., 2008), several StUbls (Elrouby et al., 2013), and certain SUMO substrates (Ballesteros et al., 2001; Khan et al., 2014; Kim et al., 2016; Mazur et al., 2017). In general, these bodies represent micron-scale compartments that lack a surrounding membrane and self-organize due to liquid-liquid phase separations (Banani et al., 2016, 2017). SUMO-SIM interactions orchestrate at least in vitro the formation of such bodies. Often these bodies are highly dynamic containing tens to hundreds of components, but only a small set of these components are essential for their structural integrity.
The E3 ubiquitin ligase COP1 (CONSTITUTIVE PHOTOMORPHOGENIC1) is also a SUMO substrate that aggregates in NBs, called photobodies (Van Buskirk et al., 2012; Kim et al., 2016; Lin et al., 2016). COP1 function is essential for skotomorphogenesis and thermomorphogenesis (Stacey et al., 1999; Lau and Deng, 2012; Park et al., 2017; Hammoudi et al., 2018). Both darkness and elevated temperature cause COP1 translocation from the cytosol to nucleus where it colocalizes with photobodies (Stacey and von Arnim, 1999; Seo et al., 2004; Van Buskirk et al., 2012; Park et al., 2017). Photobodies take their name from the presence of photoreceptors in these NBs during the daytime, including PHYTOCHROME A (phyA), phyB, CRYPTOCHROME 1 (CRY1), CRY2, and UV-B RESISTANCE 8. In addition, photobodies contain photomorphogenesis-promoting transcription regulators such as PIFs (PHYTOCHROME INTERACTING FACTORs), HFR1 (LONG HYPOCOTYL IN FAR RED 1), HY5 (ELONGATED HYPOCOTYL 5), and LAF1 (LONG AFTER FAR1-RED LIGHT 1). Light of a wavelength matching the photoreceptor triggers the aggregation of these photobodies. In darkness, many photobody components become COP1 substrates for polyubiquitination resulting in their degradation. The general notion is that the presence of COP1 in NBs denotes degradation of COP1 substrates (Van Buskirk et al., 2012). A mutation in the substrate-binding pocket of COP1, cop1-9 (G524Q), disrupts COP1 recruitment to NBs (Stacey and von Arnim, 1999), implying that substrate binding is pivotal for the presence of COP1 in NBs. In line with this, many photobody components contain a two-residue peptide motif, Val-Pro, that is directly recognized by the COP1 substrate pocket (Holm et al., 2001, 2002; Uljon et al., 2016).
Ubiquitin ligase activity of COP1 is stimulated by the SUMO E3 ligase SIZ1, and correspondingly both skoto- and thermomorphogenesis are strongly compromised in the Arabidopsis SIZ1 loss-of-function mutant siz1-2 and the Arabidopsis SIZ1 knockdown mutant sumo1-1;amiR-SUMO2 (Lin et al., 2016; Park et al., 2017; Hammoudi et al., 2018). Hypocotyl elongation under blue, red, or far-red light is also compromised to some extent in siz1-2, whereas hypocotyl elongation due to COP1 overexpression (OE) is strongly suppressed in siz1-2 (Lin et al., 2016). Importantly, COP1 interacts directly with SIZ1, and it is SUMO-modified in a SIZ1-dependent manner at a single acceptor site (Lys-193; Kim et al., 2016; Lin et al., 2016). This Lys is important for COP1 function, as mutating this site in COP1 (OE-K193R) reduces hypocotyl elongation in comparison to that in wild-type COP1-OE lines.
In turn, SIZ1 acts as a polyubiquitination substrate of COP1, resulting in SIZ1 degradation (Lin et al., 2016). Consequently, SIZ1 protein levels are increased when COP1 function is compromised in planta (Kim et al., 2016). As SIZ1 is the main SUMO E3 ligase linked to the SUMO stress pathway, suppression of COP1 function leads to an additional rise in stress-induced SUMO adduct levels (Kim et al., 2016). This signifies that COP1 in turn controls the SUMO stress response via SIZ1. Biochemical assays showed that COP1 sumoylation stimulates the ubiquitination and degradation of HY5, a positive regulator of photomorphogenesis, again confirming that sumoylation promotes COP1 activity. Genetically, the siz1-2 mutation strongly suppresses the long hypocotyl phenotype of the hy5-215 mutant in different light conditions, and HY5 ubiquitination is also reduced in siz1-2, resulting in HY5 hyperaccumulation (Lin et al., 2016; Hammoudi et al., 2018).
At the transcriptional level, both siz1-2 and sumo1-1;amiR-SUMO2 showed a delayed and reduced transcriptional response to a shift to high temperature (Hammoudi et al., 2018). Importantly, the differentially expressed genes overlapped significantly with the genomic targets of the transcription factors PIF4 and BRASSINAZOLE RESISTANT 1, two key positive regulators of thermomorphogenesis downstream of COP1 and HY5 function (Koini et al., 2009; Quint et al., 2016; Ibañez et al., 2018). Combined, these data indicate that SIZ1 and COP1 jointly control abiotic stress responses, skoto- and thermomorphogensis, while both proteins are recruited to NBs.
As the sequestering of SUMO in NBs is poorly understood in planta, we examined by which mechanism SUMO aggregates in NBs and how SUMO and COP1 then physically interact in NBs. In line with the hypothesis of phase-separated liquid protein compartments (Banani et al., 2017), we find that formation of SUMO1∙SCE1 NBs is dynamic and requires catalytic activity of the SUMO E1 and E2 enzymes in planta. Likewise, only the conjugation-competent form of SUMO1 (SUMOGG) can stimulate formation of the SUMO1∙SCE1 (SUMO∙E2) and SUMO1∙SIZ1 (SUMO∙E3) NBs, whereas the noncovalent SUMO1∙SCE1 interaction via the SIM has apparently a dual role in their formation. Colocalization of these SUMO1∙SCE1/SIZ1 NBs with COP1 depends on the SUMO acceptor site in COP1. Conversely, we reveal that SIZ1 is a COP1-dependent ubiquitination substrate due to two valine-proline (VP) motifs that can directly bind to the COP1 substrate-binding pocket. Our data thus provide a mechanistic link between the subcellular localization of the SUMO conjugation complex and COP1 in common NBs and that recruitment to these bodies depends on the intrinsic properties of the proteins involved, i.e. SCE1 conjugation activity for SUMO NBs and substrate selection for COP1 recruitment to photobodies. Moreover, we show that SIZ1 connects these two processes.
RESULTS
Arabidopsis SUMO1 Interacts via Its SIM-Binding Site with SCE1 and SIZ1
SIMs bind to SUMO by forming an alien β-strand in the β-sheet of SUMO (Song et al., 2005; Hecker et al., 2006; Sekiyama et al., 2008). Thus far, it has remained largely undefined whether Arabidopsis SUMO1 and -2 interact with their partners via SIMs. Based on homology between Arabidopsis and the human SUMOs, we mutated two conserved hydrophobic residues (Phe-32, Ile-34) in the β2-strand of Arabidopsis SUMO1 to test in the yeast two-hybrid (Y2H) assay if—in analogy to the yeast and mammalian systems—these two residues determine binding of SIM-containing proteins (Supplemental Fig. S1A). Wild-type SUMO1 and the F32A+I34A mutant (SUMO1SIM; Supplemental Table S1) were expressed as bait fusions with the GAL4 binding domain (BD), whereas four human proteins with a known SIM were used as preys (GAL4 activation domain [AD] fusion; Hecker et al., 2006). To only test for noncovalent interactions between SUMO1 and these SIM-containing proteins, we expressed a conjugation-deficient variant of SUMO1 that lacks the C-terminal Gly-Gly (diGly) motif needed for SUMO attachment to the acceptor Lys (SUMO1ΔGG). SUMO1ΔGG interacted with three of the four SIM-containing proteins, and these interactions were suppressed by the F32A+I34A mutation except for protein inhibitor of activated STAT 1 (SUMO1ΔGG+SIM; Supplemental Fig. S1A). Deletion of the diGly motif (“ΔGG”) or introduction of the F32A+I34A double mutation (“SIM”) did not reduce SUMO1 protein accumulation in yeast (Supplemental Fig. S1G). Thus, the β2-strand of Arabidopsis SUMO1 apparently facilitates SIM binding, similar to that of its human and yeast counterparts.
To assess if Arabidopsis SUMO1 interacts via this SIM interface with SCE1 or SIZ1, the BD-SUMO1 fusions were expressed together with SCE1 or SIZ1 fused to the GAL4 AD in a Y2H assay. Both SCE1 and SIZ1 interacted with SUMO1GG and SUMO1ΔGG. The SUMO1-SCE1 interaction was impaired when the SIM-binding pocket was mutated (Supplemental Fig. S1, B and C; GG+SIM and ΔGG+SIM). The interaction between SUMO1 and SIZ1 was less strong for each SUMO1 mutant tested (Supplemental Fig. S1, B and C; ΔGG, GG+SIM, and ΔGG+SIM). As the different SUMO1 variants all accumulated at least to similar protein levels as that of wild-type SUMO1 in yeast (Supplemental Fig. S1G), we conclude that the SIM(-like) interaction also plays a role in the SUMO-SCE1 and SUMO-SIZ1 interaction in Arabidopsis. In agreement, we recently demonstrated that a conserved SIM-like motif in the N terminus of SCE1 is essential for SUMO1 binding (Mazur et al., 2017), whereas others reported the presence of SIMs in the close homologous of SIZ1 from yeast and mammals (Cheong et al., 2010; Mascle et al., 2013; Kaur et al., 2017); however, the SIM motif of Arabidopsis SIZ1 still needs further characterization.
SUMO1 Conjugation Activity Causes SCE1 and SIZ1 to Relocalize to Nuclear Bodies
Next, we examined if these SIM(-like) interactions affect the subcellular localization of SCE1 and SIZ1 in planta. We first analyzed the localization of the individual proteins expressing them as YFP/GFP fusions in Nicotiana benthamiana. YFP-SUMO1, GFP-SCE1, and GFP-SIZ1 localized to the nucleus, cytosolic pockets near the FM4-64-marked plasma membrane, and in cytoplasmic strands (Supplemental Fig. S2, A–C). GFP-SUMO1 accumulated in the cytoplasm and nucleus independent of its diGly motif and SIM-binding site (Supplemental Fig. S1D). Likewise, SCE1 resided both in the cytoplasm and nucleus (Supplemental Fig. S2, B and D), whereas SIZ1 accumulated primarily in the nucleus with a small residual signal observed in the cytoplasm (Supplemental Fig. S2C).
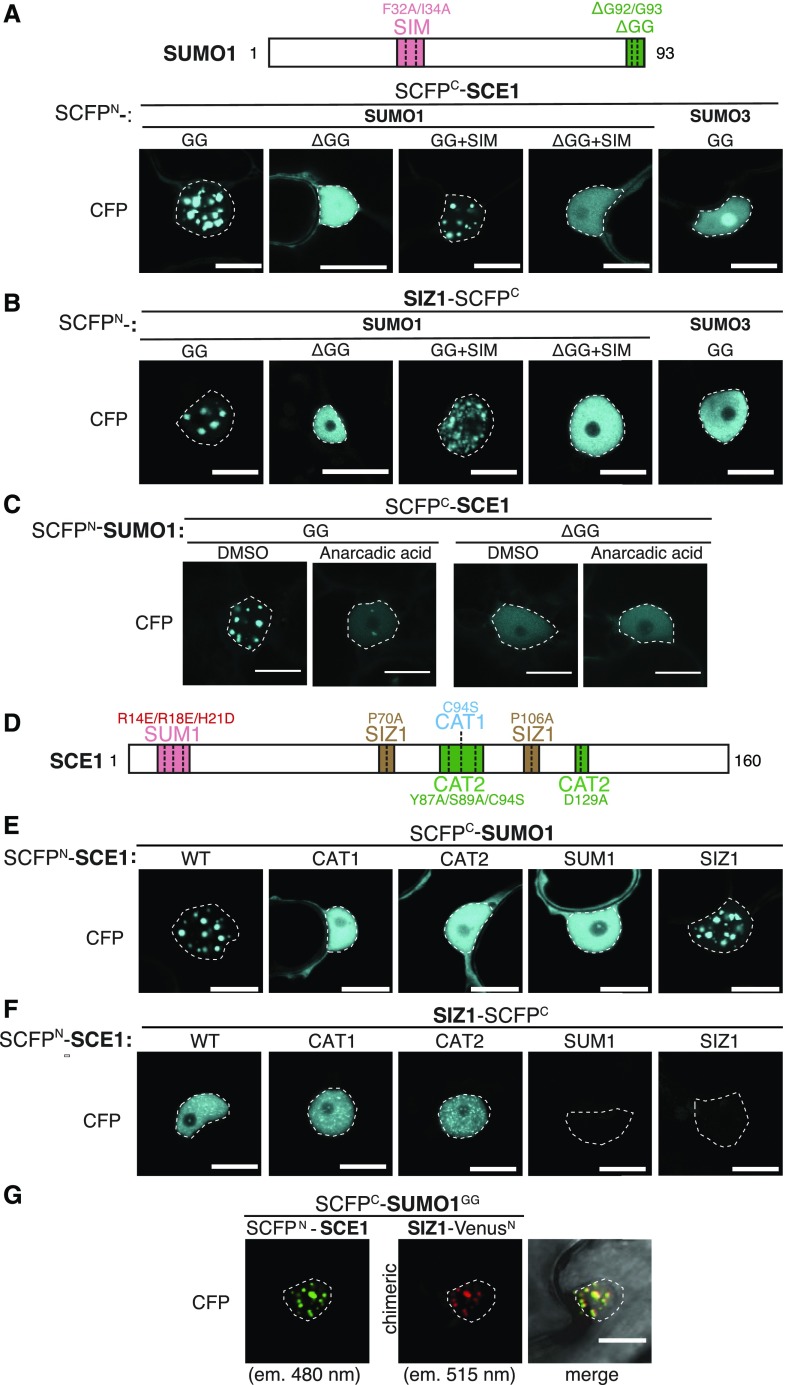
To determine if the interaction with SUMO changes the subcellular localization of SCE1 or SIZ1, we expressed the proteins as bimolecular fluorescence complementation (BiFC) pairs. To this end, the N terminus of SUMO1 was fused to super-CFPN (fragment1–173), whereas the N terminus of SCE1 was fused to SCFPC (fragment SCFP156–239). The C terminus of SIZ1 was fused to SCFPC. Expression of both SCFPN-tagged SUMO1GG and SUMO1GG+SIM with SCFPC-tagged SCE1 or SIZ1 yielded exclusively CFP reconstitution in large NBs in the nucleus for each of the four combinations (as marked with Hoechst dye in Supplemental Figure S3A.2; Fig. 1, A and B; Supplemental Fig. S3). NBs were absent for BiFC combinations with SUMO1ΔGG or SUMO1ΔGG+SIM. Instead, in combination with SCE1, the BiFC signals were found evenly spread both in the nucleus and cytoplasm (Supplemental Fig. S3A′), whereas in combination with SIZ1, the BiFC signals were found exclusively in the nucleus without any NB formation (Supplemental Fig. S3B′). These findings suggest that loading of mature SUMO is essential for NB assembly of the BiFC pairs SUMO1∙SCE1 and SUMO1∙SIZ1.
Figure 1.
SUMO nuclear body formation depends on SCE1 conjugation activity. A, Nuclear localization pattern of SUMO1∙SCE1 BiFC pairs, including mutant variants of SUMO1 and the SUMO3∙SCE1 BiFC pair. Included above is a schematic representation of residues mutated and/or deleted in SUMO1 (GG, mature SUMO; SIM, F32A+I34A; ΔGG, deletion of the C-terminal diGly motif). B, Similar to A; nuclear localization pattern of SUMO1/3-SIZ1 BiFC pairs including mutant variants of SUMO1. C, Nuclear localization pattern of the SUMO1∙SCE1 complex in response to anacardic acid inhibition of SUMO conjugation (100 μm in 1% [v/v] DMSO) 1.5 h postinfiltration. D, Schematic representation of SCE1 mutants and their substitutions. CAT1, catalytic Cys residue mutated; CAT2, binding pocket for the ψKxE SUMO acceptor motif mutated; SUM1, noncovalent association of SUMO disrupted; SIZ1, SIZ1-binding disrupted. E, Nuclear localization pattern of SUMO1∙SCE1 BiFC pairs, including mutant variants of SCE1: SCE1CAT1, SCE1CAT2, SCE1SUM1, and SCE1SIZ1. F, Nuclear localization pattern of SCE1∙SIZ1 BiFC pairs, including mutant variants of SCE1: SCE1CAT1, SCE1CAT2, SCE1SUM1, and SCE1SIZ1. G, Multicolor BiFC of SUMO1GG, SCE1, and SIZ1 showing nuclear localization pattern. The micrographs show the nuclear signal of the reconstituted SCFPN/SCFPC (SCE1∙SUMO1GG) and VenusN/SCFPC (SIZ∙SUMO1GG) fluorophores and their merged signals. The two chimeric BiFC combinations differ in their excitation and emission spectra. The BiFC pairs in A to F were fused to two halves of SCFP (SCFPN + SCFPC) with the orientation of the fusions indicated. Scale bars, 20 µm. All micrographs were taken in N. benthamiana epidermal leaf cells 2 to 3 d post-agro-infiltration with strains expressing the indicated constructs; nuclei are outlined with white lines. Supplemental Figure S3 depicts for A to F an overlay of the DIC and CFP images of the nuclei shown and a zoom-out (A′–F′) depicting the BiFC signal in the entire cell.
To confirm this notion, we examined if another Arabidopsis SUMO paralogue, SUMO3, could trigger NB assembly in a BiFC interaction with SCE1 or SIZ1. We reasoned that SUMO3 would not trigger NB assembly, as (1) compared to SUMO1 it is a poor substrate for SUMO conjugation in vitro (Lois et al., 2003), (2) it interacts weakly with SCE1 in the Y2H assay (Supplemental Fig. S1E), and (3) its overexpression in Arabidopsis fails to increase the global SUMO1/2 conjugate levels, whereas OE of SUMO1/2GG or SUMO1/2ΔGG massively increased these levels (van den Burg et al., 2010). Indeed, SUMO3GG failed to recruit SCE1 or SIZ1 in NBs (Fig. 1, A and B; Supplemental Fig. S3), whereas GFP-tagged SUMO3GG localized to the nucleus and cytoplasm similar to that observed for GFP-SUMO1GG (Supplemental Fig. S1, D and F). Additional proof that SUMO NB aggregation requires enzymatic activity came from blocking the E1 enzyme SAE1/2 with the chemical inhibitor anacardic acid. Anacardic acid binds directly to SAE1/2 and prevents formation of the activated E1 ∼ SUMO intermediate (Fukuda et al., 2009); consequently, it also prevents SUMO transfer onto the E2 (SCE1 ∼ SUMO thioester complex). In the presence of anacardic acid, the CFP signal of the SUMO1GG∙SCE1 pair disappeared from preexisting NBs within 90 min after addition of the inhibitor (Fig. 1C; Supplemental Fig. S3C). Thus, active E1 ∼ SUMO complexes are needed to retain high levels of SUMO1∙SCE1 NBs, meaning that they are dynamic.
Not Only SUMO Loading (E2 ∼ SUMO), but Also the SUMO Binding Site (E2∙SUMO) Is Critical for SCE1 to Assemble in SUMO1∙SCE1 NBs
To further discern how these NBs are formed, different Arabidopsis SCE1 variants were used that either (1) cannot interact with SUMO or SIZ1, (2) had lost their catalytic activity (SCE1CAT1, SCE1CAT2), or (3) no longer recognized the SUMO acceptor motif ΨKxE in SUMO substrates (SCE1CAT2; Mazur et al., 2017; Fig. 1D; Supplemental Table S1). Previous studies on the human and yeast SUMO E2 enzymes had identified that the residues Arg-14, Arg-18, and His-21 of SCE1 together determine binding of the second SUMO to the site distant from the catalytic pocket (E2∙SUMO complex; Bencsath et al., 2002). Likewise, the residues Pro-69 and Pro-106 of SCE1 are essential for the human Ubc9 (SUMO E2) to interact with the PIAS family of SUMO E3 ligases (closest homolog of Arabidopsis SIZ1; Mascle et al., 2013). These five residues are strictly conserved in Arabidopsis SCE1 and other plant homologs of SCE1 (Supplemental Fig. S4), and also for Arabidopsis SCE1 these residues are essential for it to interact with SUMO1 or SIZ1 in the Y2H assay (Mazur et al., 2017). The SCE1CAT1 mutant still interacted with SUMO1; however, SCE1CAT2 had lost both its catalytic activity and its noncovalent interaction with SUMO1 (see Supplemental Table S1 for an overview of the mutants used; Mazur et al., 2017).
Introduction of these mutations in SCE1 did not change its subcellular localization in planta, i.e. each variant showed a uniform distribution in both the nucleus and cytoplasm when transiently expressed as a GFP-fusion in N. benthamiana (Supplemental Fig. S2D). Both SCE1CAT1 and SCE1SUM1 appeared to accumulate to higher protein levels in planta (Supplemental Fig. S2E), possibly indicating that the stability of wild-type SCE1 is negatively impacted by its own enzymatic activity and/or its noncovalent association with SUMO1. Importantly, SCE1SIZ1 still interacted with SUMO1GG in NBs, but SCE1CAT1, SCE1CAT2, and SCE1SUM1 were not recruited to NBs, whereas they still interacted with SUMO1GG in the BiFC assay (Fig. 1E; Supplemental Fig. S3E). These findings mark that besides SUMO loading (E2 ∼ SUMO thioester complex), the second SUMO interaction (E2∙SUMO) is also critical for SCE1 to assemble in SUMO1∙SCE1 NBs.
The SUMO∙SCE1 Interaction Contributes to the SCE1-SIZ1 Interaction
In contrast to the SUMO1GG∙SCE1/SIZ1 complexes, we noted that the SCE1∙SIZ1 BiFC complex did not aggregate in NBs but rather resided in small nuclear speckles spread across the nucleus (Fig. 1F; Supplemental Fig. S3F). We already showed that the SCE1 residues Pro-70 and Pro-106 (Supplemental Table S1; SCE1SIZ1) are essential for SCE1 and SIZ1 to interact in the Y2H assay, whereas mutating the catalytic site (SCE1CAT1 and SCE1CAT2) did not impair their Y2H interaction (Mazur et al., 2017). Also in the BiFC assay, both SCE1SUM1 and SCE1SIZ1 failed to interact with SIZ1, whereas the catalytic-dead variants (SCE1CAT1 and SCE1CAT2) still interacted with SIZ1. This suggests that SUMO loading in the catalytic site of SCE1 (E2 ∼ SUMO1) is not required for SCE1 and SIZ1 to interact, whereas the noncovalent interaction (E2∙SUMO1) strengthens this interaction in both the Y2H and BiFC assays (Fig. 1F; Mazur et al., 2017). Apparently, the second SUMO acts as glue in the SCE1-SIZ1 complex or it alters the SCE1 conformation such that it enhances the interaction between SCE1 and SIZ1.
Next, we examined whether the BiFC pairs SUMO1GG∙SCE1 and SUMO1GG∙SIZ1 physically colocalized in the same NB. For this, we performed multicolored BiFC (Gehl et al., 2009), where SUMO1GG was expressed as a fusion protein with SCFPC, SCE1 was fused at its N terminus to SCFPN, and SIZ1 was fused at its C terminus to residues 1 to 173 of the Venus fluorophore (VenusN). Reconstitution of both fluorophores was examined using optical filters that separate CFP (for SCFPC-SUMO1GG with SCFPN-SCE1) from the chimeric VenusN-SCFPC signal (for SCFPC-SUMO1GG with VenusN-SIZ1). This multicolored BiFC experiment revealed that the signals for SUMO1GG∙SCE1 and SUMO1GG∙SIZ1 completely overlapped in the NBs (Fig. 1G).
As the SCE1∙SIZ1 BiFC pair did not form large NBs (Fig. 1F), we tested if the levels of free SUMO1GG were limiting, thus preventing aggregation of the SCE1∙SIZ1 pair in enlarged nuclear structures when expressed alone. We expressed the SCE1∙SIZ1 pair together with YFP-tagged SUMO1GG or SUMO1ΔGG (negative control). The YFP-SUMO1GG signal overlapped with the SCFP signal, but only in a few cells the SCE1∙SIZ1 BiFC signal shifted from speckles/puncta to enlarged NBs. YFP-SUMO1ΔGG localized as well to the nucleus, but overall it did not colocalize with the SCE1∙SIZ1 BiFC pair in nuclear specks (Supplemental Fig. S5). Thus, the SUMO1-dependent NB enlargement is foremost seen when SUMO is trapped in a BiFC interaction with SCE1 or SIZ1. Possibly, the BiFC system stabilizes a transient protein-protein interaction, which then allows formation of enlarged SUMO NBs. Alternatively, tagging of SUMO1GG with an intact YFP protein might cause steric hindrance in the ternary complex.
SUMO Chain Formation Potentially Stimulates Formation and Enlargement of SUMO NBs
As SUMO chain formation in mammalian cells promotes formation of promyelocytic leukemia NBs (Hattersley et al., 2011), we assessed whether formation of SUMO NBs depends on SUMO chain formation in plants. To block SUMO chain formation, we engineered SUMO1 variants in which Lys-9, Lys-10, or all seven Lys residues were replaced by Arg (SUMO1K9R, SUMO1K10R, SUMO1K9R+K10R, and SUMO1K7ØR; Supplemental Table S1). Although Lys-10 is the main site for SUMO chain elongation, Lys-9 can serve as an alternative site (Colby et al., 2006). Therefore, we also created the K9R+K10R double mutant. To test if these KtoR mutants can still recruit SCE1 to NBs, they were expressed together with SCE1 as BiFC pairs. Two days post-agro-infiltration, NBs were formed, but they were reduced in size and number for the KtoR mutants compared to that for wild-type SUMO1, whereas concomitantly the diffuse nuclear signal for SCFP increased (Supplemental Fig. S6A). The SUMO1K7Ø∙SCE1 pair failed entirely to form NBs 2 d postinfiltration, whereas the SUMO1K9R+K10R∙SCE1 pair yielded less NBs at this time point. These data agree with the proposed role of SUMO chains in NB formation. However, 3 d post-infiltration, NBs were found for each KtoR SUMO1 mutant (Supplemental Fig. S6A), albeit the number of cells that contained NBs was less for SUMO1K7Ø∙SCE1 (40%–50% for SUMO1K7Ø compared to 70%–100% for the other combinations). Even though we cannot rule out that these Lys mutations influence protein translation, these data suggest that SUMO chain formation potentially stimulates the targeting of the SUMO1∙SCE1 complex to NBs, affecting both their initial formation and subsequent enlargement.
SUMO1GG∙SCE1 NBs Colocalize Completely with COP1 NBs
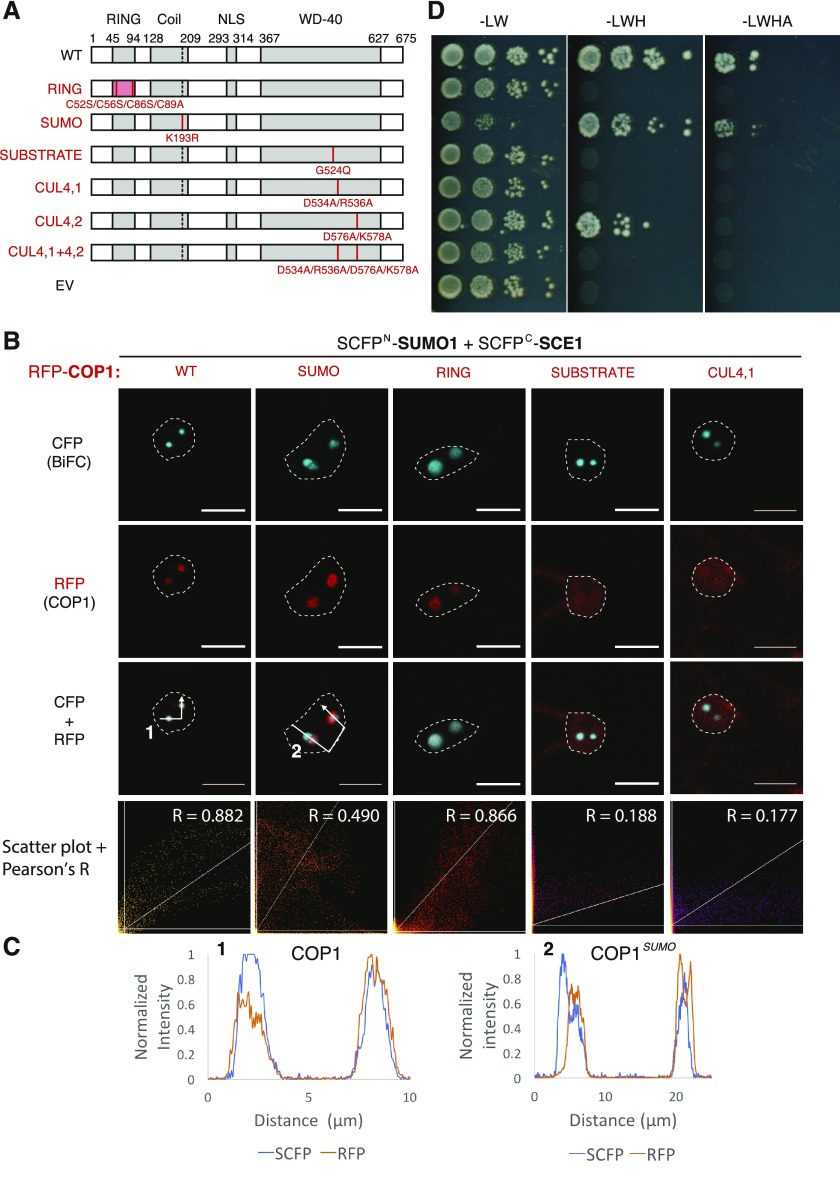
Importantly, COP1 colocalizes with SIZ1 in NBs (Kim et al., 2016). Using the Y2H assay, we confirmed that COP1 and SIZ1 interact directly, whereas COP1 does not interact with SCE1, SUMO1, or SUMO3 (Fig. 2A). We then assessed whether COP1 sumoylation controls its colocalization with SUMO1GG∙SCE1 in NBs. As for the SIZ1-COP1 combination (Kim et al., 2016), RFP-COP1 colocalized strongly with the SUMO1GG∙SCE1 BiFC pair in NBs (Fig. 2, B and C; Supplemental Fig. S6C). Importantly, both the BiFC pairs SUMO1ΔGG∙SCE1 and SUMO1GG∙SCE1CAT1 (Fig. 2, B and C) still failed to localize to NBs regardless of RFP-COP1 OE. Thus, colocalization of SCE1 and COP1 in NBs requires formation of the E2 ∼ SUMO thioester. Coexpression of RFP-COP1 with different SUMO1KtoR∙SCE1 BiFC pairs revealed that all these SUMO1KtoR∙SCE1 variants strongly colocalized with COP1 in NBs irrespective of which Lys was mutated (Supplemental Fig. S6B). Next, we tested if the COP1 SUMO acceptor site (Lys-193) is important for this colocalization. Introduction of K193R (COP1SUMO) reduced the overlap between COP1 and SUMO1∙SCE1 NBs (Fig. 3, B and C; Supplemental Fig. S7A), but it did not suppress the COP1-SIZ1 protein-protein interaction in the Y2H assay (Fig. 3, A and D), suggesting that residues other than Lys-193 promote the interaction between COP1 and SIZ1.
Figure 2.
The SUMO1∙SCE1 BiFC pair colocalizes with COP1 in nuclear bodies when catalytically active. A, Y2H analysis of the interaction between COP1 as a GAL4 BD fusion protein and the SUMO (machinery) proteins fused to the GAL4 AD domain. Yeast growth was scored 3 d after incubation on selective media at 30°C (−Leu [L] and Trp [W], −LW and His [H], −LWH + 1 mm 3-Amino-1,2,4-triazole [3AT], −LWH and Adenine [A]). B and C, Nuclear localization pattern of the SUMO1∙SCE1 BiFC pair in cells overexpressing RFP-COP1. B, ΔGG, conjugation-deficient SUMO variant; C, CAT1, catalytically inactive SCE1. Micrographs show from top-to-bottom the reconstituted BiFC signal, RFP-COP1, and their merged signals. Conditions were identical to those in Figure 1. Scale bars, 10 µm.
Figure 3.
The COP1-SIZ1 interaction in NBs depends on both the substrate binding pocket and the SUMO acceptor site in COP1. A, Schematic representation of COP1 variants and the protein-protein interaction domain disrupted (left side, red text) with their mutations shown: RING, RING Zn2+-finger binding domain mutant; SUMO, loss of SUMO acceptor site; SUBSTRATE, mutation in the substrate binding groove; CUL4, mutations in the CUL4-binding “WDRX” motifs. WT, Wild type. B, Nuclear localization pattern of the SUMO1∙SCE1 BiFC pair in cells overexpressing functional mutants of RFP-COP1. Micrographs depict from top to bottom the BiFC CFP signal, RFP-COP1, and their combined signals. Bottom row depicts a scatter plot of the RFP/CFP signal intensities per pixel and their Pearson’s correlation coefficient (R). Conditions were identical to those in Figure 1. Scale bars, 10 µm. C, Normalized intensity profiles depict the fluorescence signal intensities for SCFP: (BiFC pair) and RFP-COP1: coexpression of (1) wild-type COP1 or (2) COP1SUMO; the profiles follow the white arrows depicted in B. Note the profiles of RFP-COP1SUMO (A) and SUMO∙SCE1 CFP signals poorly overlap in (2). D, Mapping of the SIZ1 interaction site in COP1 using Y2H analysis of the COP1 variants depicted in B. The COP1 variants were fused to the GAL4 BD, whereas SIZ1 was fused to the GAL4 AD-fusion. Yeast growth was scored 3 d after incubation on selective medium at 30°C.
Structural studies had revealed that COP1 substrates interact via a VP motif with COP1 by docking in the central groove of the COP1 WD-40 propeller head (Uljon et al., 2016). The mutation Q529E (the causal mutation in the cop1-9 allele, hereafter referred to as COP1SUBSTRATE) is positioned in this VP-binding groove and was shown to disrupt the binding of various COP1 substrates (Holm et al., 2002). Importantly, COP1SUBSTRATE fails to form NBs when expressed as a GFP-fusion protein (Stacey et al., 1999). COP1 also interacts with the DNA damage-binding protein 1-Cullin 4 (CUL4) E3 ubiquitin ligase complex via two WDRX motifs; these WDRX motifs are also located in the WD-40 domain near the VP-binding site (Chen et al., 2010).
We used these COP1 variants to test whether the capacity of COP1 to bind substrates or CUL4 determines (1) its targeting to NBs and (2) in particular to SUMO NBs. First, we tested if these COP1 variants still interacted with SIZ1 in the Y2H assay (Fig. 3D). Except for COP1SUMO, all the other mutants failed to interact with SIZ1 (Fig. 3D). When fused to RFP, COP1SUMO and COP1RING still formed NBs in N. benthamiana, whereas the variants COP1SUBSTRATE, COP1CUL4,1, and COP1CUL4,1 and 4,2 failed to localize to NBs in planta (Fig. 3C; Supplemental Fig. S7B). Mutations in the WD-40 domain thus suppressed formation of COP1 NBs. To quantify the degree of colocalization between these COP1 variants and the SUMO1GG∙SCE1 BiFC signal, the RFP/CFP pixel intensities were depicted in a scatter plot. This yielded a strong positive correlation between the signal intensities of wild-type RFP-COP1 and the SUMO1GG∙SCE1 CFP signal (Pearson’s R = 0.882). The localization of RFP-COP1RING also correlated strongly with the SUMO1GG∙SCE1 CFP signal (Pearson’s R = 0.866). The degree of colocalization was less for RFP-COP1SUMO (Pearson’s R = 0.490), whereas localization of the variants RFP-COP1SUBSTRATE, -COP1CUL4,1, and -COP1CUL4,1 and 4,2 did not correlate with the SUMO1GG∙SCE1 signal (Pearson’s R = 0.188, 0.177 and 0.148, respectively; Fig. 3B; Supplemental Fig. S7B).
Likewise, RFP-COP1 colocalized with the SUMO1∙SIZ1 pair in NBs (Fig. 4A). Their colocalization in NBs required an intact substrate-binding pocket in COP1 (COP1SUBSTRATE). Similar to that of the SUMO1∙SCE1 BiFC pair, the SUMO acceptor site in COP1 (COP1SUMO) contributed to the overlap between the RFP-COP1 and SUMO1∙SIZ1 signals in NBs, whereas disruption of the COP1 ubiquitin ligase activity (COP1RING) had no apparent effect on targeting of the SUMO1∙SIZ1 complex to these COP1 NBs (Fig. 4A). Thus, the COP1 substrate binding pocket appears to be the main determinant for COP1 recruitment to NBs and its binding to SIZ1.
Figure 4.
Formation of COP1 + SUMO1∙SIZ1-containing NBs depends on the COP1 substrate-binding pocket that apparently recruits SIZ1 via VP motifs. A, Nuclear localization pattern of the SUMO1∙SIZ1 BiFC pair in cells overexpressing RFP-COP1 variants: COP1SUMO, COP1RING, and COP1SUBSTRATE. See Figure 3A for details on the COP1 variants. Micrographs show from top to bottom the BiFC signal, RFP-COP1, and their merged signals. Conditions were identical to those in Figure 1. Scale bars, 10 µm. B, Schematic representation of SIZ1 variants and the protein-protein interaction domain disrupted (left side) by the mutations introduced: plant homeodomain (PHD) and Pro-Ile-Asn-Ile-Thr (PINIT), both reduced substrate binding; SP-RING, no interaction with SCE1; VP1 and VP2, putative interacting motifs for the COP1 substrate binding groove. C, Mapping of the COP1 interaction site in SIZ1 using Y2H analysis of the SIZ1 variants depicted in B. Similar to that in Figure 3, SIZ1 variants were fused to GAL4 AD-fusion, whereas COP1 was fused to GAL4 BD. Yeast growth was scored after 3 d at 30°C.
SIZ1 Contains Two VP Domains Important for COP1 Binding
To map the COP1-binding site in SIZ1, a series of SIZ1 mutants was prepared. In yeast, both the plant homeodomain and Pro-Ile-Asn-Ile-Thr domain are essential for ScSIZ1-directed sumoylation of the substrate PCNA (proliferating cell nuclear antigen; Yunus and Lima, 2009). Loss-of-function mutations in these two domains in Arabidopsis SIZ1 did not compromise the interaction with COP1 in the Y2H assay. Disruption of the Siz/PIAS (SP)-really interesting new gene (RING) domain, which is needed to recruit the E2 ∼ SUMO thioester into a complex with its substrate proliferating cell nuclear antigen (Yunus and Lima, 2009), impaired the interaction between SIZ1 and COP1 (Fig. 4, B and C). SIZ1 also contains two VP motifs (VP1, Val-251, and VP2, Val-725/728). We tested if COP1 recognizes SIZ1 via these two VP motifs. Both motifs were mutated by replacing the Val residues with Asp. Mutating VP1 did not suppress the SIZ1-COP1 interaction, whereas mutating VP2 reduced this interaction. Furthermore, mutating both VP motifs disrupted the interaction entirely (Fig. 4, B and C). Thus, the SP-RING domain of SIZ1 contributes to the interaction with COP1, but the VP motifs combined are essential for this interaction.
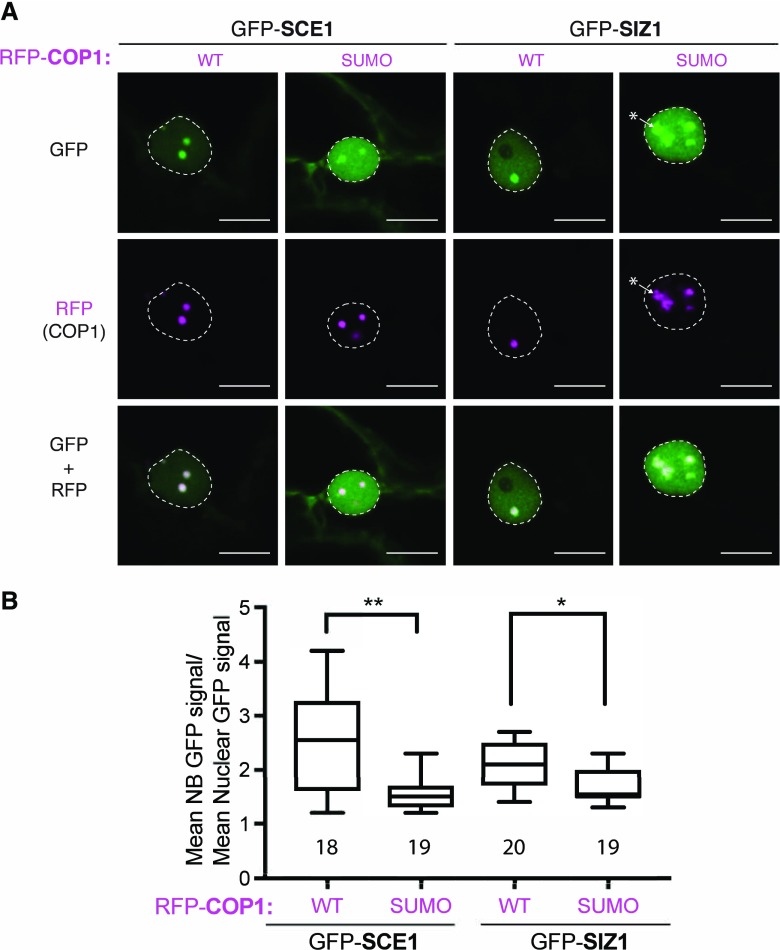
To determine if SCE1 and SIZ1 can already reside in COP1 NBs independent of their SUMO-BiFC interaction, we coexpressed GFP-tagged SCE1 or SIZ1 together with RFP-tagged COP1 or COP1SUMO. The GFP signal of both proteins (SCE1 and SIZ1) was enriched in RFP-COP1 NBs (Fig. 5A). Moreover, their targeting to COP1 NBs was significantly less when the COP1 SUMO acceptor site was mutated (Fig. 5, A and B) and these GFP-SIZ1/RFP-COP1SUMO NBs also had an amorphous shape (asterisk in Fig. 5A). Combined, these data suggest that the COP1 SUMO acceptor site controls recruitment of SCE1 and SIZ1 to COP1 NBs.
Figure 5.
Colocalization of GFP-tagged SCE1/SIZ1 with RFP-COP1 NBs is compromised following mutation of the COP1 SUMO acceptor site. A, Nuclear localization pattern of GFP-tagged SCE1 and SIZ1 in the presence of RFP-COP1 or the RFP-COP1SUMO SUMO acceptor site mutant (Lys-193Arg). * marks amorphous NBs in the GFP-SIZ1, RFP-COP1SUMO combination. Micrographs from top to bottom: GFP, RFP, and their merged signals. Scale bars, 10 µm; WT, wild type. B, Quantification of the average GFP signal intensity in the NBs per nucleus divided by the average fluorescence signal in the nucleus (with the data of three biological replicates pooled with at least five nuclei per replica). Total number of nuclei analyzed is shown. Significant differences were detected using an unpaired Student’s t test assuming unequal variances; **P < 0.01, *P < 0.05. Conditions were identical to those in Figure 1.
Recruitment of SCE1 to COP1 NBs Requires the SIZ1 Gene in Arabidopsis
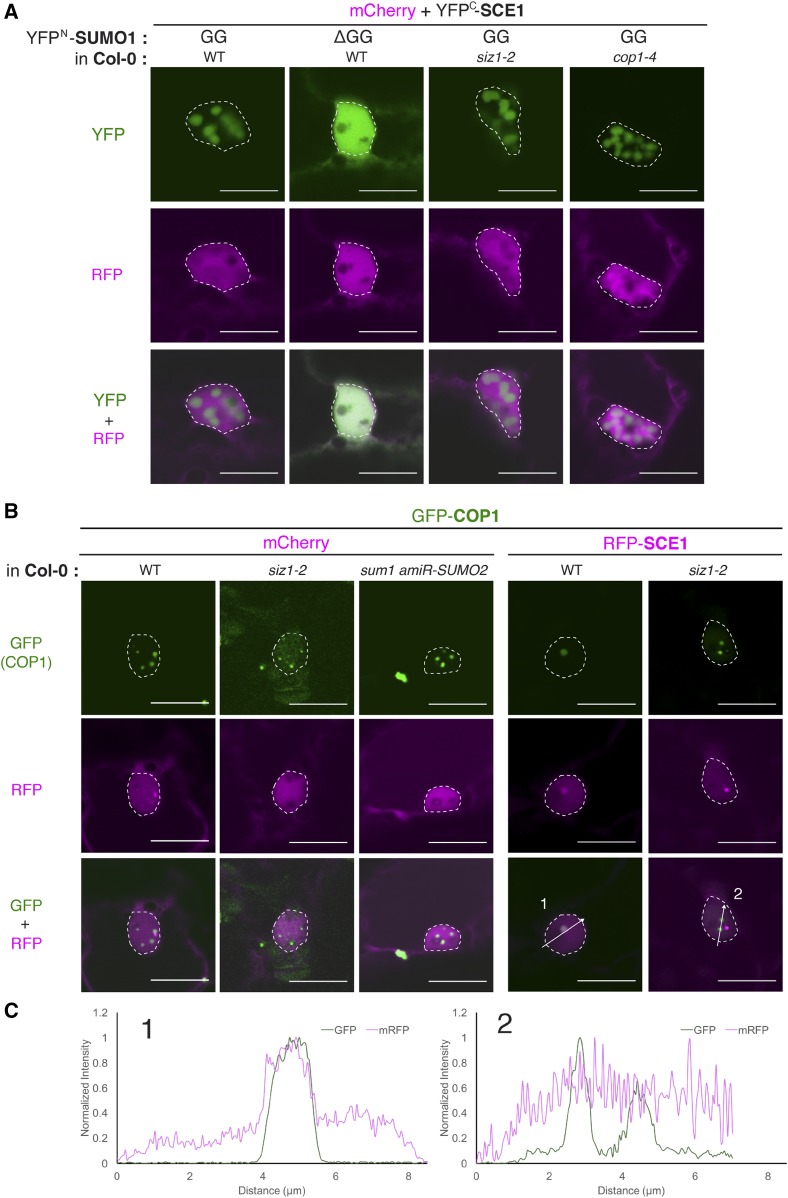
As the native proteins are still present in the N. benthamiana BiFC experiments, we shifted our experimental method to particle bombardments in Arabidopsis leaf epidermal cells. Using the Arabidopsis mutants cop1-4, siz1-2, and sumo1;amiR-SUMO2, we genetically inferred whether these endogenous proteins are required for (co)localization of SCE1 and COP1 in NBs. The different constructs were introduced in high-level expression vectors suitable for particle bombardment (Walter et al., 2004), in which SCE1 and SUMO1GG/SUMO1ΔGG were tagged at their N-termini with the BiFC halves NYFP and CYFP, respectively. Similar to the N. benthamiana data, the SUMO1GG∙SCE1 combination formed NBs in Arabidopsis, whereas SUMO1∆GG∙SCE1 interacted but this combination yielded a uniform BiFC signal in the nucleus (Fig. 6A). Targeting of SUMO1GG∙SCE1 to NBs did not change when the BiFC protein complex was expressed in the genotypes cop1-4 or siz1-2, confirming that SUMO conjugation is the main force behind SUMO NB formation. To assess genetically if sumoylation activity is essential for COP1 NB formation, GFP-COP1 was expressed in wild-type Arabidopsis (Col-0), siz1-2, and sumo1;amiR-SUMO2 using particle bombardment. Irrespective of these three genetic backgrounds, GFP-tagged COP1 localized to NBs in Arabidopsis cells (Fig. 6B). This corroborates our notion that SUMO conjugation activity is not a key driving force for COP1 aggregation in NBs. To demonstrate that the colocalization of SCE1 and COP1 in NBs depends on SIZ1, RFP-SCE1, and GFP-COP1 were transiently coexpressed in wild-type and siz1-2 Arabidopsis cells using particle bombardment. As expected, in wild-type cells, RFP-SCE1 was enriched in GFP-COP1 NBs, whereas in siz1-2 cells, RFP-SCE1 was absent from these COP1 NBs (Fig. 6, B and C). Thus, recruitment of SCE1 and COP1 the same NB requires the presence of the endogenous SIZ1 protein.
Figure 6.
SUMO conjugation also triggers NB formation in Arabidopsis, which depends genetically on SIZ1 for recruitment to COP1 NBs. A, Nuclear localization pattern of the SCE1∙SCE1 BiFC pair in Arabidopsis cells (wild type [WT], siz1-2, or cop1-4) transformed using particle bombardment. The BiFC constructs and the Arabidopsis genotypes used are indicated at the top. To detect the transformed cells, tissue was cobombarded with mCherry. The micrographs show from top to bottom the YFP, RFP, and the merged signal; nuclei are outlined with a white line. Four-week-old Arabidopsis rosettes were bombarded. Two days postbombardment, plant material was transferred to a dark box, and the fluorescence signals were examined after a further day. B, Nuclear localization pattern of GFP-COP1 and RFP-SCE1 in Arabidopsis cells (wild type, siz1-2, or sumo1;amiR-SUMO2) transformed using particle bombardment. To detect the transformed cells, tissue was either cobombarded with mCherry or RFP-SCE1. Micrographs with nuclei with GFP-COP1 containing NBs are shown. Scale bars, 10 μm. C, Normalized intensity profiles depict the fluorescence signal intensities of GFP-COP1 and RFP-SCE1 co-expressed in wild type and the siz1-2 mutant, along arrows 1 and 2 in the right two panels of (B).
Simultaneous Recruitment of CRY1/2 and SUMO1∙SCE1 to COP1 NBs
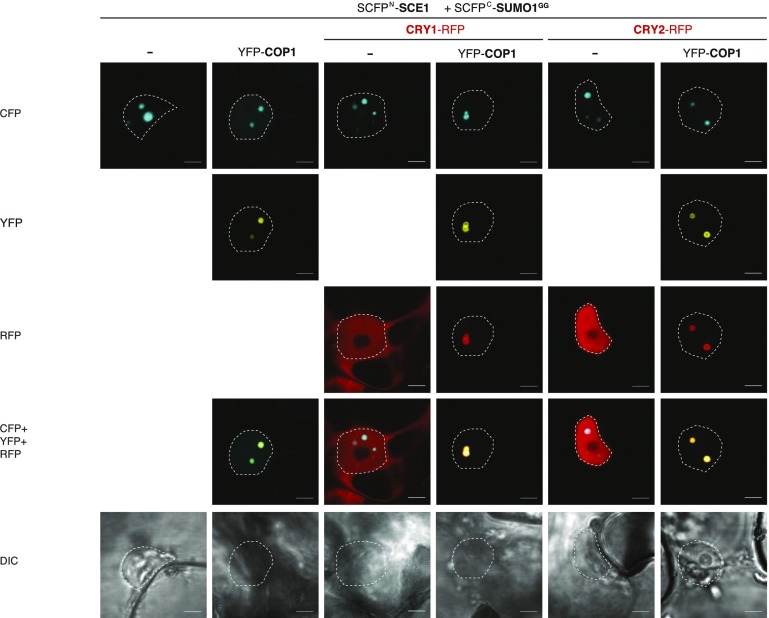
CRY1 and CRY2 are blue light receptors that inhibit COP1 activity indirectly via SUPPRESSOR OF PHYA-105 (SPA) proteins or directly, respectively, after blue light exposure (Wang et al., 2001; Liu et al., 2011; Holtkotte et al., 2017). Fluorescent protein fusions of CRY2 localize to NBs after blue light exposure (Más et al., 2000). As, at least to our knowledge, neither CRY1 nor CRY2 are sumoylated, their subcellular localization might correlate with COP1, but not with the SUMO conjugation enzymes. CRY1-GFP and CRY2-GFP were coexpressed with RFP-COP1, RFP-SCE1, and SIZ1-RFP in N. benthamiana cells. CRY2-GFP localized in our system in NBs and CRY1-GFP localized to the whole nucleus. As expected, both CRY1 and CRY2 were recruited to mRFP-COP1 NBs (Supplemental Fig. S8, A and B). SIZ1-mRFP or mRFP-SCE1 coexpressed with CRY1/CRY2-GFP were evenly distributed in the nucleus and did not alter CRY1- or CRY2-GFP localization. Importantly, SCE1 and SIZ1 were not recruited to CRY2-GFP NBs. Next, the SUMO1GG∙SCE1 BiFC pair was coexpressed with CRY1-mRFP or CRY2-mRFP. Neither CRY1-mRFP nor CRY2-mRFP was recruited to the SUMO1GG∙SCE1 NBs (Fig. 7). However, when YFP-COP1 was coexpressed in the same cells, all components colocalized to common NBs. Thus, CRY2 NBs and SUMO1GG∙SCE1 NBs are different entities that coincide in COP1 NBs.
Figure 7.
CRY1-mRFP or CRY2-mRFP and the SUMO1∙SCE1 BiFC complex colocalize in COP1 NBs. Micrographs of cells transiently expressing SCFPN-SCE1 and SCFPC-SUMO1GG with or without YFP-COP1 and either CRY1-mRFP or CRY2-mRFP, imaged 3 d post-agro-infiltration. Scale bars, 10 µm.
DISCUSSION
Here, we reveal the mechanism for condensation of SUMO1, SCE1, and SIZ1 in NBs. Their condensation in NBs requires both SCE1 catalytic activity and an intact binding site for the second SUMO on SCE1 (Fig. 1, A and E). Strikingly, COP1 fully colocalizes with these NBs. COP1 is the master regulator of skoto- and thermomorphogenesis (Hoecker, 2017; Park et al., 2017). Once present in the nucleus, COP1 targets positive regulators of light signaling in photobodies for degradation (Van Buskirk et al., 2012). We found that mutating the COP1 SUMO acceptor site (Lys-193) reduces the overlap between COP1 NBs and SUMO1GG-SCE1 BiFC NBs, albeit both still predominantly localize to NBs (Fig. 3, B and C; Supplemental Fig. S7A). In support, colocalization of SCE1 or SIZ1 alone (GFP-tag) with COP1 NBs was also reduced when both proteins were coexpressed with the COP1 SUMO acceptor mutant (K193R; Fig. 5). In the Y2H assay, COP1 interacts physically only with SIZ1 and not with other components of the SUMO conjugation machinery (SCE1, SUMO1, or SUMO3). In line with this, COP1 sumoylation is SIZ1 dependent and stimulates COP1 biochemical activity, resulting in ubiquitylation and increased degradation of SIZ1 and HY5, another COP1 substrate (Lin et al., 2016). We identified and characterized two VP motifs in SIZ1, which represent a known recognition motif found in COP1 substrates (Uljon et al., 2016), that together were essential for SIZ1 to interact with COP1 (Fig. 4, B and C). Targeting of SCE1 to COP1 NBs required, genetically, also a functional SIZ1 gene in Arabidopsis, whereas aggregation of the SUMO1GG∙SCE1 BiFC pair in NBs was independent of a functional SIZ1 or COP1 allele (Fig. 6, B and C). Thus, COP1 likely recognizes SIZ1 as a substrate via these two VP motifs and in turn SIZ1 brings SCE1 activity to COP1. Previously, we noted that the short hypocotyl phenotype of a cop1-4 mutant is further enhanced by introducing a SIZ1 loss-of-function mutation (cop1-4;siz1-2; Lin et al., 2016; Hammoudi et al., 2018), which implies that SIZ1 promotes the residual activity of the cop1-4 isoform. Notably, cop1-4 plants express a truncated COP1 protein consisting of its N-terminal half including Lys-193 (Stacey et al., 1999), suggesting that it can still be sumoylated by SIZ1 even though the WD-40 domain is missing. Possibly, the SPA proteins also have a role in recruiting SIZ1 to the truncated cop1-4 isoform. Moreover, aggregation of COP1 and SCE1 to NBs was in both cases independent of the SIZ1 gene, again confirming that SIZ1 merely acts as a bridge protein. As many photobody components are SUMO substrates (e.g. phyB, HFR1, LAF1, HY5, HYL [HY5 HOMOLOG], PIF4, and PIF5; Ballesteros et al., 2001; Seo et al., 2003; Conti et al., 2014; Sadanandom et al., 2015; Tan et al., 2015; Mazur et al., 2017; Rytz et al., 2018), it is thus likely that SIZ1 resides in photobodies prior to COP1 recruitment and that once COP1 is present in these photobodies, its activity is then stimulated by SIZ1. However, this idea needs further study.
Sumoylation and Blue Light Signaling Coincide in COP1 Bodies
The blue light photoreceptors CRY1 and CRY2 inhibit COP1 activity in a blue-light-dependent manner (Wang et al., 2001). The interaction of CRY2 and COP1 is direct, whereas CRY1 uses SPA1 (and other SPA proteins) as a bridge protein (Wang et al., 2001; Liu et al., 2011; Holtkotte et al., 2017). Upon blue light exposure, CRY2 is rapidly degraded. COP1 is only partially responsible for this light-dependent degradation of CRY2 (Shalitin et al., 2002). We observed that CRY1 and CRY2 are recruited to SCE1, SIZ1, and the SUMO-SCE1 BiFC complex in a COP1-dependent manner (Fig. 7; Supplemental Fig. S8). This suggests that the first steps in the blue light response are sumoylation independent and that thereafter all components are apparently localized to the same NBs for downstream responses. As CRY1 is not and CRY2 is partially dependent on COP1 for its degradation, their action might, however, affect or be affected respectively by COP1 sumoylation. As sumoylation enhances COP1 activity (Lin et al., 2016), CRY1 or CRY2 might either prevent COP1 sumoylation or make COP1 insensitive to CRY1 or CRY2 interference under light conditions.
Both SUMO Loading in SCE1 and Binding of a Second SUMO by SCE1 Drives SUMO1∙SCE1 NB Formation
Our data on the molecular interactions among Arabidopsis SUMO1, SCE1, and SIZ1 support that they adopt a ternary complex as reported for their yeast and human counterparts (Bencsath et al., 2002; Bernier-Villamor et al., 2002; Reverter and Lima, 2005; Mascle et al., 2013; Sekhri et al., 2015). Recruitment of this complex to NBs depends tightly on their intermolecular interactions in N. benthamiana and Arabidopsis. We established that the SIM-binding cleft around Phe-32 (Colby et al., 2006) is functionally conserved in Arabidopsis SUMO1 and that it increases the interaction strength between SUMO and SCE1/SIZ1 (Fig. 1A). Mutations in this SIM-binding pocket of SUMO did not suppress aggregation of the SUMO∙SCE1 and SUMO∙SIZ1 BiFC pairs in NBs, whereas conjugation-deficient variants of SUMO1 failed to aggregate in NBs (Fig. 1, A and B). Conversely, mutations in the second SUMO-binding pocket of SCE1 (SCE1SUM1) blocked NB formation for the SUMO1-SCE1 BiFC pair. Thus, binding of a second SUMO to SCE1 is apparently a key factor for NB assembly. Three lines of evidence argue that SUMO NBs here seen are not an artifact of BiFC or caused by unspecific aggregation of (misfolded) proteins, but rather require active formation and/or maintenance. First, preexisting NBs disappear within 90 min after inhibition of the SUMO E1 enzyme by anacardic acid. Second, NB formation requires an intact SCE1 catalytic site. Third, SUMO3 failed to trigger NB formation.
Our data suggest that SUMO chain formation is not essential but might promote SUMO1∙SCE1 NB formation (Supplemental Fig. S6A). The noncovalent interaction between SUMO and SCE1 was reported to be important for SUMO chain formation (Knipscheer et al., 2007). Our data support a mechanistic model in which both thioester formation (E2 ∼ SUMO) and noncovalent binding of SUMO to SCE1 (E2∙SUMO) are essential for the redistribution of Arabidopsis SUMO1 and SCE1 to NBs. This is reminiscent of the mechanism by which SUMO drives NB formation in yeast and human cells, e.g. Polychrome2 in Polycomb group bodies and promyelocytic leukemia bodies (Yang and Sharrocks, 2010; Jentsch and Psakhye, 2013).
SIZ1 and COP1 Activity Go Hand-in-Hand in Regulating Plant Growth
As was revealed in recent years, SIZ1-mediated SUMO conjugation and COP1-mediated ubiquitination are intimately connected in regulating plant growth. This connection strongly correlates with their here-studied (co)localization in NBs. Enzyme activity and substrate recognition either due to the presence of a nonconsensus sumoylation site or a VP peptide motif are core requirements for this interaction. How SUMO stimulates COP1 activity is unknown, but the SUMO glue hypothesis (Matunis et al., 2006; Jentsch and Psakhye, 2013) states that SUMO-SIM interactions strengthen existing interactions within protein complexes ultimately concentrating the proteins involved in micron-size bodies (Banani et al., 2016). As observed in other eukaryotic systems, OE of mature SUMO1 with SCE1 or SIZ1 resulted in NB formation (in our BiFC assay). This transient expression system apparently yields sufficient protein levels for SUMO NB formation. At sufficiently high concentrations, SUMO-SIM interactions facilitate a fluid phase separation yielding droplets (Banani et al., 2016). These droplets consist of a protein network including many client proteins (Banani et al., 2016). In this light, COP1 recruitment should be seen as a client interaction. Our studies on the role of SUMO chains for NB formation cannot exclude a supportive role for the native SUMO pool in this protein network.
Coincidental or Functional: The Overlap between COP1 and SIZ1 Substrates?
Our study strengthens the notion that COP1 and sumoylation go “hand-in-hand” in controlling each other’s activities while targeting a shared set of substrates. Many photobody components are COP1 substrates/interactors while also being (putative) SUMO conjugation targets, e.g. phyB, DELLAs, HFR1, LAF1, HY5, and HYL (Ballesteros et al., 2001; Seo et al., 2003; Conti et al., 2014; Sadanandom et al., 2015; Tan et al., 2015; Mazur et al., 2017). Jointly, these COP1 substrates control PIF activity at the transcriptional and posttranslational level, which in turn directly controls the transcriptional growth response (Paik et al., 2017). Likewise, the TF ABI5 (ABA-INSENSITIVE 5), a known interactor of SIZ1 and a SUMO substrate, was found to translocate to COP1-containing NBs in the presence of the regulator ABI5 INTERACTING PROTEIN (Lopez-Molina et al., 2003; Miura et al., 2009). In a similar way, disruption of a putative SUMO acceptor site in the TF LAF1 prevented LAF1 translocation to nuclear speckles (Ballesteros et al., 2001). In another case, the TF HFR1 was shown to interact with the bacterial SUMO protease Xanthomonas outer protein D from the bacterium Xanthomonas in nuclear speckles (Tan et al., 2015). Translocation of HFR1 and ABI5 to NBs was in both cases associated with their degradation, which again provides a functional link between COP1 and sumoylation in the regulation of nuclear processes. Considering that SIZ1 controls both skoto- and thermomorphogenesis (Lin et al., 2016; Hammoudi et al., 2018), COP1 recruitment to SUMO bodies points to the existence of a (transient) SUMO conjugation wave in photobodies with unknown physiological and biochemical consequence. As previously shown, SIZ1 is needed for hypocotyl elongation in both darkness and/or high-temperature conditions, and the siz1-2 mutation delays and weakens the transcription response of a substantial proportion of the genomic targets of PIF4 and BRZ1 (Lin et al., 2016; Hammoudi et al., 2018), two master regulators of thermomorphogenesis (Quint et al., 2016; Ibañez et al., 2018). Thus, in addition to having a role in plant immunity, SIZ1 rapidly interprets light as an important positive regulator of the dark- and high-temperature-induced growth response.
MATERIALS AND METHODS
Construction Mutants and Vectors for Y2H Analysis
All molecular techniques were performed using standard methods (Sambrook and Russel, 2001). Primers (synthesized by Eurofins genomics) are listed in Supplemental Table S2. Information on clones with primers can be found in Supplemental Table S3. Primers containing the attB1 and attB2 recombination sites were used to amplify the coding sequences (see Supplemental Table S2). The PCR products were cloned into the pDONR207 or pDONR221 using BP Clonase II (Thermo Fisher) and checked by sequencing. The inserts were transferred to destination vectors using Gateway LR Clonase II (Thermo Fisher) and the clones were resequenced. For the GAL4 BD/AD-fusion Y2H constructs, the cDNA clones were introduced in pDEST22/pDEST32 (Thermo Fisher). As the SUMO1∙SCE1 interaction was weak in the pDEST system (due to low expression levels; Supplemental Fig. S1B), these proteins were expressed with pGBKT7/pGADT7 (Clontech). CRY1 (G12079) and CRY2 (G19559) cDNA clones were obtained from the Arabidopsis Biological Research Centre. For in planta protein localization, the coding sequences were introduced in the destination plasmids pGWB452 (N-terminal GFP tagging), pGWB442 (N-terminal YFP tagging) or pGWB655 (N-terminal mRFP-tagging; Nakagawa et al., 2007). For bimolecular fluorescence complementation (BiFC) studies, we used the destination vectors pSCYNE(R) (N terminus SCFP3A), pSCYCE(R) (C terminus of SCFP3A), and pVYNE (N terminus of Venus; Gehl et al., 2009) or pESPYNE-Gateway (N terminus of YFP)/pESPYCE-Gateway (C terminus of YFP; Walter et al., 2004; Schütze et al., 2009).
Plant Protein Isolation and Detection Using Immunoblotting
To detect GFP-tagged proteins in Nicotiana benthamiana, total protein was extracted from ground leaf material in 2× (v/w) extraction buffer (8 m urea, 100 mm Tris, pH 6.8, 2% [w/v] SDS, 10 mm dithiothreitol), incubated on ice for 15 min, centrifuged at 13,000g for 20 min at 4°C, and then separated on 10% (w/v) SDS-PAGE, blotted to polyvinylidene difluoride membranes, and, after blocking with 5% (w/v) milk in phosphate-buffered saline, detected using monoclonal antibodies (diluted 1:1,000) directed against GFP (Chromotec #029762). The secondary antibody goat anti-rat IgG conjugated to horseradish peroxidase (Thermo Fisher #31470) was used at a dilution of 1:10,000. The proteins were visualized using enhanced chemiluminescence (homemade recipe). Equal loading of the protein samples was confirmed by Ponceau staining of the blots.
Protein Isolation from Yeast and Immunoblot Analysis
To obtain a total protein lysate from yeast, the protocol from Yeast Protocols Handbook was followed (Clontech; Yeast Protocols Handbook,http://www.takara.co.kr/file/manual/pdf/PT3024-1.pdf). Additional details are described in Mazur et al. (2017).
Transient Expression of Proteins in N. benthamiana Using Agro-infiltration
Transient expression of proteins was performed as described by Ma et al. (2012). In brief, Agrobacterium tumefaciens GV3101 cells containing the desired constructs were infiltrated into 4- to 5-week-old N. benthamiana leaves (OD600 = 1.0 for each construct). To suppress gene silencing, an A. tumefaciens strain GV3101 carrying pBIN61 containing the P19 silencing suppressor of tomato busy shunt virus (Cao et al., 2018) was coinfiltrated with the samples (OD600 = 0.5). Protein accumulation was examined 2 to 3 d post-agro-infiltration.
Transient Expression of Proteins in N. benthamiana with Anacardic Acid Treatment
Four-week-old N. benthamiana plants were used for agro-infiltration. Three days post-agro-infiltration, 100 μm anacardic acid in 1% (v/v) dimethyl sulfoxide (DMSO) or only 1% (v/v) DMSO (as a negative control) were directly injected into N. benthamiana leaves transiently expressing the BiFC constructs. One and a half hours after anacardic acid infiltration, the treated leaf discs were collected and fluorescence was analyzed using a Zeiss LSM510 confocal laser microscope. Two independent biological experiments were carried out, and a minimum of 50 nuclei for each sample and treatment were observed.
Transient Expression of Proteins in Arabidopsis Using Particle Bombardment
Complete Arabidopsis (Arabidopsis thaliana) rosettes of 4- to 5-week-old plants grown in 11 h light/13 h dark (22°C) were placed on 1% (w/v) agar containing 85 μm benzimidazole (Sigma-Aldrich) and kept in the growth chamber until bombardment. Transformation by particle bombardment was performed as described previously (Schweizer et al., 1999; Shirasu et al., 1999). In brief, 1-μm-diameter gold particles were coated with 2.5 μg of each type of plasmid. The PDS1000/HE particle gun with Hepta-adaptor (Bio-Rad) was used according to manufacturer’s protocol using a 900-psi rupture disk (Bio-Rad). After bombardment, petri dishes were sealed with medical tape and returned to the growth chamber for 2 to 3 d before inspection. Plates were kept in the dark from the evening before inspection till loading on the confocal microscopy to retain COP1 in the nucleus.
GAL4 Y2H Protein-Protein Interaction Assay
The protocol for Y2H assays was followed as described in de Folter and Immink (2011) and further details are described in Mazur et al. (2017).
Confocal Microscopy
N. benthamiana and Arabidopsis leaves were analyzed 2 to 3 d post-agro-infiltration or particle bombardment, respectively. The Hoechst 33342 (Sigma-Aldrich) chromatin stain and FM4-64 (Thermo Fisher) membrane dye and endocytic tracer were syringe infiltrated into the leaves at a final concentration of 1 μg/mL and 50 µm in water, respectively, and infiltrated leaves were stored in the dark in a petri dish on wet paper. FM4-64 could be directly imaged, Hoechst 33342 was incubated for at least an hour before imaging. Accumulation of the tagged proteins was examined in leaf epidermal cells using a Zeiss LSM510 or Nikon A1 confocal laser-scanning microscope. Images at the LSM510 were taken with C-Apochromat 40× water immersive objective with a numerical aperture of 1.2. For the Nikon A1, images were taken with a Plan Fluor 40× oil immersive differential interference contrast lens (numerical aperture = 1.3). SCFP-, GFP-, chimeric VenusN-SCFPC-, YFP-, and RFP-labeled samples were excited with 458-, 488-, 488-, 514-nm, or 568-nm diode lasers, respectively. For the Zeiss LSM510, GFP and the VenusN-SCFPC chimera were detected using a 520- to 555-nm BP filter (BP520–555), SCFP (BiFC) with BP470–500, RFP with BP585–615. For the Nikon A1, SCFP was detected with BP468–502; YFP/GFP with BP500–550, and RFP with BP570–620. Also for the Nikon A1, for simultaneous Hoechst 33342, SCFP, and mRFP imaging, Hoechst 33342 was excited with the 402-nm diode laser and detected at 425 to 475 nm, SCFP was excited with the 488-nm diode laser and detected at 500 to 550 nm, and mRFP was excited with the 561-nm diode laser and detected at 570 to 620 nm. FM4-64 was excited with the 514-nm diode laser and detected at 570 to 620 nm.
Bright-field images were recorded with the transmitted light photomultiplier detector. For both microscopes, coexpressed fluorophores or dyes were excited consecutively to limit bleed through of emission signals between detection channels. For all observations, the pinhole was set at 1 Airy unit. Images were processed using ImageJ (NIH).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank database under the following accession numbers: PIASx (Human Gene ID 9063), Senataxin (Human Gene ID: 23064), TOPORS (Human Gene ID: 10210) and SP100 (Human Gene ID: 6672) and the Arabidopsis Information Resource under the following accession numbers: SUMO1 (AT4G26840), SUMO3 (AT5G55170), SCE1 (AT3G57870), SIZ1 (AT5G60410.2), COP1 (AT2G32950), CRY1 (AT4G08920), and CRY2 (AT1G04400).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Arabidopsis SUMO1 interacts specifically with SCE1 via a noncovalent SIM-like interaction
Supplemental Figure S2. The ternary complex between Arabidopsis SUMO1, SCE1, and SIZ1 is stabilized by the substrate-binding pocket (CAT2) and noncovalent interactions between the different proteins
Supplemental Figure S3. The SUMO1GG-SCE1 and SUMO1GG-SIZ1 BiFC complexes localize to the nucleus in nuclear bodies
Supplemental Figure S4. Alignment of SCE1 protein sequences from different monocot and dicot plant species showing that SCE1 protein displays high-level conservation at the sequence level
Supplemental Figure S5. Coexpression of SCE1∙SIZ1 as a BiFC pair with an excess of YFP-SUMO1GG drives an increased pool of the SCE1∙SIZ1 BiFC pair to small puncta
Supplemental Figure S6. SUMO chain formation accelerates localization of the SUMO1∙SCE1 BiFC pair in nuclear bodies
Supplemental Figure S7. Quantification of the colocalization of the SUMO1∙SCE1 BiFC pair with COP1
Supplemental Figure S8. Neither mRFP-SCE1 nor SIZ1-mRFP are recruited to CRY2 NBs
Supplemental Table S1. SUMO1, COP1, SIZ1 and SCE1 mutants used in this study
Supplemental Table S2. Sequences of oligonucleotides used in this study
Supplemental Table S3. Clone information
Acknowledgments
The authors kindly acknowledge L. Tikovsky and H. Lemereis for horticultural assistance, the van Leeuwenhoek Centre for Advanced Microscopy, Section Molecular Cytology (SILS-University of Amsterdam) for use of their microscopy facilities, V. Hammoudi for SIZ1SP-RING. We thank D. Baulcome (University of Cambridge, UK), I. Dikic (Goethe University, Frankfurt am main, Germany), Dongqing Xu (Peking University, Beijing), J. Haas (Ludwig-Maximilians-University of Munich, Germany), J. Kudla (WWU Muenster, Germany), Klaus Harter (ZMBP Tübingen, Germany), T. Nagakawa (Shimane University, Japan), Seong Wook Yang (University of Copenhagen, Denmark), Jin Bo Lin (Chinese Academy of Sciences, Beijing, China), Dae-Jin Yun (Gyeongsang National University, Korea), and Ute Höcker (Universität zu Köln) for sharing plasmids and seeds. We thank the ABRC for sending us the CRY1 and CRY2 cDNA clones.
Footnotes
Senior author.
The Netherlands Scientific Organisation (ALW-VIDI grant 864.10.004 to H.A.v.d.B.) and the Topsector T&U program Better Plants for Demands (grant 1409-036 to H.A.v.d.B.), including the partnering breeding companies, supported this work; F.M. is financially supported by Keygene N.V. (the Netherlands).
Articles can be viewed without a subscription.
References
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK (2016) Compositional Control of Phase-Separated Cellular Bodies. Cell 166: 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA (2002) Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem 277: 47938–47945 [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 [DOI] [PubMed] [Google Scholar]
- Cao L, Blekemolen MC, Tintor N, Cornelissen BJC, Takken FLW (2018) The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol Plant 11: 691–705 [DOI] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MS, Park HC, Hong MJ, Lee J, Choi W, Jin JB, Bohnert HJ, Lee SY, Bressan RA, Yun DJ (2009) Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol 151: 1930–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MS, Park HC, Bohnert HJ, Bressan RA, Yun DJ (2010) Structural and functional studies of SIZ1, a PIAS-type SUMO E3 ligase from Arabidopsis. Plant Signal Behav 5: 567–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby T, Matthäi A, Boeckelmann A, Stuible HP (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142: 318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Nelis S, Zhang C, Woodcock A, Swarup R, Galbiati M, Tonelli C, Napier R, Hedden P, Bennett M, et al. (2014) Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev Cell 28: 102–110 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG (2011) Yeast protein-protein interaction assays and screens. Methods Mol Biol 754: 145–165 [DOI] [PubMed] [Google Scholar]
- Elrouby N, Bonequi MV, Porri A, Coupland G (2013) Identification of Arabidopsis SUMO-interacting proteins that regulate chromatin activity and developmental transitions. Proc Natl Acad Sci USA 110: 19956–19961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho A, Melchior F (2013) Sumoylation: A regulatory protein modification in health and disease. Annu Rev Biochem 82: 357–385 [DOI] [PubMed] [Google Scholar]
- Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura K, Sodeoka M, Yoshida M (2009) Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol 16: 133–140 [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD (2010) The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Guo L, Giasson BI, Glavis-Bloom A, Brewer MD, Shorter J, Gitler AD, Yang X (2014) A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol Cell 55: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoudi V, Fokkens L, Beerens B, Vlachakis G, Chatterjee S, Arroyo-Mateos M, Wackers PFK, Jonker MJ, van den Burg HA (2018) The Arabidopsis SUMO E3 ligase SIZ1 mediates the temperature dependent trade-off between plant immunity and growth. PLoS Genet 14: e1007157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N, Shen L, Jaffray EG, Hay RT (2011) The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol Biol Cell 22: 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I (2006) Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117–16127 [DOI] [PubMed] [Google Scholar]
- Hendriks IA, D’Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 21: 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U. (2017) The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr Opin Plant Biol 37: 63–69 [DOI] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkotte X, Ponnu J, Ahmad M, Hoecker U (2017) The blue light-induced interaction of cryptochrome 1 with COP1 requires SPA proteins during Arabidopsis light signaling. PLoS Genet 13: e1007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C, Delker C, Martinez C, Bürstenbinder K, Janitza P, Lippmann R, Ludwig W, Sun H, James GV, Klecker M, et al. (2018) Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr Biol 28: 303–310.e3 [DOI] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I (2013) Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet 47: 167–186 [DOI] [PubMed] [Google Scholar]
- Kaur K, Park H, Pandey N, Azuma Y, De Guzman RN (2017) Identification of a new small ubiquitin-like modifier (SUMO)-interacting motif in the E3 ligase PIASy. J Biol Chem 292: 10230–10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Rozhon W, Unterholzner SJ, Chen T, Eremina M, Wurzinger B, Bachmair A, Teige M, Sieberer T, Isono E, et al. (2014) Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat Commun 5: 4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Jang IC, Seo HS (2016) COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 Ubiquitin ligase activity. Front Plant Sci 7: 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK (2007) Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J 26: 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Lamoliatte F, McManus FP, Maarifi G, Chelbi-Alix MK, Thibault P (2017) Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat Commun 8: 14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lin XL, Niu D, Hu ZL, Kim DH, Jin YH, Cai B, Liu P, Miura K, Yun DJ, Kim WY, et al. (2016) An Arabidopsis SUMO E3 ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet 12: e1006016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, Lima CD, Chua NH (2003) Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lukasik E, Gawehns F, Takken FL (2012) The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol Biol 835: 61–74 [DOI] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- Mascle XH, Lussier-Price M, Cappadocia L, Estephan P, Raiola L, Omichinski JG, Aubry M (2013) Identification of a non-covalent ternary complex formed by PIAS1, SUMO1, and UBC9 proteins involved in transcriptional regulation. J Biol Chem 288: 36312–36327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Zhang XD, Ellis NA (2006) SUMO: The glue that binds. Dev Cell 11: 596–597 [DOI] [PubMed] [Google Scholar]
- Mazur MJ, Spears BJ, Djajasaputra A, van der Gragt M, Vlachakis G, Beerens B, Gassmann W, van den Burg HA (2017) Arabidopsis TCP Transcription Factors Interact with the SUMO Conjugating Machinery in Nuclear Foci. Front Plant Sci 8: 2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107: 16512–16517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim JI, Huq E (2017) Expanding Roles of PIFs in Signal Integration from Multiple Processes. Mol Plant 10: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Ha JH, Kim JY, Park CM (2017) COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol 215: 269–280 [DOI] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN (2008) A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33: 201–208 [DOI] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz TC, Miller MJ, McLoughlin F, Augustine RC, Marshall RS, Juan YT, Charng YY, Scalf M, Smith LM, Vierstra RD (2018) SUMOylome Profiling Reveals a Diverse Array of Nuclear Targets Modified by the SUMO Ligase SIZ1 during Heat Stress. Plant Cell 30: 1077–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Ádám É, Orosa B, Viczián A, Klose C, Zhang C, Josse EM, Kozma-Bognár L, Nagy F (2015) SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 11108–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual, Vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007) Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- Schweizer P, Christoffel A, Dudler R (1999) Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J 20: 541–552 [DOI] [PubMed] [Google Scholar]
- Sekhri P, Tao T, Kaplan F, Zhang XD (2015) Characterization of amino acid residues within the N-terminal region of Ubc9 that play a role in Ubc9 nuclear localization. Biochem Biophys Res Commun 458: 128–133 [DOI] [PubMed] [Google Scholar]
- Sekiyama N, Ikegami T, Yamane T, Ikeguchi M, Uchimura Y, Baba D, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M (2008) Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem 283: 35966–35975 [DOI] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Nielsen K, Piffanelli P, Oliver R, Schulze-Lefert P (1999) Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J 17: 293–299 [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem 280: 40122–40129 [DOI] [PubMed] [Google Scholar]
- Stacey MG, von Arnim AG (1999) A novel motif mediates the targeting of the Arabidopsis COP1 protein to subnuclear foci. J Biol Chem 274: 27231–27236 [DOI] [PubMed] [Google Scholar]
- Stacey MG, Hicks SN, von Arnim AG (1999) Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11: 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC Jr., Lima CD (2016) Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 536: 304–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CM, Li MY, Yang PY, Chang SH, Ho YP, Lin H, Deng WL, Yang JY (2015) Arabidopsis HFR1 is a potential nuclear substrate regulated by the Xanthomonas type III effector XopD(Xcc8004). PLoS One 10: e0117067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uljon S, Xu X, Durzynska I, Stein S, Adelmant G, Marto JA, Pear WS, Blacklow SC (2016) Structural Basis for Substrate Selectivity of the E3 Ligase COP1. Structure 24: 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk EK, Decker PV, Chen M (2012) Photobodies in light signaling. Plant Physiol 158: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg HA, Kini RK, Schuurink RC, Takken FLW (2010) Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22: 1998–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma L-G, Li J-M, Zhao H-Y, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD (2010) The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol Cell Biol 30: 2193–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, Lima CD (2006) Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol 13: 491–499 [DOI] [PubMed] [Google Scholar]
- Yunus AA, Lima CD (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell 35: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, Matunis MJ (2008) Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem 283: 29405–29415 [DOI] [PMC free article] [PubMed] [Google Scholar]