Key Points
Question
Which hospitalized adult patients have an increased risk of carbapenem-resistant or extensive β-lactam–resistant Pseudomonas aeruginosa infections?
Findings
In this cohort study of 7775 hospitalized patients with 11 052 P aeruginosa infections, 20.2% of infections were carbapenem resistant and 9.0% were extensive β-lactam resistant. Risk scores that included history of carbapenem-resistant P aeruginosa infection, tracheostomy, and carbapenem use in the prior 30 days were associated with a diagnosis of antibiotic-resistant P aeruginosa infections.
Meaning
These risk scores appear to help identify patients at highest risk of antibiotic-resistant P aeruginosa infections and consequently support physicians in the selection of appropriate empirical treatment in the hospital setting.
This cohort study develops 2 clinical risk scores based on variables available at clinical presentation to estimate the risk of carbapenem resistance or extensive β-lactam resistance among hospitalized adult patients with Pseudomonas aeruginosa infections.
Abstract
Importance
Treatment of patients with infections due to Pseudomonas aeruginosa has been complicated by increased antibiotic resistance rates, which contribute to delayed appropriate treatment and deleterious outcomes.
Objective
To develop 2 clinical risk scores based on variables available at clinical presentation to estimate the risk of carbapenem resistance (CR) or extensive β-lactam resistance (EBR) among hospitalized, adult patients with P aeruginosa infections.
Design, Setting, and Participants
This retrospective cohort study included adult (age, ≥18 years) members of Kaiser Permanente Southern California (KPSC) with a P aeruginosa infection during hospitalization from September 1, 2011, through August 31, 2016, who received antibiotic therapy within 2 days of the culture date. Data were analyzed from July 2, 2017, through August 15, 2018.
Exposures
Demographic, clinical, and laboratory covariates 1 year before the index culture date were evaluated.
Main Outcomes and Measures
Pseudomonas aeruginosa was categorized as antibiotic susceptible, CR, or EBR (nonsusceptibility to carbapenems, ceftazidime, and combined piperacillin sodium and tazobactam sodium). Patients were randomly split (1:1) into training and validation data sets. The training data set was used to develop 2 prediction models using high-performance logistic regression with variable selection by Schwarz-Bayesian criterion. The models were translated into risk scores, with risk score points equaling the weighted sums of regression coefficients from the prediction model. The patient’s risk was estimated as the inverse logit of the risk score.
Results
Of the 7775 patients with 11 502 P aeruginosa infections included in the analysis, most were male (4308 [55.4%]) and non-Hispanic white (3927 [50.5%]). The mean (SD) age was 70.3 (15.5) years. Among 11 502 P aeruginosa infections, 2324 (20.2%) were CR, 9178 (79.8%) were non-CR, 1033 (9.0%) were EBR, and 10 469 were non-EBR (91.0%). The strongest predictors of resistance in the CR and EBR models were history of CR P aeruginosa infection (odds ratios [ORs], 8.80 [95% CI, 6.74-11.49] and 5.04 [95% CI, 3.88-6.54], respectively), tracheostomy (ORs, 3.49 [95% CI, 2.92-4.16] and 3.13 [95% CI, 2.50-3.91], respectively), and carbapenem use in the prior 30 days (ORs, 4.18 [95% CI, 3.29-5.31] and 2.26 [95% CI, 1.74-2.93], respectively). The models for CR and EBR performed well, with areas under the receiver operating characteristics curve of 0.81 or greater for the training and validation data sets.
Conclusions and Relevance
The findings of this study suggest that parsimonious risk scores can aid physicians in appropriate treatment selection during the critical period when P aeruginosa infection is suspected but antibiotic susceptibility results are not yet available.
Introduction
Pseudomonas aeruginosa is a common nosocomial pathogen that has a remarkable capacity to exhibit resistance to commonly used antipseudomonal antibiotics, for example carbapenems, aminoglycosides, and fluoroquinolones.1,2,3,4 Two of the most concerning resistance patterns exhibited by P aeruginosa are carbapenem resistance (CR) and extensive β-lactam resistance (EBR). Patients with CR and EBR P aeruginosa infections are at an increased risk for delayed receipt of appropriate antimicrobial therapy, resulting in prolonged hospitalization, increased risk for subsequent antibiotic-resistant infections, morbidity, and mortality.5,6,7,8,9 These outcomes ultimately lead to further antibiotic use and can exacerbate existing antibiotic resistance in an institution.
To promote appropriate antibiotic use within a health care institution, institutions are recommended to create tools and policies informed by real-world evidence to increase the probability that patients receive early, appropriate therapy.10 To facilitate such tool and policy creation, we developed and internally validated 2 clinical risk scores to estimate the probabilities of CR and EBR based on covariates available on clinical presentation. We focused on these 2 P aeruginosa nonsusceptible phenotypes because they represent 2 of the major treatment challenges among patients with P aeruginosa infections. Because carbapenems are common first-line therapies for P aeruginosa infections, we developed the CR model to identify situations where carbapenems should be avoided. Further, because cross-resistance to β-lactams is commonplace among P aeruginosa infections, we developed the EBR model to identify patients for whom use of common antipseudomonal β-lactam antibiotics would likely contribute to delayed appropriate therapy.
Methods
Setting
Kaiser Permanente Southern California (KPSC) is an integrated health care organization with more than 4.2 million members.11 Kaiser Permanente Southern California consists of 14 medical centers that use electronic health records to integrate medical information, including diagnostic, medication, procedure codes, and laboratory results from outpatient, emergency department, and hospital settings. The study protocol was reviewed and approved by the KPSC institutional review board, which waived requirement for informed consent for these internal data that met the criteria for very low risk to study participants. This study followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guideline.
Study Design and Population
We conducted a retrospective cohort study among adult (age, ≥18 years) KPSC members who had a P aeruginosa infection (identified by culture growth and a series of ≥1 of the following tests: biochemical testing, oxidase, odor, morphologic characterization, mass spectrometry [Vitek MS; BioMerieux Vitek, Inc], and Vitek GN identification cards [BioMerieux Vitek] based on established biochemical methods and newly developed substrates measuring carbon source utilization, enzymatic activities, and resistance) during a hospitalization from September 1, 2011, through August 31, 2016, and who received antibiotic therapy within 2 days of the P aeruginosa index culture date. We defined the index date for all patients as the first date on which they had a positive culture for P aeruginosa. Patients were included if they had 6 months of continuous KPSC enrollment before the index date, allowing for 45-day enrollment gaps, and had the KPSC pharmacy benefit. We excluded patients with a diagnosis of cystic fibrosis (based on codes from International Classification of Diseases, Ninth Revision, or International Statistical Classification of Diseases and Related Health Problems, Tenth Revision) at any point in their medical history.
We then defined P aeruginosa infections by their antibiotic susceptibility using MicroScan panel NM45 (Beckman Coulter, Inc), which consists of miniaturizations of the broth dilution susceptibility test that have been dehyrated. We categorized resistant P aeruginosa infections as (1) CR (nonsusceptible to meropenem or imipenem) or (2) EBR (nonsusceptible to carbapenem [meropenem or imipenem], ceftazidime [the only cephalosporin routinely tested for resistance at KPSC], and combined piperacillin sodium and tazobactam sodium). These 2 groups were not mutually exclusive; those categorized as EBR were also included in the CR group. We defined nonsusceptibility as having resistant or intermediate in vitro test results to the drugs of interest based on Clinical and Laboratory Standards Institute break points. We defined the susceptible comparison groups by subtracting the CR or EBR infections from the total population with P aeruginosa infection, creating groups with carbapenem nonresistance (non-CR) and extensive β-lactam nonresistance (non-EBR), respectively.
A patient could have more than 1 hospitalization during the study period, and a patient could have more than 1 P aeruginosa infection per hospitalization; however, on identification of a resistant P aeruginosa culture, we would only look for another resistant isolate after a 3-day window following the preceding culture. We included the most resistant P aeruginosa culture (EBR > CR > neither) recovered during each 3-day window in the analyses.
Data Collection
We collected demographic, clinical, and microbiological data from electronic health record documentation of inpatient and outpatient care. eTable 1 in the Supplement provides a complete list of covariates and definitions. Mutually exclusive race/ethnicity categories were assigned to members from the KPSC electronic health records. Priority was given to self-reported race/ethnicity data; where not available, these data were imputed from available spoken language or detailed ethnic group information.
Development of the CR and EBR P aeruginosa Infection Risk Scores
We used the same methods to develop the risk scores for CR P aeruginosa infection and EBR P aeruginosa infection. First, we randomly split (1:1) our study populations into training and validation data sets (eTables 2 and 3 in the Supplement). We then developed the 2 (CR and EBR) prediction models in the training data set. Before inclusion in the multivariable model, we assessed variables for collinearity using the variance inflation factor and correlation. We defined a lack of multicollinearity between predictors as a variance inflation factor of less than 2.5 or a correlation coefficient of less than 0.6. When 2 or more variables were found to be collinear, we selected variables for inclusion based on magnitude of effect and clinical relevance. Comorbidities were collinear with skilled nursing facility and were removed.
In the training data set, the variables for our final models were selected by the Schwarz-Bayesian criterion,12,13 a penalized measure of fit by high-performance logistic regression used to select a parsimonious model while avoiding overfitting. We assessed the performance of our final models by comparing the area under the receiver operating characteristic curves (AUCs) and sensitivity against 1 − specificity plots to models developed using standard logistic regression as well as LASSO (eFigure 1 in the Supplement).12,14,15
Using our multivariable models, we developed our risk scores based on methods previously described by Sullivan et al.16 Briefly, risk scores were calculated as weighted sums of regression coefficients from the prediction model; the patient’s risk was estimated as the inverse logit of the risk score. Higher risk scores are associated with greater predicted risk of CR or EBR.
Evaluation of Risk Score Performance
We assessed the performance of the final prediction models in the validation data sets. We assessed the ability of the model to discriminate between patients with and without CR or EBR P aeruginosa infections through the AUC. An AUC of 1.00 corresponds to perfect discrimination, whereas an AUC of 0.50 corresponds to no discriminating ability. We also assessed the calibration of the point-based risk score system by comparing the risk observed with the risk predicted by the risk score within the validation data set. The comparison is visualized in plots of observed risk and the mean point-based predicted risk by quintiles, where the quintiles were generated according to the model-based predicted risk. All analyses were performed using SAS software (version 9.3; SAS Institute, Inc).
Sensitivity Analyses
We performed sensitivity analyses using only the first infection per patient. We used the same model selection process and model performance assessment as described earlier.
Statistical Analysis
Data were analyzed from July 2, 2017, through August 15, 2018. We summarized all categorical variables and examined differences between patients with susceptible P aeruginosa infections and those with CR or EBR P aeruginosa infections, using the χ2 test or Fisher exact test when appropriate. All tests were 2-tailed, and we considered P < .05 to be statistically significant.
Results
Of the 7775 patients (4308 men [55.4%] and 3467 women [44.6%]) with 11 502 P aeruginosa infections included in the analysis, most were non-Hispanic white (3927 [50.5%]), Hispanic (1968 [25.3%]), or non-Hispanic black (1246 [16.0%]) (eTable 4 in the Supplement). The mean (SD) age was 70.3 (15.5) years. No data were missing for this study. Age was a requirement for inclusion; sex and Charlson comorbidity index data were complete; and unknown race/ethnicity was included as a stratum. In the total study population, 5979 patients (76.9%) had 1 P aeruginosa infection; 1017 (13.1%), 2 infections; 376 (4.8%), 3 infections; and 403 (5.2%), 4 or more infections. Among 11 502 P aeruginosa infections that met inclusion criteria, we observed 2324 CR infections and 1033 EBR infections. The susceptibility profiles of the P aeruginosa infections, stratified by CR and EBR status, are shown in eTable 5 in the Supplement. Among those with a CR P aeruginosa infection, 1094 (47.1%) were susceptible to piperacillin-tazobactam, and 1209 (52.0%) were susceptible to ceftazidime. Details on the selection of the final study population can be found in eFigure 2 in the Supplement. Compared with non-CR infections, CR P aeruginosa infections were more likely to occur in men (1381 [59.4%]) and non-Hispanic white patients (1043 [44.9%]), and patients had a mean (SD) age of 67.5 (15.4) years. Compared with non-EBR infections, EBR P aeruginosa infections were also more likely to occur in men (637 [61.7%]) and non-Hispanic white patients (429 [41.5%]), and patients had a mean (SD) age of 67.4 (14.9) years (Table 1). Of the 3357 patients with resistant infection, 2460 (73.3%) had a Charlson comorbidity index of 3 or greater (Table 1).
Table 1. Selected Characteristics of Patients Antibiotic-Resistant and Nonresistant Pseudomonas aeruginosa Infections.
| Characteristic | All Infections, No. (%) (N = 11 502) | Infections by CR Status, No. (%) | Infections by EBR Status, No. (%) | ||||
|---|---|---|---|---|---|---|---|
| CR (n = 2324) | Non-CR (n = 9178) | P Valuea | EBR (n = 1033) | Non-EBR (n = 10 469) | P Valuea | ||
| Demographic | |||||||
| Male | 6554 (57.0) | 1381 (59.4) | 5173 (56.4) | .008 | 637 (61.7) | 5917 (56.5) | .001 |
| Age at index date, y | |||||||
| 18-39 | 667 (5.8) | 143 (6.2) | 524 (5.7) | <.001 | 50 (4.8) | 617 (5.9) | <.001 |
| 40-64 | 2979 (25.9) | 687 (29.6) | 2292 (25.0) | 326 (31.6) | 2653 (25.3) | ||
| 65-84 | 6137 (53.4) | 1236 (53.2) | 4901 (53.4) | 545 (52.8) | 5592 (53.4) | ||
| ≥85 | 1719 (14.9) | 258 (11.1) | 1461 (15.9) | 112 (10.8) | 1607 (15.4) | ||
| Race/ethnicity | |||||||
| Non-Hispanic white | 5543 (48.2) | 1043 (44.9) | 4500 (49.0) | <.001 | 429 (41.5) | 5114 (48.8) | <.001 |
| Asian, Hawaiian, or Pacific Islander | 969 (8.4) | 234 (10.1) | 735 (8.0) | 108 (10.4) | 861 (8.2) | ||
| Non-Hispanic black | 2003 (17.4) | 469 (20.2) | 1534 (16.7) | 219 (21.2) | 1784 (17.0) | ||
| Hispanic | 2891 (25.1) | 557 (24.0) | 2334 (25.4) | 268 (25.9) | 2623 (25.1) | ||
| Native American, Alaska Native, multiple, other, or unknownb | 96 (0.8) | 21 (0.9) | 75 (0.8) | 9 (0.9) | 87 (0.8) | ||
| Index admission | |||||||
| Specimen site of index infection | |||||||
| Other | 6306 (54.8) | 897 (38.6) | 5409 (58.9) | <.001 | 340 (32.9) | 5966 (57.0) | <.001 |
| Respiratory tract | 5196 (45.2) | 1427 (61.4) | 3769 (41.1) | 693 (67.1) | 4503 (43.0) | ||
| Source of infection | |||||||
| Present at hospital admission | 7520 (65.4) | 1274 (54.8) | 6246 (68.1) | <.001 | 502 (48.6) | 7018 (67.0) | <.001 |
| Hospital-onset | 3982 (34.6) | 1050 (45.2) | 2932 (31.9) | 531 (51.4) | 3451 (33.0) | ||
| Coinfecting pathogen | 3345 (29.1) | 705 (30.3) | 2640 (28.8) | .14 | 321 (31.1) | 3024 (28.9) | .14 |
| ICU before index culture | 3164 (27.5) | 924 (39.8) | 2240 (24.4) | <.001 | 461 (44.6) | 2703 (25.8) | <.001 |
| SNF transfer | 1004 (8.7) | 435 (18.7) | 569 (6.2) | <.001 | 231 (22.4) | 773 (7.4) | <.001 |
| Invasive devices and/or procedures from admission to culture date | |||||||
| Endotracheal tube | 2085 (18.1) | 556 (23.9) | 1529 (16.7) | <.001 | 256 (24.8) | 1829 (17.5) | <.001 |
| Tracheostomy | 2142 (18.6) | 1068 (46.0) | 1074 (11.7) | <.001 | 582 (56.3) | 1560 (14.9) | <.001 |
| Hemodialysis | 670 (5.8) | 286 (12.3) | 384 (4.2) | <.001 | 160 (15.5) | 510 (4.9) | <.001 |
| Gastric or jejunal feeding tube | 2692 (23.4) | 1026 (44.1) | 1666 (18.2) | <.001 | 529 (51.2) | 2163 (20.7) | <.001 |
| Indwelling urinary catheter | 5165 (44.9) | 1410 (60.7) | 3755 (40.9) | <.001 | 688 (66.6) | 4477 (42.8) | <.001 |
| CVC, port, or PICC | 2473 (21.5) | 816 (35.1) | 1657 (18.1) | <.001 | 431 (41.7) | 2042 (19.5) | <.001 |
| Surgery before index culture | 1615 (14.0) | 423 (18.2) | 1192 (13.0) | <.001 | 209 (20.2) | 1406 (13.4) | <.001 |
| LOS from admission to index culture, d | |||||||
| 0-3 | 7772 (67.6) | 1316 (56.6) | 6456 (70.3) | <.001 | 519 (50.2) | 7253 (69.3) | <.001 |
| 4-10 | 1752 (15.2) | 336 (14.4) | 1416 (15.4) | 154 (14.9) | 1598 (15.3) | ||
| ≥11 | 1978 (17.2) | 672 (28.9) | 1306 (14.2) | 360 (34.8) | 1618 (15.4) | ||
| P aeruginosa infection 30 d before index admission | |||||||
| None | 8622 (75.0) | 1294 (55.7) | 7328 (79.8) | <.001 | 464 (44.9) | 8158 (77.9) | <.001 |
| Carbapenem-susceptible | 2001 (17.4) | 308 (13.2) | 1693 (18.4) | 155 (15.0) | 1846 (17.6) | ||
| CR | 879 (7.6) | 722 (31.1) | 157 (1.7) | 414 (40.1) | 465 (4.4) | ||
| Health care exposure in 6 mo before index admission | |||||||
| Previous hospitalization | 7293 (63.4) | 1883 (81.0) | 5410 (58.9) | <.001 | 869 (84.1) | 6424 (61.4) | <.001 |
| Previous ICU admission | 2508 (21.8) | 908 (39.1) | 1600 (17.4) | <.001 | 461 (44.6) | 2047 (19.6) | <.001 |
| Comorbidity | |||||||
| Myocardial infarction | 1936 (16.8) | 455 (19.6) | 1481 (16.1) | <.001 | 224 (21.7) | 1712 (16.4) | <.001 |
| Congestive heart failure | 3396 (29.5) | 839 (36.1) | 2557 (27.9) | <.001 | 413 (40.0) | 2983 (28.5) | <.001 |
| Peripheral vascular disease | 6324 (55.0) | 1414 (60.8) | 4910 (53.5) | <.001 | 626 (60.6) | 5698 (54.4) | <.001 |
| Cerebrovascular disease | 2394 (20.8) | 700 (30.1) | 1694 (18.4) | <.001 | 327 (31.7) | 2067 (19.7) | <.001 |
| Dementia | 795 (6.9) | 211 (9.1) | 584 (6.4) | <.001 | 98 (9.5) | 697 (6.6) | .001 |
| Chronic pulmonary disease | 4714 (41.0) | 1037 (44.6) | 3677 (40.1) | <.001 | 474 (45.9) | 4240 (40.5) | .001 |
| Peptic ulcer disease | 336 (2.9) | 106 (4.6) | 230 (2.5) | <.001 | 55 (5.3) | 281 (2.7) | <.001 |
| Mild liver diseasec | 990 (8.6) | 259 (11.1) | 731 (8.0) | <.001 | 127 (12.3) | 863 (8.2) | <.001 |
| Diabetes | 4882 (42.4) | 1114 (47.9) | 3768 (41.1) | <.001 | 516 (50.0) | 4366 (41.7) | <.001 |
| Hemiplegia or paraplegia | 1106 (9.6) | 342 (14.7) | 764 (8.3) | <.001 | 166 (16.1) | 940 (9.0) | <.001 |
| Malignant disease, including leukemia and lymphoma (not malignant neoplasm of skin) | 2070 (18.0) | 395 (17.0) | 1675 (18.2) | .16 | 169 (16.4) | 1901 (18.2) | .15 |
| Moderate or severe liver diseased | 169 (1.5) | 55 (2.4) | 114 (1.2) | <.001 | 18 (1.7) | 151 (1.4) | .44 |
| Metastatic solid tumor | 661 (5.7) | 109 (4.7) | 552 (6.0) | .01 | 53 (5.1) | 608 (5.8) | .37 |
| Organ transplant | 304 (2.6) | 78 (3.4) | 226 (2.5) | .02 | 30 (2.9) | 274 (2.6) | .58 |
| AIDS/HIV | 66 (0.6) | 17 (0.7) | 49 (0.5) | .26 | 2 (0.2) | 64 (0.6) | .09 |
| Charlson comorbidity index | |||||||
| 0 | 1079 (9.4) | 136 (5.8) | 943 (10.3) | <.001 | 54 (5.2) | 1025 (9.8) | <.001 |
| 1 | 1330 (11.6) | 189 (8.1) | 1141 (12.4) | 68 (6.6) | 1262 (12.1) | ||
| 2 | 1842 (16.0) | 317 (13.6) | 1525 (16.6) | 133 (12.9) | 1709 (16.3) | ||
| ≥3 | 7251 (63.0) | 1682 (72.4) | 5569 (60.7) | 778 (75.3) | 6473 (61.8) | ||
| Antibiotic use in 30 d before index date | |||||||
| Aminoglycosides | 392 (3.4) | 147 (6.3) | 245 (2.7) | <.001 | 79 (7.6) | 313 (3.0) | <.001 |
| Carbapenems | 884 (7.7) | 542 (23.3) | 342 (3.7) | <.001 | 277 (26.8) | 607 (5.8) | <.001 |
| Ceftazidime, cefepime, or ceftolozane-tazobactam | 2498 (21.7) | 759 (32.6) | 1739 (18.9) | <.001 | 385 (37.3) | 2113 (20.2) | <.001 |
| Other cephalosporinse | 4726 (41.1) | 613 (26.4) | 4113 (44.8) | <.001 | 238 (23.0) | 4488 (42.9) | <.001 |
| Monobactams | 138 (1.2) | 53 (2.3) | 85 (0.9) | <.001 | 31 (3.0) | 107 (1.0) | <.001 |
| Glycylcyclines and tetracyclines | 901 (7.8) | 107 (4.6) | 794 (8.7) | <.001 | 41 (4.0) | 860 (8.2) | <.001 |
| Penicillin | 3699 (32.2) | 864 (37.2) | 2835 (30.9) | <.001 | 418 (40.5) | 3281 (31.3) | <.001 |
| Quinolones | 2608 (22.7) | 801 (34.5) | 1807 (19.7) | <.001 | 353 (34.2) | 2255 (21.5) | <.001 |
| Piperacillin-tazobactam | 2982 (25.9) | 746 (32.1) | 2236 (24.4) | <.001 | 374 (36.2) | 2608 (24.9) | <.001 |
| Hospital facility | |||||||
| A | 729 (6.3) | 132 (5.7) | 597 (6.5) | <.001 | 57 (5.5) | 672 (6.4) | <.001 |
| B | 900 (7.8) | 171 (7.4) | 729 (7.9) | 69 (6.7) | 831 (7.9) | ||
| C | 1138 (9.9) | 215 (9.2) | 923 (10.1) | 76 (7.4) | 1062 (10.1) | ||
| D | 1424 (12.4) | 285 (12.3) | 1139 (12.4) | 121 (11.7) | 1303 (12.4) | ||
| E | 663 (5.8) | 127 (5.5) | 536 (5.8) | 55 (5.3) | 608 (5.8) | ||
| F | 680 (5.9) | 157 (6.8) | 523 (5.7) | 63 (6.1) | 617 (5.9) | ||
| G | 649 (5.6) | 167 (7.2) | 482 (5.2) | 86 (8.3) | 563 (5.4) | ||
| H | 542 (4.7) | 137 (5.9) | 405 (4.4) | 68 (6.6) | 474 (4.5) | ||
| I | 768 (6.7) | 140 (6.0) | 628 (6.8) | 64 (6.2) | 704 (6.7) | ||
| J | 1333 (11.6) | 194 (8.3) | 1139 (12.4) | 85 (8.2) | 1248 (11.9) | ||
| K | 946 (8.2) | 144 (6.2) | 802 (8.7) | 56 (5.4) | 890 (8.5) | ||
| L | 903 (7.9) | 248 (10.7) | 655 (7.1) | 143 (13.8) | 760 (7.2) | ||
| M | 827 (7.2) | 207 (8.9) | 620 (6.8) | 90 (8.7) | 737 (7.0) | ||
Abbreviations: CR, carbapenem resistant; CVC, central venous catheter; EBR, extensive β-lactam (carbapenem, ceftazidime, and combined piperacillin sodium and tazobactam sodium) resistant; ICU, intensive care unit; LOS, length of stay; PICC, peripherally inserted central catheter; SNF, skilled nursing facility.
The χ2 test and 2-tailed Fisher exact test were used to calculate P values for categorical and continuous variables, respectively.
Other race was defined by self-report of other or Bahamian, Guatemalan, Honduran, Mestizo, Nicaraguan, or Panamanian.
Includes International Classification of Diseases, Ninth Revision (ICD-9), or International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD-10), codes for alcoholic cirrhosis, cirrhosis without mention of alcohol, biliary cirrhosis, and chronic hepatitis.
Includes ICD-9 and ICD-10 codes for hepatic coma, portal hypertension, other sequelae of chronic liver disease, and esophageal varices.
Includes cefaclor, cefazolin sodium, cefixime, cefotaxime sodium, cefotetan disodium, cefoxitin sodium, cefpodoxime proxetil, ceftriaxone sodium, cefuroxime sodium, and cephalexin and excludes ceftazidime, cefepime hydrochloride, and a combination of ceftolozane sulfate and tazobactam sodium.
Compared with non-CR and non-EBR infections, the CR and EBR infections were more likely to be isolated in patients with an intensive care unit stay before the index culture (CR, 924 of 2324 [39.8%]; non-CR, 2240 of 9178 [24.4%]; EBR, 461 of 1033 [44.6%]; non-EBR, 2703 of 10 469 [25.8%]) or among those transferred from a skilled nursing facility (CR, 435 of 2324 [18.7%]; non-CR, 569 of 9178 [6.2%]; EBR, 231 of 1033 [22.4%]; non-EBR, 773 of 10 469 [7.4%]). The CR and EBR infections were much more likely to occur in patients with invasive devices or procedures and longer length of stay between admission and index culture than susceptible infections (Table 1). Carbapenem-resistant P aeruginosa was isolated in the 30 days before the index admission among 722 of 2324 (31.1%) with an index CR infection vs 157 of 9178 (1.7%) of those with an index non-CR infection. Among patients with index EBR infections, 414 of 1033 (40.1%) had a prior CR culture in the previous 30 days, compared with only 465 of 10 469 (4.4%) among those with index non-EBR infections. Finally, compared with carbapenem-susceptible or extensively β-lactam–susceptible infections, CR and EBR infections were much more likely to occur among patients who received carbapenems (542 of 2324 [23.3%] and 277 of 1033 [26.8%], respectively), ceftazidime, cefepime hydrochloride, and combined ceftolozane sulfate and tazobactam sodium (759 of 2324 [32.7%] and 385 of 1033 [37.3%], respectively), quinolones (801 of 2324 [34.5%] and 353 of 1033 [34.2%], respectively), and piperacillin-tazobactam (746 of 2324 [32.1%] and 374 of 1033 [36.2%], respectively) in the 30 days before the index date (Table 1).
Among 11 502 P aeruginosa infections, 2324 (20.2%) were CR, 9178 (79.8%) were non-CR, 1033 (9.0%) were EBR, and 10 469 were non-EBR (91.0%). The strongest predictors of resistance in the CR and EBR models were history of CR P aeruginosa infection (odds ratios [ORs], 8.80 [95% CI, 6.74-11.49] and 5.04 [95% CI, 3.88-6.54], respectively), tracheostomy (ORs, 3.49 [95% CI, 2.92-4.16] and 3.13 [95% CI, 2.50-3.91], respectively), and carbapenem use in the prior 30 days (ORs, 4.18 [95% CI, 3.29-5.31] and 2.26 [95% CI, 1.74-2.93], respectively). The models for CR and EBR performed well, with areas under the receiver operating characteristics curve of 0.81 or greater for the training and validation data sets.
Development and Performance of the CR P aeruginosa Infection Risk Score
The training and validation data sets consisted of 5834 and 5668 infections, respectively. The final multivariable model covariate risk estimates and points are shown in Table 2; the higher the risk score points, the greater the risk of resistant infection The risk score contained 8 variables; the variables that conferred the greatest number of points in the risk score were history of CR P aeruginosa infection in the 30 days before index admission (7 points) and carbapenem use in the 30 days before the index date (5 points) (Table 2). Cumulative patient scores ranged from −3 points to 20 points. Patients with 20 points had a predicted risk of 0.98, whereas patients with a point total of −3 had a predicted risk of 0.03 (eFigure 3 in the Supplement).
Table 2. Final Covariates in Multivariable Model and Associated Point Values for CR Pseudomonas aeruginosa Infectiona.
| Variable | OR (95% CI) | P Value | Pointsb |
|---|---|---|---|
| Intercept | NA | NA | NA |
| SNF transfer | 1.36 (1.08-1.72) | .009 | 1 |
| Tracheostomy (from admission to culture date) | 3.49 (2.92-4.16) | <.001 | 4 |
| P aeruginosa infection 30 d before index admission | |||
| None | 1 [Reference] | <.001 | NA |
| Carbapenem-susceptible | 0.58 (0.47-0.71) | −2 | |
| CR | 8.80 (6.74-11.49) | 7 | |
| Previous hospitalization 6 mo before index admission | 1.80 (1.51-2.15) | <.001 | 2 |
| Carbapenems 30 d before the index date | 4.18 (3.29-5.31) | <.001 | 5 |
| Other cephalosporins 30 d before the index datec | 0.77 (0.66-0.91) | .002 | −1 |
| Quinolones | 1.53 (1.29-1.81) | <.001 | 1 |
Abbreviations: CR, carbapenem resistant; NA, not applicable; OR, odds ratio; SNF, skilled nursing facility.
Includes 5834 patients.
Calculated as weighted sums of regression coefficients from the prediction model; patient’s risk was estimated as the inverse logit of the risk score. Scores range from −3 to 20; higher risk scores are associated with greater predicted risk of resistant infection. Constant for the point system, Β = 0.3109.
Includes cefaclor, cefazolin sodium, cefixime, cefotaxime sodium, cefotetan disodium, cefoxitin sodium, cefpodoxime proxetil, ceftriaxone sodium, cefuroxime sodium, and cephalexin and excludes ceftazidime, cefepime hydrochloride, and a combination of ceftolozane sulfate and tazobactam sodium.
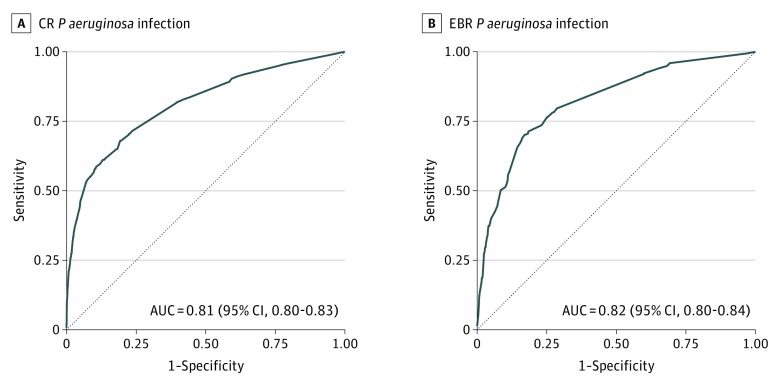
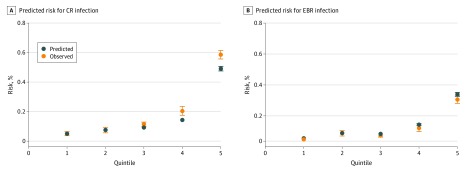
The score differentiated between patients who did and did not have a CR P aeruginosa infection, with an AUC of 0.82 (95% CI, 0.80-0.83) in the training data set. The score performed comparably in the validation data set, with an AUC of 0.81 (95% CI, 0.80-0.83) (Figure 1A). In the validation data set, predicted risk for CR infection agreed closely with observed risk (Figure 2A and eTable 6 in the Supplement). The predicted risk was most closely aligned with observed events among the lowest-risk patients, with some divergence at the higher quintiles of risk.
Figure 1. Performance of the Prediction Models for Carbapenem-Resistant (CR) or Extensive β-Lactam–Resistant (EBR) Pseudomonas aeruginosa Infection.
Diagonal line indicates no discrimination (ie, area under the receiver operating characteristic curve [AUC] = 0.5).
Figure 2. Comparison of Observed and Point-Based Predicted Risk by Deciles According to Model-Based Predicted Risk in Validation Data Set.
Includes 5668 patients. Error bars represent 95% CIs. CR indicates carbapenem resistant; EBR, extensive β-lactam (carbapenem, ceftazidime, and combined piperacillin sodium and tazobactam sodium) resistant.
Development and Performance of the EBR P aeruginosa Infection Risk Score
The sample sizes of the training and validation data sets are the same as stated for the CR model. The final EBR P aeruginosa risk score contained 7 characteristics (Table 3). The variables that conferred the greatest number of points were history of CR P aeruginosa infection in the 30 days before the index date (5 points) and tracheostomy (4 points). Patient scores ranged from 0 to 19 points. Patients with 19 points had a predicted risk of 0.88, whereas patients with a total of 0 had a predicted risk of 0.02 (eFigure 3 in the Supplement).
Table 3. Final Covariates in Multivariable Model and Associated Point Values for Excessive β-Lactam (Carbapenem, Ceftazidime, and Combined Piperacillin Sodium and Tazobactam Sodium)–Resistant Pseudomonas aeruginosa Infectiona.
| Variable | OR (95% CI) | P Value | Pointsb |
|---|---|---|---|
| Intercept | NA | NA | NA |
| SNF transfer | 1.73 (1.33-2.26) | <.001 | 2 |
| Tracheostomy (from admission to culture date) | 3.13 (2.50-3.91) | <.001 | 4 |
| CVC, port, or PICC (from admission to culture date) | 1.74 (1.40-2.17) | <.001 | 2 |
| P aeruginosa infection 30 d before index admission | <.001 | ||
| None | 1 [Reference] | ||
| Carbapenem-susceptible | 0.92 (0.70-1.21) | 0 | |
| CR | 5.04 (3.88-6.54) | 5 | |
| Previous hospitalization 6 mo before index admission | 2.24 (1.73-2.91) | <.001 | 3 |
| Carbapenems 30 d before index date | 2.26 (1.74-2.93) | <.001 | 3 |
Abbreviations: CR, carbapenem resistant; CVC, central venous catheter; NA, not applicable; OR, odds ratio; PICC, peripherally inserted central catheter; SNF, skilled nursing facility.
Includes 5834 patients.
Points were calculated as weighted sums of regression coefficients from the prediction model; patient’s risk was estimated as the inverse logit of the risk score. Scores could range from 0 to 19; higher risk scores were associated with greater predicted risk of resistant infection. Constant for the point system Β = 0.3109.
The score differentiated patients who did and did not have an EBR P aeruginosa infection, with an AUC of 0.84 (95% CI, 0.82-0.86) in the training data set. The score performed comparably in the validation data set, with an AUC of 0.82 (95% CI, 0.80-0.84) (Figure 1B). Predicted risk for EBR infection in the validation data set agreed closely with observed risk (Figure 2B and eTable 6 in the Supplement).
Sensitivity Analyses
We found no differences in final variable selection for the CR and EBR models when we included only the first infection for patients. Further, the AUCs for the sensitivity analyses were similar to those for our main analyses (eTable 7 and eFigure 4 in the Supplement).
Discussion
Although risk factors for resistant P aeruginosa infections have been previously identified in the literature, this study takes risk estimation further by developing 2 clinical risk scores to estimate the probabilities of CR and EBR among hospitalized adult patients with P aeruginosa infections based on covariates available at clinical presentation. Patients with these infections are at high risk of delayed appropriate therapy owing to resistance against commonly prescribed empirical antipseudomonal antibiotics. These resistant phenotypes are now commonplace, and clinical prediction tools of this nature are vitally needed. Consistent with other studies, we found that more than 20% of P aeruginosa infections among adult hospitalized patients were CR.17,18 More alarmingly, nearly 10% of P aeruginosa infections were resistant to most of the β-lactam antibiotics commonly used to empirically treat patients with P aeruginosa infections. Our risk scores can help physicians identify patients who would benefit the most from tailored treatment regimens in the critical period when a P aeruginosa infection is suspected and antibiotic susceptibility results are not yet available. With this information, physicians can make more informed empirical antibiotic selections and thereby increase the likelihood of timely appropriate antibiotic therapy. The CR model will identify situations where a carbapenem should be avoided and other antipseudomonal β-lactams should be used with extreme caution, because nearly 50% of CR P aeruginosa infections were nonsusceptible to piperacillin-tazobactam and ceftazidime. The EBR model identifies situations where a patient requires an individualized empirical regimen that does not include a carbapenem, ceftazidime, or piperacillin-tazobactam. Conversely, these models can identify situations where commonly used antipseudomonal β-lactams are appropriate, thereby reducing overuse of unnecessary combinations of broad-spectrum antibiotics for P aeruginosa infections.
Our main findings corroborate a recent meta-analysis that found prior carbapenem use, prior quinolone use, and presence of medical devices at baseline to have the highest pooled estimates for CR P aeruginosa infection.19 History of a hospitalization and transfer from a skilled nursing facility were also found to be important risk factors across both multivariable models, which is consistent with findings of several other studies.19,20,21,22 Presence of a prior CR P aeruginosa infection was found to be the strongest predictor for CR and EBR. This finding was not surprising owing to the chronic, recurrent nature of many antibiotic-resistant P aeruginosa infections, especially among patients undergoing prolonged mechanical ventilation.23 In contrast, history of a non-CR P aeruginosa infection was found to be protective in the CR model. It is not uncommon for patients to be reinfected with the same organisms; however, the exact reasons for the protective effect observed with a prior non-CR P aeruginosa infection are unclear and merit further inquiry.
In the conversion of these risk factors into flexible tools that can be used at the bedside, we generated risk scores that allow all potential combinations of variables that a patient may have to be translated into a predicted probability of risk. Risk scores of both models ranged from negative risk scores that reflect protective variables to scores as high as 20 points that show the high risk associated with the presence of all risk variables and no protective variables. Although, to our knowledge, no critical threshold values exist in the literature that strongly predict a CR or EBR P aeruginosa infection, a greater than 20% probability of CR or EBR infections was estimated when the risk scores were greater than 4 and 5, respectively. A CR risk score of greater than 4 was achieved among individuals with a P aeruginosa infection in the presence of any of the following final model covariates: prior receipt of a carbapenem, presence of a tracheostomy, or a prior CR P aeruginosa infection. For EBR, several of the risk variables were necessary to predict a greater than 20% probability of EBR (risk score >10).
Strengths and Limitations
Important strengths of our study include the availability of a large data set with 10 136 hospitalizations and 11 502 P aeruginosa infections among a diverse patient population. Our integrated care environment includes inpatient, outpatient, and emergency services, allowing us to assess wide-ranging and comprehensive covariates, including those present before hospital admission. The models performed well in the validation data sets because the predicted risks were consistent with the observed risk, with only slight divergence at very high predicted risk levels for the CR model.
Although our risk score was found to perform adequately using a split-sample approach to validation, an important next step will be external validation of the risk score in other populations and other hospital systems, which also may help to identify clinically meaningful cutoff values for this tool. We omitted risk factors that are not readily available at bedside on admission and may miss factors that are predictive of CR and/or EBR infection. Further, additional prediction methods may improve the prediction modeling in future work, such as neural networks, random forest, and SuperLearner, that allow the incorporation of several algorithms simultaneously to deliver the strongest prediction model.24 We leveraged multiple prediction methods in this analysis to maximize performance of the final model and found only minimal differences in the predictive performance between the approaches. In addition, although susceptibility of P aeruginosa to cefepime is highly correlated with that of ceftazidime, resistance between the two can vary depending on the resistant mechanisms. Therefore, the score may not be perfectly applicable to prediction of P aeruginosa resistance to cefepime. Whether our findings are applicable to patient populations not included in this study, such as adults with cystic fibrosis and those younger than 18 years, is unclear. As with all clinical tools of this nature, caution and appropriate clinical judgment should be exercised when applying the risk score, particularly before validation at external institutions.
Conclusions
We developed parsimonious clinical risk scores to aid physicians in appropriate treatment selection in the critical period between the suspicion of P aeruginosa infections and the availability of antibiotic susceptibility results. The prediction models created in this study are consistent with the priorities identified by the World Health Organization and serve as models for institutions seeking mechanisms to improve antibiotic treatment protocols and minimize delays in delivery of timely appropriate therapy. Conversely, this tool may help inform appropriate use of combinations of broad-spectrum antibiotics among patients with susceptible P aeruginosa infections.
eTable 1. Definition of Covariates
eTable 2. Comparison of Training and Validation Data Sets for Patients With CR and Non-CR Pseudomonas aeruginosa Infections
eTable 3. Comparison of Training and Validation Data Sets for Patients With EBR and Non-EBR Pseudomonas aeruginosa Infections
eTable 4. Selected Characteristics of Patients With CR, Non-CR, EBR, and Non-EBR Pseudomonas aeruginosa Infections
eTable 5. Antibiotic Susceptibility Results, Stratified by CR and EBR Status
eTable 6. Comparison of Observed and Point-Based Predicted Risk by Quintile for CRe and EBR Pseudomonas aeruginosa (n = 5668)
eTable 7. Sensitivity Analyses Including only the First Infection per Patient
eFigure 1. Comparison of Model Performance by Variable Selection Approach
eFigure 2. Flowchart of Study Population Selection
eFigure 3. Estimated Risk of Carbapenem-Resistant (CR) and Carbapenem, Ceftazidime, and Piperacillin-Tazobactam–Resistant (EBR) Pseudomonas aeruginosa Infection by Point Total
eFigure 4. Models in Training and Validation Data Sets
References
- 1.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):-. doi: 10.1016/j.tim.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 2.Jacoby GA, Munoz-Price LS. The new β-lactamases. N Engl J Med. 2005;352(4):380-391. doi: 10.1056/NEJMra041359 [DOI] [PubMed] [Google Scholar]
- 3.Walsh TR. Clinically significant carbapenemases: an update. Curr Opin Infect Dis. 2008;21(4):367-371. doi: 10.1097/QCO.0b013e328303670b [DOI] [PubMed] [Google Scholar]
- 4.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34(5):634-640. doi: 10.1086/338782 [DOI] [PubMed] [Google Scholar]
- 5.Kang CI, Kim SH, Kim HB, et al. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis. 2003;37(6):745-751. doi: 10.1086/377200 [DOI] [PubMed] [Google Scholar]
- 6.Kang CI, Kim SH, Park WB, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005;49(2):760-766. doi: 10.1128/AAC.49.2.760-766.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kollef MH, Sherman G, Ward S, Fraser VJ. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest. 1999;115(2):462-474. doi: 10.1378/chest.115.2.462 [DOI] [PubMed] [Google Scholar]
- 8.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med. 1998;244(5):379-386. doi: 10.1046/j.1365-2796.1998.00379.x [DOI] [PubMed] [Google Scholar]
- 9.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582-610. doi: 10.1128/CMR.00040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization, Department of Communicable Disease Surveillance and Response WHO global strategy for containment of antimicrobial resistance. http://www.who.int/csr/resources/publications/drugresist/en/EGlobal_Strat.pdf. Accessed December 30, 2016.
- 11.Koebnick C, Langer-Gould AM, Gould MK, et al. Sociodemographic characteristics of members of a large, integrated health care system: comparison with US Census Bureau data. Perm J. 2012;16(3):37-41. doi: 10.7812/TPP/12-031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund B. Logistic Model Selection with SAS PROC's LOGISTIC, HPLOGISTIC, HPGENSELECT: Paper AA02. Paper presented at: MidWest SAS Users Group; 2017; St Louis, MO. [Google Scholar]
- 13.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6(2):461-464. doi: 10.1214/aos/1176344136 [DOI] [Google Scholar]
- 14.Johnston G, Rodriguez RN. Introducing the HPGENSELECT Procedure: Model Selection for Generalized Linear Models and More: Paper SAS1742-2015. https://support.sas.com/resources/papers/proceedings15/SAS1742-2015.pdf. Accessed January 2017.
- 15.Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc B. 1996;58(1):267-288. [Google Scholar]
- 16.Sullivan LM, Massaro JM, D’Agostino RB Sr. Presentation of multivariate data for clinical use: the Framingham Study risk score functions. Stat Med. 2004;23(10):1631-1660. doi: 10.1002/sim.1742 [DOI] [PubMed] [Google Scholar]
- 17.Lautenbach E, Synnestvedt M, Weiner MG, et al. Imipenem resistance in Pseudomonas aeruginosa: emergence, epidemiology, and impact on clinical and economic outcomes. Infect Control Hosp Epidemiol. 2010;31(1):47-53. doi: 10.1086/649021 [DOI] [PubMed] [Google Scholar]
- 18.Gaynes R, Edwards JR; National Nosocomial Infections Surveillance System . Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41(6):848-854. doi: 10.1086/432803 [DOI] [PubMed] [Google Scholar]
- 19.Voor In’t Holt AF, Severin JA, Lesaffre EM, Vos MC. A systematic review and meta-analyses show that carbapenem use and medical devices are the leading risk factors for carbapenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(5):2626-2637. doi: 10.1128/AAC.01758-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furtado GH, Gales AC, Perdiz LB, Santos AE, Wey SB, Medeiros EA. Risk factors for hospital-acquired pneumonia caused by imipenem-resistant Pseudomonas aeruginosa in an intensive care unit. Anaesth Intensive Care. 2010;38(6):994-1001. [DOI] [PubMed] [Google Scholar]
- 21.Zavascki AP, Barth AL, Gaspareto PB, et al. Risk factors for nosocomial infections due to Pseudomonas aeruginosa producing metallo-β-lactamase in two tertiary-care teaching hospitals. J Antimicrob Chemother. 2006;58(4):882-885. doi: 10.1093/jac/dkl327 [DOI] [PubMed] [Google Scholar]
- 22.Ohmagari N, Hanna H, Graviss L, et al. Risk factors for infections with multidrug-resistant Pseudomonas aeruginosa in patients with cancer. Cancer. 2005;104(1):205-212. doi: 10.1002/cncr.21115 [DOI] [PubMed] [Google Scholar]
- 23.Chien JY, Hsueh PR, Yu CJ, Yang PC. The evolution of drug-resistant microorganisms in patients with prolonged mechanical ventilation. Am J Infect Control. 2009;37(3):231-236. doi: 10.1016/j.ajic.2008.05.016 [DOI] [PubMed] [Google Scholar]
- 24.Rose S. Mortality risk score prediction in an elderly population using machine learning. Am J Epidemiol. 2013;177(5):443-452. doi: 10.1093/aje/kws241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Definition of Covariates
eTable 2. Comparison of Training and Validation Data Sets for Patients With CR and Non-CR Pseudomonas aeruginosa Infections
eTable 3. Comparison of Training and Validation Data Sets for Patients With EBR and Non-EBR Pseudomonas aeruginosa Infections
eTable 4. Selected Characteristics of Patients With CR, Non-CR, EBR, and Non-EBR Pseudomonas aeruginosa Infections
eTable 5. Antibiotic Susceptibility Results, Stratified by CR and EBR Status
eTable 6. Comparison of Observed and Point-Based Predicted Risk by Quintile for CRe and EBR Pseudomonas aeruginosa (n = 5668)
eTable 7. Sensitivity Analyses Including only the First Infection per Patient
eFigure 1. Comparison of Model Performance by Variable Selection Approach
eFigure 2. Flowchart of Study Population Selection
eFigure 3. Estimated Risk of Carbapenem-Resistant (CR) and Carbapenem, Ceftazidime, and Piperacillin-Tazobactam–Resistant (EBR) Pseudomonas aeruginosa Infection by Point Total
eFigure 4. Models in Training and Validation Data Sets