Abstract
The aims of this study were to analyze the incidence and morphology of cyclin D1+ DLBCL and cases of Richter transformation (RT), and to elucidate possible molecular mechanisms of cyclin D1 overexpression. Seventy-two cases of de novo DLBCL and 12 cases of RT were included in this study. Cyclin D1 positivity was found in 10/66 (15%) cases of unselected de novo DLBCL and in 2/11 (18%) cases of RT. Seven independently identified cases of cyclin D1+ DLBCL, including one RT, were added to the study. Centroblastic morphology was found in 17/19 (89%) cases of cyclin D1+, most with a post-germinal center phenotype (CD10−, BCL6+, MUM1+ ). No alterations in the CCND1 gene indicative for a translocation t(11;14) were identified by FISH. Analysis of the MYC locus yielded gene copy alterations in five cases and no disruption of the gene locus in any case, suggesting an alternative mechanism of cyclin D1 deregulation.
Keywords: Cytogenetics, non-Hodgkin lymphoma, molecular genetics, cyclin D1
Introduction
The CCND1 gene localized on chromosome 11q13 encodes the cyclin D1 oncogene product, a major regulator of the G1–S transition of the cell cycle [1,2]. Aberrant expression of cyclin D1 protein has been implicated in the pathogenesis of several types of human neoplasms [3–5]. In lymphoid malignancies, cyclin D1 mRNA and protein overexpression is usually the consequence of the chromosomal translocation t(11;14)(q13;q32), which juxtaposes the immunoglobulin heavy chain (IgH) gene on 14q32 to the CCND1 gene on 11q13. This translocation is the genetic hallmark of mantle cell lymphoma (MCL) [6], and can be detected in more than 90% of cases. Of interest, cyclin D1 overexpression can also be found in about half of the cases of plasma cell myeloma (PCM) [7], a finding correlated with the presence of the t(11;14) or extra copies of chromosome 11 [7]. Hairy cell leukemia also shows abnormal expression of cyclin D1. However, in contrast to PCM and MCL, it occurs in the absence of t(11;14) or other genetic abnormalities of the 11q13 region, pointing toward a different molecular mechanism of cyclin D1 overexpression [8]. Of note, cyclin D1 is expressed in a cell-type specific fashion, and normal lymphocytes do not express cyclin D1, but instead cyclin D2 or cyclin D3 [9]. Besides the mentioned lymphoma subtypes, cyclin D1 is believed to be negative in other B-cell lymphomas [10].
Diffuse large B-cell lymphomas (DLBCLs) constitute a heterogeneous group of aggressive lymphomas with respect to morphology, clinical presentation and course, immunophenotype, and gene expression [11]. DLBCLs are generally believed to be cyclin D1 negative; however, in recent years there have been isolated case reports of DLBCL expressing cyclin D1 [12,13]. In a recent comprehensive study, Ehinger and colleagues [14] found cyclin D1 expression in 4.3% of cases in a large cohort of 231 CD5 negative cases of DLBCL. Interestingly, we recently observed a case of chronic lymphocytic leukemia (CLL) with clonally related Richter transformation (RT) in the tonsil, exhibiting strong nuclear cyclin D1 staining in the DLBCL component. Nevertheless, the association of cyclin D1 expression with specific histological subtypes of DLBCL and/or in the setting of RT has not been studied systematically.
The aims of this study were (1) to analyze the incidence of cyclin D1 overexpression in an un-selected series of 77 cases of DLBCL of different histological subtypes, including 11 cases of RT, and (2) to perform a comprehensive immunophenotypic analysis (germinal center B-cell type vs. activated B-cell type) in the cyclin D1 positive cases.
Materials and methods
Lymphoma cases
Seventy-seven cases of unselected DLBCL, including 11 cases of RT and 66 cases of de novo DLBCL of different histological subtypes (centroblastic, immunoblastic, T-cell/histiocyte-rich large B-cell lymphoma) from the files of the Institute of Pathology, Technical University of Munich (Germany) were included in the study. Seven additional cases of cyclin D1+ DLBCL, including one case of RT in CLL, were added to the study cohort from four different institutions (National Cancer Institute, National Insitutes of Health (NIH), Bethesda, USA; Massachusetts General Hospital, Boston, USA; Institute of Pathology, University of Tu¨ bingen, Germany; and Newcastle University, Newcastle Upon Tyne, UK). All cases were formalin-fixed and paraffin-embedded biopsy specimens. Diagnoses were confirmed during the diagnostic workup with standard stains (hematoxylin and eosin [H&E], Giemsa), and with immunohistochemistry with a panel of antibodies to assess lymphoid phenotype and cell of origin of the DLBCL. All cases were evaluated and classified according to the guidelines of the World Health Organization: WHO classification of tumours of hematopoietic and lymphoid tissues [11].
The study was approved by the local ethical committee.
Immunohistochemistry
Staining for cyclin D1 was performed using a monoclonal rabbit antibody (clone SP4; Lab Vision, Fremont, CA, USA). Three cases of t(11;14) + MCL were used as positive controls. Nuclear expression in more than 10% of tumor cells was considered positive. Endothelial cells and histiocytes were used as an internal positive control. To assign a case to germinal center (GCB) or non-GCB (activated B-cell [ABC] or unclassified) phenotype, the algorithm proposed by Hans and colleagues [15] was used.
Immunohistochemistry was performed on an automated immunostainer (Ventana Medical Systems, Tucson, AZ, USA) following the manufacturer’s protocols, with slight modifications, as previously reported [16].
Immunofluorescence double staining
In selected cases double staining for cyclin D1 expression and CD20 was performed by immunofluorescence. Sections were washed in phosphate buffered saline (PBS) and blocked in 5% normal goat serum (NGS) for 30 min in a moist chamber at 37°C. The primary antibody, anti-cyclin D1 (Dako, Hamburg, Germany), was diluted 1:100 and incubated at room temperature for 1 h, followed by incubation with a secondary antibody conjugated with Cy5 (Invitrogen, Karlsruhe, Germany), and diluted 1:100 in PBS with 5% NGS (final concentration 10 µg/mL) at room temperature for 1 h. Subsequently, slides were incubated with anti-CD20 antibody (DakoCytomation, Hamburg, Germany) diluted 1:100 at room temperature for 1 h. An appropriate secondary antibody was conjugated with fluorescein isothiocyanate (FITC; Invitrogen). Sections were rinsed and counterstained with Hoechst 33342 (15 μg/mL), dried, and coverslipped with an antifade solution. Stains were evaluated using a confocal laser scanning microscope (TCS SP2; Leica, Heidelberg, Germany).
Fluorescence in situ hybridization
The status of CCND1 and MYC gene loci was examined in cyclin D1 positive DLBCL using a bicolor fluorescence in situ hybridization (FISH)-based detection system with a CCND1 or MYC two color break-apart probe and a CCND1 locus specific probe (Figure 1). Additionally, a specific IgH/MYC dual fusion translocation probe was applied.
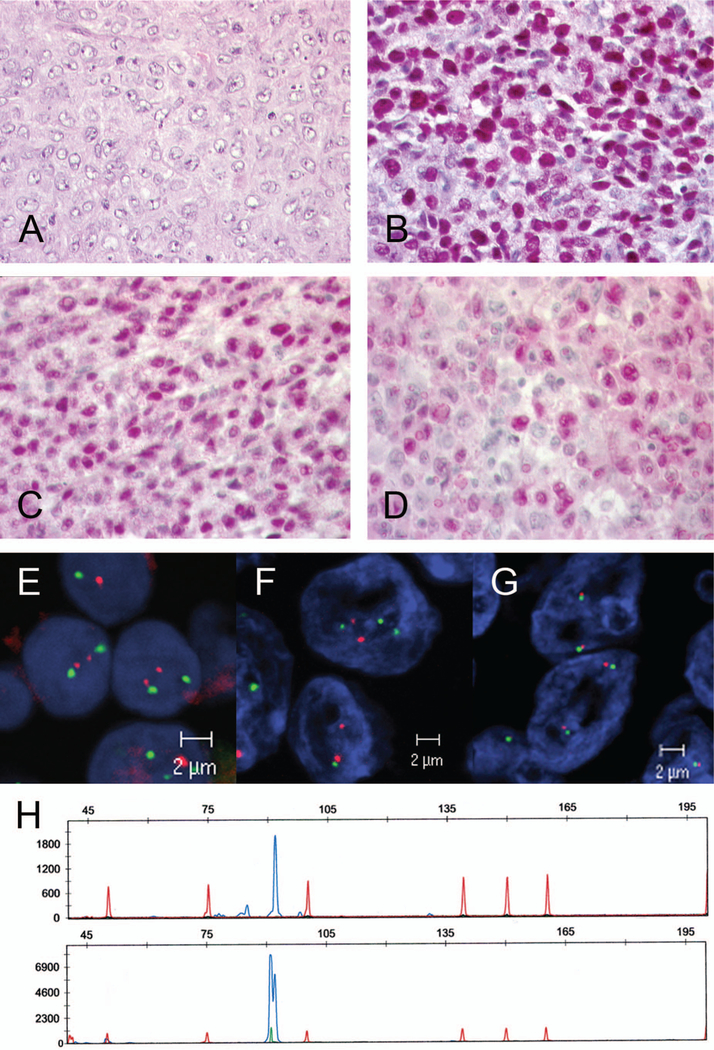
Figure 1.
Case 17: Richter transformation of B-CLL. (A) Diffuse large B-cell lymphoma with centroblastic morphology (H&E, original magnification × 400). (B) MUM1 strong nuclear positivity in the majority of tumor cells (immunoperoxidase, original magnification × 400). (C) BCL6 positivity in the majority of tumor cells (immunoperoxidase, original magnification × 400). (D) Cyclin D1 expression in the tumor cells. Note that cyclin D1 was detectable in a majority of cells with weak to moderate intensity (original magnification × 400). (E, F) Interphase FISH analysis with dual color fusion probe for t(11;14)(q13)(q32). Note that neither B-CLL (E) nor transformed DLBCL (F) harbored a t(11;14) chromosomal translocation involving the CCND1 gene locus. (G) FISH analysis using a dual color break-apart probe for the MYC gene locus was not indicative for a chromosomal break involving the MYC gene locus. (H) Fragment length analysis of the CDR3 region of the immunoglobulin heavy chain gene revealed a monoclonal peak of the same size in B-CLL (upper panel) and in Richter transformation (lower panel), demonstrating that both tumors were clonally related.
Normal lymphoid tissue, and mantle cell lymphoma and Burkitt lymphoma samples with known translocations, were included as controls. Interphase FISH and signal evaluation was performed as described previously [17]. Briefly, serial 5 µm sections were deparaffinized with xylene and rehydrated with decreasing concentrations of ethanol. Antigen retrieval was performed by microwave treatment at 350 W for 40 min in citrate buffer (pH 6.0). DNA was denatured in 70% formamide/2 × saline–sodium citrate (SSC) at 75°C for 15 min, and after dehydration with graded ethanol the slides were dried. The probe, a Vysis LSI CCND1 (11q13) dual color break-apart rearrangement probe, Vysis LSI MYC (8q24) dual color break-apart rearrangement probe, or Vysis LSI IgH/MYC CEP8 tri-color dual fusion translocation probe (Abbott Molecular, Wiesbaden, Germany), respectively, was denatured separately for 5 min at 75°C and placed on the slides. The slides were coverslipped, sealed with rubber cement, and incubated at 37°C overnight in a moist chamber. After in situ hybridization, slides were washed in 2 × SSC/0.1% NP-40 (pH 7.4) at 73°C for 2 min and counterstained with diamidino-2-phenylindole (DAPI; Sigma, Hannover, Germany). Slides were analyzed using a Zeiss Axio Imager (Carl Zeiss MicroImaging GmbH, Go¨ttingen, Germany).
In every case, 100 cells were evaluated for a split of the physiologically co-localized signals, indicative of the presence of chromosomal translocation involving the CCND1 or MYC gene loci, or for numerical chromosomal alterations. The cut-offs for all probe sets were determined on 10 reactive samples (mean ± 3 × standard deviation [SD]).
Results
Cyclin D1 protein expression in de novo DLBCL and RT
Of the 66 unselected cases of de novo DLBCL initially studied, 10 (15%) were considered positive for cyclin D1 expression. In most of these 10 cases, neoplastic cells (˃10%) with weak positivity for cyclin D1 were identified (Figure 2). To confirm the localization of the cyclin D1 reactivity in the CD20 positive neoplastic cells, immunofluorescence double stains were performed (Figure 2). In all cases these analyses unequivocally demonstrated the co-localization of CD20 and cyclin D1 in the tumor cells. Two of 11 cases of RT (18%) were positive for cyclin D1. These cases showed a strong nuclear positivity for cyclin D1 in the DLBCL component, whereas the CLL component was negative for cyclin D1. The low and high grade components were clonally related in the two cases (Figure 1).
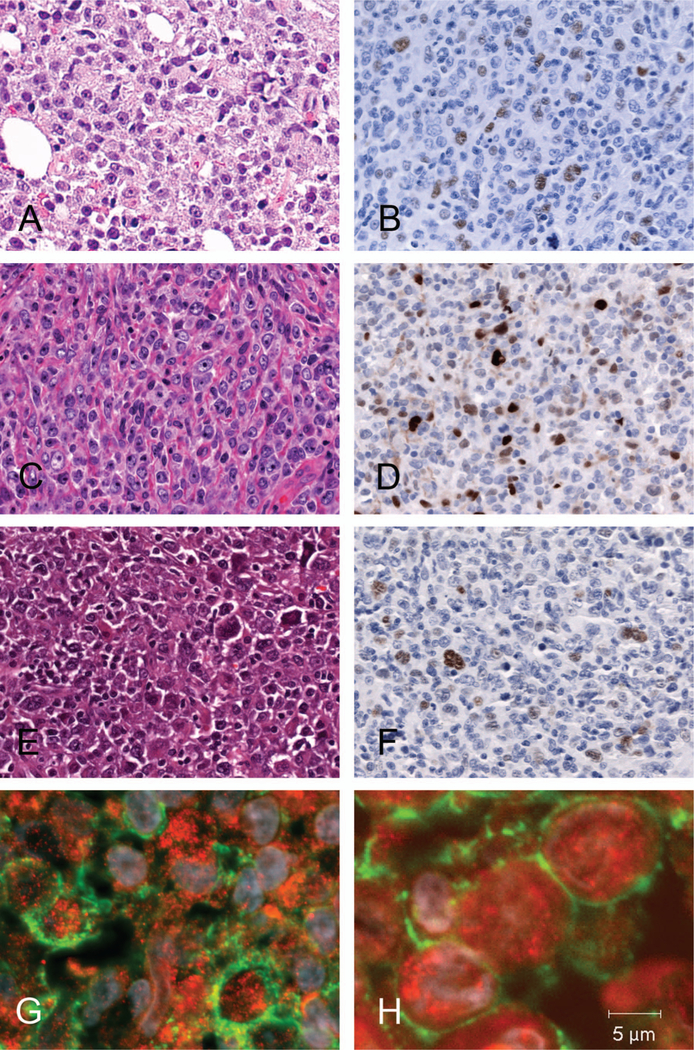
Figure 2.
Morphological spectrum and immunophenotype of cyclin D1+ DLBCL. (A) Most cases featured a centroblastic morphology (H&E, original magnification × 200). (C) One case presented with immunoblastic morphology (H&E, original magnification × 400). (B, D, F) In immunohistochemical analysis in most cases nuclear cyclin D1 positivity was weak to moderate in tumor cells with strong positivity in macrophages and endothelial cells serving as endogenous control (immunoperoxidase, original magnification of B × 200, D, F × 400, respectively). (E) In a case of T-cell/histiocyte-rich B-cell lymphoma (H&E, original magnification × 400) cyclin D1 reactivity was accentuated to the large B-cells (F, immunoperoxidase, original magnification × 400). (G, H) Immunofluorescence double staining unequivocally co-localized nuclear cyclin D1 expression (red) in CD20 positive B-cells (green membrane) (original magnification × 1000).
Morphology and immunophenotype of cyclin D1+ DLBCL
In order to characterize better the cyclin D1+ DLBCL, in addition to the 10 positive cases described above, six cases of de novo cyclin D1+ DLBCL from other institutions were included in the analysis. These six cases also showed cyclin D1 positivity in more than 10% of the tumor cells. Although an arbitrary cut-off of 10% was used in this study, most cases showed cyclin D1 expression in above 20% of the cells. The 16 cases of cyclin D1+ DLBCL included in the study displayed rather homogeneous morphological patterns. Most cases revealed centroblastic morphology (14 cases), only one case showed an immunoblastic morphology, and one other case corresponded to a T-cell/histiocyte-rich large B-cell lymphoma. Table I summarizes the results of the immunophenotypic studies. CD20 was positive in all cases. All cases were consistently negative for CD10, BCL6 was positive in 15 of 16 cases (94%), and MUM1 was positive in 13/16 cases (81%). Only one case stained weakly positive for CD5 (case 16). Proliferation rate, as assessed by MIB1 antibody, yielded a wide range of positivity. All cases showed a proliferation rate above 40%. The immunophenotype of most cases (CD10−, MUM1+, and/or BCL6+ ) indicated an activated B-cell phenotype.
Table I.
Immunohistochemical analysis of 16 cases of DLBCL (de novo) and three cases of Richter transformation.
| Case | Immunohistochemistry |
Morphology | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CD20 | CD5 | MIB1* | BCL2 | CD10 | MUM1 | BCL6 | Cyclin D1* | ||
| De novo | |||||||||
| 1 | + | − | 50 | + | − | + | + | 20 | IB |
| 2 | + | − | 40 | + | − | +/− | − | 15 | CB |
| 3 | + | − | 45 | − | − | + | + | 15 | CB |
| 4 | + | − | 70 | + | − | + | + | 10 | CB |
| 5 | + | − | 80 | + | − | + | + | 20 | CB |
| 6 | + | − | 70 | − | − | + | + | 20 | CB |
| 7 | + | − | 70 | − | − | − | + | 20 | CB |
| 8 | + | − | 75 | − | − | − | + | 20 | CB |
| 9 | + | − | 40 | + | − | + | + | 20 | CB |
| 10 | + | − | 80 | 7 | − | + | + | 30 | CB |
| 11† | + | − | 60 | +/− | − | − | + | 30 | CB |
| 12† | + | − | 70 | NA | − | + | −/+ | 20 | CB |
| 13† | + | − | 60 | NA | − | + | + | 20 | T/HRLBCL |
| 14† | + | − | 80 | NA | − | + | + | 10 | CB |
| 15† | + | − | 90 | + | − | + | + | 20 | CB |
| 16† | + | Weak | 50 | + | − | + | −/+ | 20 | CB |
| RT | |||||||||
| 17 | − | − | 90 | NA | − | + | + | 80 | CB |
| 18† | + | − | 70 | + | − | + | + | 70 | CB |
| 19 | + | − | 70 | + | − | + | + | 20 | CB |
For cyclin D1 staining and proliferation fraction (MIB1) the percentage of positive cells is given.
Cases 11–16 and 18 were not part of the original cohort of 66 cases, but independently identified.
DLBCL, diffuse large B-cell lymphoma; RT, Richter transformation; NA, not available; IB, immunoblastic; CB, centroblastic; T/HRLBCL:,T-cell/histiocyte-rich large B-cell lymphoma.
Morphology and immunophenotype of cyclin D1+ RT
In order to characterize better the cases of RT, a third case, from a different institution, was added to the study cohort. The three cases of RT showed the strongest cyclin D1 positivity, with two cases expressing cyclin D1 in the majority of tumor cells. All three cases revealed centroblastic morphology. Table I summarizes the results of the immunophenotypic studies. Two cases showed expression of CD20 whereas one case was negative; in this case the CLL component was CD20 positive. The three cases had a very homogeneous immunophenotype, being consistently negative for CD10 and positive for BCL6 and MUM1. In contrast to the CLL counter- part, the RT cases were CD5 and CD23 negative. All three cases were highly proliferative (70–90%). The immunophenotype of the RT cases (CD107, MUM1+, and/or BCL6+ ) was similar to de novo DLBCL, indicating an activated B-cell phenotype.
Fluorescence in situ hybridization
FISH analysis for the translocation t(11;14) (q13;q32) was negative in all cases. Copy number gains in the CCND1 locus were not detected. One case of RT was tetraploid (case 18). Analysis of the MYC locus yielded three gene copies in four cases (cases 3, 4, 7, and 9), two of which were associated with trisomy of chromosome 8 (cases 4 and 9). Case 5 harbored four MYC gene copies. Disruption of the gene locus indicative for a genetic translocation involving MYC was not found in any case.
Discussion
In this study, the incidence of cyclin D1 expression in de novo DLBCL, as well as in the setting of RT, was investigated. Ten of 66 unselected cases of de novo DLBCL were positive for cyclin D1 (15%) and two out of 11 RT cases (18%) showed cyclin D1 expression. FISH analyses demonstrated that the expression of cyclin D1 protein in these tumors was not associated with the characteristic translocation t(11;14), nor with numerical aberrations of chromosome 11. FISH analysis of the MYC locus, as an important upstream regulator of CCND1, did not reveal translocations. However, increased MYC signals resulting from trisomy 8 were identified in a minority of the cases.
The incidence of cyclin D1 overexpression in DLBCL in our series was more than three times higher (15% vs. 4%) than that of Ehinger et al., who analyzed 231 cases of de novo DLBCL [14]. This difference is most probably due to the higher sensitivity of the cyclin D1 antibody clone SP4 used in our study, as compared to the clone P2D11F11 (Novocastra) used in a proportion of the cases in the Swedish study. However, another study using the same antibody [18] found a cyclin D1 positivity in only 2% of DLBCL. Our findings suggest that overexpression of cyclin D1 in DLBCL is not as infrequent as previously thought.
As described by Ehinger et al. [14], we also found a predominantly centroblastic morphology in cases of cyclin D1+ DLBCL, with rare cases having either an immunoblastic morphology (one case) or corresponding to T-cell/histiocyte-rich large B-cell subtype (one case). Interestingly, all cases of cyclin D1 positive DLBCL were negative for CD10, but most were positive for BCL6, and the majority of cases showed strong expression of MUM1, thus exhibiting a post-germinal center or activated B-cell phenotype. This observation may be a hint to the mechanism of cyclin D1 overexpression in these cases, which is clearly different from the mechanism in MCL where cyclin D1 is overexpressed as a consequence of the t(11;14). Furthermore, MCL is generally believed to originate from a naive pregerminal center B-cell, whereas cyclin D1+ DLBCL features a post-germinal center B-cell phenotype.
The cyclin D1 DLBCL described by Ehinger et al. [14] did not harbor a t(11;14); however, two out of 10 cases displayed CCND1 gene copy number gains and one case showed loss of genetic material telomeric of the CCND1 gene locus. These findings further confirm the concept of alternative deregulation mechanisms of CCND1 and/or post-translational alterations rather than the classical t(11;14) in DLBCL [19]. The importance of recognizing the existence of cyclin D+1 DLBCL is to avoid a possible misdiagnosis of MCL. MCL is a neoplasia derived from pregerminal or naive lymphocytes. The classical phenotype of MCL is CD20+, CD5+, BCL2+ and strong expression of cyclin D1 in the majority of tumor cells, distinct from our cases. In addition, the expression of BCL6 and/or MUM1 in practically all the cases of DLBCL is a further argument against MCL. Although both MUM1 and BCL6 can be observed in a minority of classical MCL [20,21], coexpression of both markers is infrequent. On the other hand, a point to keep in mind to avoid overdiagnosis of cyclin D1+ DLBCL is the expression of cyclin D1+ by reactive histiocytes, which may pose a problem in DLBCL with a high content of histiocytes.
In summary, the differential diagnosis of the blastoid variant of MCL versus cyclin D1+ DLBCL should be based on morphology, on the immunophenotype of the lymphoid cells, and on the generally rather weak and heterogeneous positivity of cyclin D1, in only a proportion of the tumor cells in DLBCL.
Cyclin D1 is a common downstream effector of several growth pathways, among which MYC plays an important role. MYC is an oncogene that functions both in the stimulation of cell proliferation and in apoptosis [22]. MYC elicits its oncogenic activity by causing immortalization [22], and to a lesser extent transformation of cells, in addition to several other mechanisms. Coordination of MYC with cyclin D1 or its upstream activators not only accelerates tumor formation, but also may drive tumor progression to a more aggressive phenotype [22]. Moreover, one previously reported case of cyclin D1 DLBCL contained a MYC translocation [13]. Therefore, alterations in the MYC gene locus, as a possible mechanism underlying the overexpression of cyclin D1, were investigated in the present series. FISH analysis demonstrated in four cases (cases 3, 4, 7, and 9) three MYC gene copies, two of which were associated with trisomy 8 (cases 4 and 9). The two other cases showed two signals of the centromere-specific probe for chromosome 8, indicating a true copy number gain of the MYC gene locus. One case (case 5) harbored four MYC gene copies. However, disruption of the MYC gene locus indicative of a genetic translocation was not found. Whether the increase in MYC copy numbers in some of the cases could imply a gene dosis effect is not clear. Nevertheless, our data suggest that deregulation of MYC through translocation is not responsible for the expression of cyclin D1 in DLBCL.
Because cyclin D1 is an important regulator of the G1 to S phase progression of the cell cycle [1,2], we also wanted to see whether the overexpression of cyclin D1 was associated with a high proliferation rate. Proliferation rate as assessed with the MIB1 antibody showed a wide range of positivity in both cyclin D1+ DLBCL and cyclin D1− DLBCL, indicating that expression of cyclin D1 did not influence proliferation in DLBCL. In an attempt to investigate the mechanism through which cyclin D1 is induced in DLBCL, we analyzed the mutation status of B-RAF and K-RAS gene loci in five cases by sequencing analysis (data not shown), as important upstream regulators of CCND1 [23,24]. However, none of the cases harbored mutations in these genes, in concordance with DLBCL in general. Further analysis is warranted to elucidate the mechanism(s) of abnormal cyclin D1 expression in DLBCL.
Of particular interest is one case of RT (case 17), in which the immunophenotype of the DLBCL component changed significantly during the transformation. All B-cell markers, including CD20, as well as CD23 and CD5 were lost, whereas cyclin D1 was strongly expressed in the majority of tumor cells, raising the possibility of a second neoplasm. Nevertheless, clonal analysis of the IgH gene demonstrated an identical clone in both components (Figure 1). Neither the CLL nor the DLBCL component harbored a t(11;14) or copy number gains of the CCND1 gene locus. There is no explanation as to why the DLBCL component in this case or in the other two RT cases showed strong cyclin D1 positivity in the tumor cells. This finding raises the question of whether the expression of cyclin D1 in DLBCL might be indicative of tumor progression. As evidenced by the cases of RT, which showed the strongest positivity in the tumor cells, cyclin D1 overexpression may be associated with a particular, yet not fully understood mechanism of transformation in B-CLL. It might be speculated that the mechanism of CCND1 overexpression may be related to the described CCND1 expression in the proliferation centers of rare cases of B-CLL [25].
In conclusion, we report the abnormal expression of cyclin D1 protein in DLBCL, which is mainly associated with centroblastic morphology and reveals a postgerminal center or activated B-cell phenotype (most commonly CD10−, BCL6+, MUM1+ ). The abnormal expression of cyclin D1 is not associated with t(11;14) or alterations in chromosome 11, suggesting an alternative mechanism of cyclin D1 deregulation in both de novo DLBCL and in Richter transformation.
Acknowledgement
The authors thank Claudia Kloß and Barbara Mankel for excellent technical assistance.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev 1993;7:812–821. [DOI] [PubMed] [Google Scholar]
- 2.Quelle DE, Ashmun RA, Shurtleff SA, et al. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev 1993;7:1559–1571. [DOI] [PubMed] [Google Scholar]
- 3.Yang WI, Zukerberg LR, Motokura T, Arnold A, Harris NL. Cyclin D1 (Bcl-1, PRAD1) protein expression in low-grade B-cell lymphomas and reactive hyperplasia. Am J Pathol 1994; 145:86–96. [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi H, Saito H, Kitano K, et al. Overexpression of the PRAD1 oncogene in a patient with multiple myeloma and t(11;14)(q13;q32). Acta Haematol 1995;94:199–203. [DOI] [PubMed] [Google Scholar]
- ►5.Bosch F, Jares P, Campo E, et al. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood 1994;84:2726–2732. [PubMed] [Google Scholar]
- ►6.Banks PM, Chan J, Cleary ML, et al. Mantle cell lymphoma. A proposal for unification of morphologic, immunologic, and molecular data. Am J Surg Pathol 1992;16:637–640. [DOI] [PubMed] [Google Scholar]
- 7.Troussard X, Avet-Loiseau H, Macro M, et al. Cyclin D1 expression in patients with multiple myeloma. Hematol J 2000;1:181–185. [DOI] [PubMed] [Google Scholar]
- 8.de Boer CJ, Kluin-Nelemans JC, Dreef E, et al. Involvement of the CCND1 gene in hairy cell leukemia. Ann Oncol 1996;7:251–256. [DOI] [PubMed] [Google Scholar]
- 9.Ajchenbaum F, Ando K, DeCaprio JA, Griffin JD. Independent regulation of human D-type cyclin gene expression during G1 phase in primary human T lymphocytes. J Biol Chem 1993;268:4113–4119. [PubMed] [Google Scholar]
- 10.Specht K, Kremer M, Muller U, et al. Identification of cyclin D1 mRNA overexpression in B-cell neoplasias by real-time reverse transcription-PCR of microdissected paraffin sections. Clin Cancer Res 2002;8:2902–2911. [PubMed] [Google Scholar]
- 11.Swerdlow SH, Campo E, Harris NL, et al. , editors. WHO classification of tumours of haematopoietic and lymphoid tissues Lyon: IARC Press; 2008. [Google Scholar]
- 12.Teruya-Feldstein J, Gopalan A, Moskowitz CH. CD5 negative, cyclin D1-positive diffuse large B-cell lymphoma (DLBCL) presenting as ruptured spleen. Appl Immunohistochem Mol Morphol 2009;17:255–258. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Justo M, Huang Y, Ye H, et al. Cyclin D1-positive diffuse large B-cell lymphoma. Histopathology 2008;52:900–903. [DOI] [PubMed] [Google Scholar]
- 14.Ehinger M, Linderoth J, Christensson B, Sander B, Cavallin-Stahl E. A subset of CD5-diffuse large B-cell lymphomas expresses nuclear cyclin D1 with aberrations at the CCND1 locus. Am J Clin Pathol 2008;129:630–638. [DOI] [PubMed] [Google Scholar]
- ►15.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004;103:275–282. [DOI] [PubMed] [Google Scholar]
- 16.Quintanilla-Martinez L, Pittaluga S, Miething C, et al. NPM-ALK-dependent expression of the transcription factor CCAAT/enhancer binding protein beta in ALK-positive anaplastic large cell lymphoma. Blood 2006;108:2029–2036. [DOI] [PubMed] [Google Scholar]
- 17.Specht K, Haralambieva E, Bink K, et al. Different mechanisms of cyclin D1 overexpression in multiple myeloma revealed by fluorescence in situ hybridization and quantitative analysis of mRNA levels. Blood 2004;104:1120–1126. [DOI] [PubMed] [Google Scholar]
- 18.Metcalf RA, Zhao S, Anderson MW, et al. Characterization of D-cyclin proteins in hematolymphoid neoplasms: lack of specificity of cyclin-D2 and D3 expression in lymphoma subtypes. Mod Pathol 2010;23:420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ►19.Liu H, Wang J, Epner EM. Cyclin D1 activation in B-cell malignancy: association with changes in histone acetylation, DNA methylation, and RNA polymerase II binding to both promoter and distal sequences. Blood 2004;104:2505–2513. [DOI] [PubMed] [Google Scholar]
- 20.Camacho FI, Garcia JF, Cigudosa JC, et al. Aberrant Bcl6 protein expression in mantle cell lymphoma. Am J Surg Pathol 2004;28:1051–1056. [DOI] [PubMed] [Google Scholar]
- 21.Gualco G, Weiss LM, Harrington WJ Jr, Bacchi CE. BCL6, MUM1, and CD10 expression in mantle cell lymphoma. Appl Immunohistochem Mol Morphol 2010;18:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao DJ, Thakur A, Wu J, Biliran H, Sarkar FH. Perspectives on c-Myc, cyclin D1, and their interaction in cancer formation, progression, and response to chemotherapy. Crit Rev Oncog 2007;13:93–158. [DOI] [PubMed] [Google Scholar]
- 23.Baitei EY, Zou M, Al-Mohanna F, et al. Aberrant BRAF splicing as an alternative mechanism for oncogenic B-Raf activation in thyroid carcinoma. J Pathol 2009;217:707–715. [DOI] [PubMed] [Google Scholar]
- 24.Intini D, Agnelli L, Ciceri G, et al. Relevance of Ras gene mutations in the context of the molecular heterogeneity of multiple myeloma. Hematol Oncol 2007;25:6–10. [DOI] [PubMed] [Google Scholar]
- 25.O’Malley DP, Vance GH, Orazi A. Chronic lymphocytic leukemia/small lymphocytic lymphoma with trisomy 12 and focal cyclin d1 expression: a potential diagnostic pitfall. Arch Pathol Lab Med 2005;129:92–95. [DOI] [PubMed] [Google Scholar]