SUMMARY
Long-term epigenetic reprogramming of innate immune cells in response to microbes, also termed “trained immunity,” causes prolonged altered cellular functionality to protect from secondary infections. Here, we investigated whether sterile triggers of inflammation induce trained immunity and thereby influence innate immune responses. Western diet (WD) feeding of Ldlr−/− mice induced systemic inflammation, which was undetectable in serum soon after mice were shifted back to a chow diet (CD). In contrast, myeloid cell responses toward innate stimuli remained broadly augmented. WD-induced transcriptomic and epigenomic reprogramming of myeloid progenitor cells led to increased proliferation and enhanced innate immune responses. Quantitative trait locus (QTL) analysis in human monocytes trained with oxidized low-density lipoprotein (oxLDL) and stimulated with lipopolysaccharide (LPS) suggested inflammasome-mediated trained immunity. Consistently, Nlrp3−/−/Ldlr−/− mice lacked WD-induced systemic inflammation, myeloid progenitor proliferation, andre-programming. Hence, NLRP3 mediates trained immunity following WD and could thereby mediate the potentially deleterious effects of trained immunity in inflammatory diseases.
In Brief
Systemic inflammation induced by a Western diet is largely blunted by dietary changes, but myeloid cell-induced innate immune responses remain augmented and could potentially contribute to inflammatory disease.
INTRODUCTION
A major shift in the burden of infectious diseases has occurred over the last two centuries. Whereas approximately half of all deaths were accounted for by microbial infections in the late 19th century, today this burden has dropped to 15% (GBD 2015 Mortality and Causes of Death Collaborators, 2016). Three prominent changes driving this change include: (1) widespread sanitation improvement in the 19th century, (2) continued introduction and larger deployment of vaccines in this period, and (3) deployment of antibiotics. However, as infectious disease burden has dropped predominantly in the last century, the development of chronic non-communicable diseases has dramatically increased. Today, in Western societies, over 80% of deaths are due to non-communicable diseases including those associated with aging and diseases caused or influenced by the consumption of Western-type calorically rich diets, such as type II diabetes, obesity, and cardiovascular diseases.
For atherosclerosis development, increased circulating levels of LDL cholesterol have been linked mechanistically and genetically to clinical event risks (Mega et al., 2015). However, it is also well appreciated that disease progression is strongly associated with inflammatory processes involving cells of the innate immune system, mainly monocyte-derived macrophages (Hansson and Hermansson, 2011; Ridker et al., 2017). Hence, it is of fundamental and translational importance to understand explicitly the mechanisms that link the consumption of calorically rich diets to increased inflammation.
To respond to invading pathogens, vertebrates have evolved innate and adaptive immune systems. Contrary to the adaptive immunity arm, which can induce antigen-specific memory formation upon pathogen encounter, the innate immunity arm quickly mounts non-antigen-specific protective responses toward pathogens. Protective anti-pathogenic innate immune responses are evoked by the activation of a series of innate immune signaling receptors including among others, Toll-like receptors (TLRs) and the nucleotide-binding oligomerization (NOD)-like receptors (NLRs) (Cao, 2016). However, these receptors can further recognize “sterile” danger signals, which are thought to trigger inflammation in non-communicable diseases (Zimmer et al., 2015). These observations link activation of innate immune receptors with the development of cardiovascular diseases.
Emerging evidence has indicated that the vertebrate innate immune system has evolved elaborate adaptive mechanisms allowing them to respond more vigorously to future infections. This type of functionally adapted response after an initial trigger known as “innate immune memory” or “trained immunity” is mediated by epigenetic and metabolic reprogramming that can last for prolonged periods of time (Netea et al., 2011, 2016). Innate immune memory has likely evolved to provide non-specific protection from secondary infections, and most studies that characterized innate immune memory effects have focused on the ability of microbial triggers to induce cellular reprogramming. However, it is also conceivable that “sterile” inflammatory triggers, such as Western diet (WD), can induce trained immunity. In such a scenario, a secondary stimulus would then trigger altered and potentially pathological immune responses. Indeed, recent cell culture data suggest that oxidized low-density lipoprotein (oxLDL) particles, which are known to trigger innate immune responses, can induce memory responses via epigenetic modification of human monocytes (Bekkering et al., 2014).
Here, we have investigated whether WD feeding triggers trained immunity in the Ldlr−/− atherosclerosis mouse model. We show that WD induced systemic inflammation that subsided after shifting mice to chow diet (CD). Furthermore, WD triggered a proliferative hematopoietic cell expansion associated with functionally reprogrammed granulocyte monocyte precursor cells (GMPs). These responses were maintained over prolonged times after reversing the diet from WD to CD indicating that WD can induce trained immunity. Mechanistically, we identified the NLRP3 inflammasome as the central receptor, which mediates WD-induced systemic inflammation and myeloid precursor reprogramming, opening therapeutic opportunities to interfere with WD-associated pathologies.
RESULTS
WD Induces Systemic Inflammation and Functional Reprogramming of Myeloid Cells
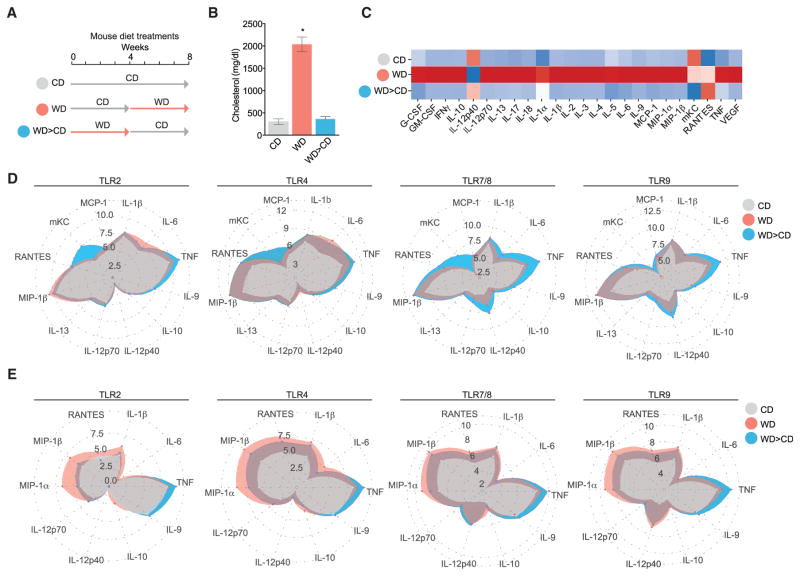
Given the inflammatory nature of a WD, we sought to test whether WD feeding in an experimental model of atherosclerosis induces innate immune memory. We chose to study the effects of WD after 4 weeks feeding since at this early time point only minimal atherosclerotic lesions can be observed (Duewell et al., 2010), and thus a contribution of plaque-derived immune cells and potential anti-inflammatory effects during plaque regression is minimized. Ldlr−/− mice were fed a regular CD, a WD for 4 weeks, or first WD for 4 weeks followed by 4 weeks CD (Figure 1A). Circulating cholesterol peaked by 4 weeks and returned to baseline in mice that were shifted back to CD after WD feeding (Figures 1B and S1A). Likewise, circulating levels of growth factors, cytokines, or chemokines, as well as acute phase proteins, which were all elevated after 4 weeks WD feeding, returned to baseline in mice fed the WD for 4 weeks followed by 4 weeks CD (Figures 1C and S1C; Table S1). Mice had minimal, non-significant weight gain over the course of 4 weeks WD and no significant weight loss was observed after changing mice back to CD for 4 weeks following the 4 weeks WD (Figure S1B). Together, these studies showed that WD feeding induces a transient hypercholesterolemia concomitant with a systemic inflammatory response. To test whether the WD provoked functional immune cell reprogramming, we isolated bone marrow cells and splenic CD11b+ monocytic cells from mice fed either regular CD, 4 weeks WD, or 4 weeks WD followed by 4 weeks CD. These cells were stimulated ex vivo with a panel of TLR ligands (LPS, PGN, R848, and CpG) and secretion of cytokines and chemokines was analyzed. Immune cells isolated from WD-fed mice had significantly enhanced TLR responses indicating a primed cell state. Intriguingly, even though the systemic cytokines had normalized upon shifting mice back to the CD after WD, the TLR responses of cells isolated from the WD-fed and then CD-rested mice remained augmented when compared to cells isolated from CD-fed mice. Of note, in addition to the quantitative changes of cytokine responses, we found qualitative changes. For example, bone marrow cells isolated from WD-fed mice that were then CD-rested had even stronger chemokine (C-X-C motif) ligand 1 (CXCL1) and tumor necrosis factor (TNF) responses, yet displayed decreased IL-6 responses when compared to cells isolated from mice on WD (Figures 1D, 1E, S1D, and S1E). These results suggest that WD feeding induced a complex myeloid cell reprogramming leading to long-lasting and qualitatively altered hyper-responsiveness even after resting mice from WD feeding.
Figure 1. WD Feeding Induces Systemic Inflammation and Functional Reprogramming.
(A) Schematic of dietary interventions. Female Ldlr−/− mice were fed either CD, WD for 4 weeks, or WD for 4 weeks followed by CD for 4 weeks (WD > CD).
(B) Systemic serum cholesterol in response to dietary intervention in Ldlr−/− mice.
(C) Heatmap representing normalized serum cytokine levels from mice fed as indicated.
(D and E) Bone marrow cells (D) or splenic CD11b+ monocytes (E), isolated from Ldlr−/− mice following dietary intervention treated ex vivo with vehicle or different TLR stimuli for 6 hr. Log 2 transformed data represented as spider plots for the following stimulations: Pam3Csk4, LPS, R848, and CpG.
For (B) n = 6–10 animals; for (C) n = 3–5 animals per group; ± SEM, p < 0.05 versus CD (B and C); versus un-stimulated cells (D and E). Experiments were performed twice independently and data are representative of a single experiment.
WD Triggers Myelopoiesis and Transcriptional Reprogramming of Myeloid Precursor Cells
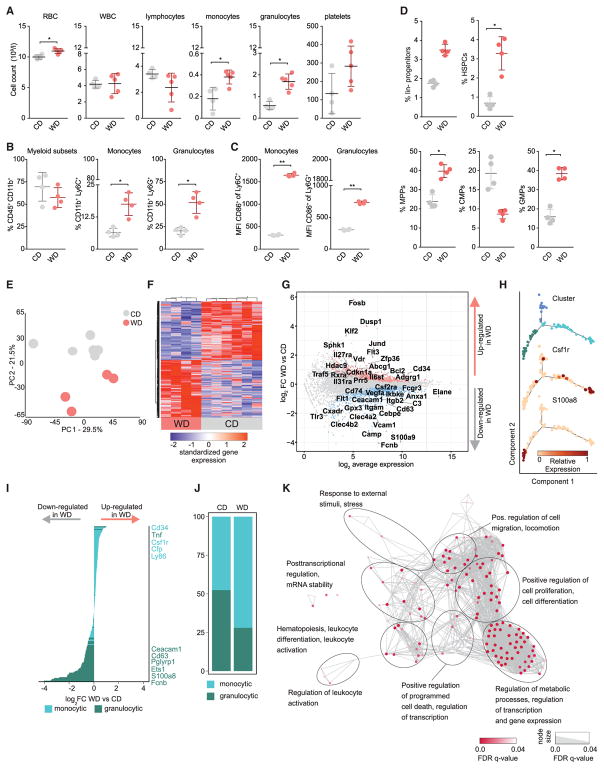
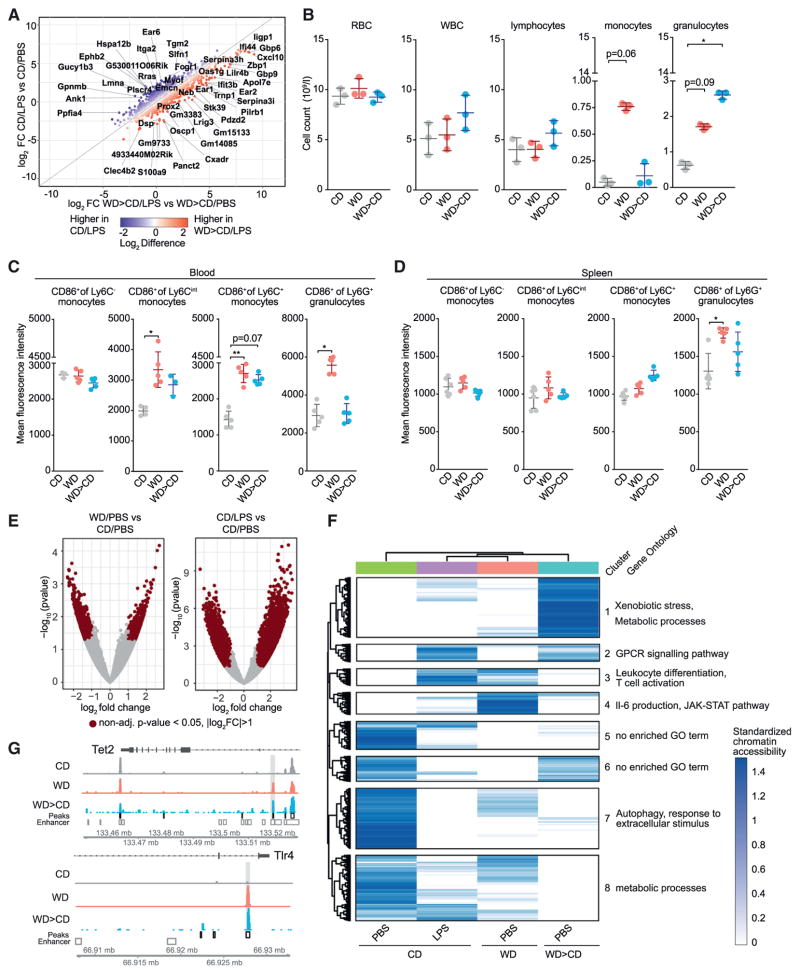
We next determined the effect of WD on circulating blood cell populations (Figure S2A). Absolute numbers of circulating red blood cells (RBCs) as well as myeloid cell subsets, including monocytes and granulocytes, were markedly increased after WD feeding (Figures 2A and 2B). Additionally, WD feeding induced an increased activation status in circulating myeloid subsets, as indicated by CD86 surface expression (Figures 2C and S3A). Splenic inflammatory monocyte and granulocyte numbers were significantly increased as well (Figure S2B), although CD86 surface expression remained unaltered (Figures S2C and S3A).
Figure 2. WD Induces Hematopoiesis and Transcriptional Reprogramming of GMPs.
(A) Total counts of the indicated blood cell populations in CD- or WD-fed (4 weeks) female Ldlr−/− mice.
(B and C) Relative numbers (%) (B) and activation status (C) of circulating myeloid subsets isolated from female Ldlr−/− mice.
(D) Percentage of hematopoietic precursor cells as indicated in female Ldlr−/− mice fed either CD or WD (4 weeks).
(E) PCA of RNA-seq data of GMPs isolated from CD- or WD-fed mice. PCA is based on variable genes (non-adjusted [adj.] p value < 0.05, n = 4,672).
(F) Gene and sample wise hierarchical clustering based on the 1,000 genes with the highest variance within the dataset of GMPs purified from WD- or CD-fed mice. Gene expression values are Z score standardized.
(G) MA-plot showing DE genes in GMPs of WD- or CD-fed mice. DE genes (|FC| > 1.5, non-adj. p value < 0.05) are colored in red (upregulated in WD) and blue (downregulated in WD) and notable genes are highlighted.
(H) Trajectory analysis of single-cell RNA-seq data (GEO: GSE70235 and GEO: GSE70240) with computational clustering (top) representing the expression of Csf1R or S100A8, overlaid onto the developmental trajectory to identify monocytic or granulocytic lineage determination. Cells of the monocytic (turquoise) and granulocytic (dark green) branches were used to determine signature genes to test for lineage potential in (I) and (J).
(I) Expression differences of monocytic (turquoise) and granulocytic (dark green) signature genes in GMPs from mice fed as indicated.
(J) Enrichment of monocytic and granulocytic signatures in GMPs isolated from CD- and WD-fed mice and computationally inferred by linear support vector regression analysis. Enrichments are significant (non-adj. p value < 0.001).
(K) GO term enrichment network analysis of differentially expressed genes from GMPs isolated from WD- and CD-fed mice. Each dot represents a significantly enriched GO term and connections indicate shared genes between GO terms. Significance (false discovery rate [FDR]) is indicated by color (lower FDR: more intense color) and size (lower FDR: bigger nodes/thicker borders) of nodes (upregulated genes) or borders (downregulated genes); ± SEM, p < 0.05 versus CD; for (A)–(D) n = 3–5 animals per group. Experiments were performed twice independently and data are representative of a single experiment.
See also Figure S2.
To test whether the observed changes in specific leukocyte subsets in the blood were also apparent at the bone marrow level, we determined the quantities of hematopoietic precursor subsets by comparing WD- to CD-fed mice. We found that the abundance of hematopoietic stem cell progenitors (HSPCs), multipotent progenitor cells (MPPs), as well as granulocyte-monocyte progenitor cells (GMPs) were all significantly increased after WD (Figure 2D). To better understand the mechanisms whereby WD induces myelopoiesis and functional reprogramming of myeloid cells, we next isolated myeloid progenitor subsets from the bone marrow compartment by fluorescence-activated cell sorting (FACS) (Figure S2D). As GMPs are the most differentiated myeloid progenitor subsets that give rise to monocytes and granulocytes, we decided to study these cells by an unbiased approach and performed transcriptional RNA profiling by RNA sequencing (RNA-seq) and subsequent computational analysis (Figure S2E).
Principal component analysis (Figure 2E) and unsupervised hierarchical clustering of the 1,000 most variable genes (Figure 2F) demonstrated that GMPs globally reprogram transcriptional responses after WD. Analysis of the most changed genes (Figure 2G) suggested that WD induced upregulation of genes involved in cell proliferation, including KLF2, DUSP1, CDKN1A, CCND1, SOX4, SMO, BCL2, and a skewing of GMP development toward the monocytic cell lineage (JUND, FOSB, FLT3, and HDAC9). To test for a WD-induced lineage bias in GMPs, we first identified specific gene signatures for monocytic or granulocytic lineage-commitment by differential expression (DE) gene analysis between the monocytic and granulocytic branches of the GMP developmental trajectory (Figure 2H) generated from publicly available single-cell RNA-seq datasets (GEO: GSE70235 and GEO: GSE70240). Monocytic signature genes were highly enriched (CD34, CSF1R, CFP, and LY86), while most of the granulocytic signature genes were downregulated (S100a8 and S100a9, ETS1, PGLYRP1, CD63, and CEA-CAM1) in GMPs isolated from WD-fed as compared to CD-fed mice (Figure 2I). These data were in accordance with a linear support vector regression analysis indicating an increase in monocytic lineage potential after WD (Figure 2J). We next analyzed which Gene Ontology (GO) terms and KEGG pathways were enriched in the DE genes after WD (Figure S2F). GO enrichment analysis using the genes that were upregulated after WD, was further visualized as a biological GO enrichment network (Figure 2K). Together, these analyses identified that genes upregulated in GMPs from WD-fed mice were associated with regulatory processes involved in hematopoiesis, cell proliferation, programmed cell death, metabolic processes, and cell migration, as well as immune cell differentiation, leukocyte activation, and immune processes involved in cellular stress responses and WNT receptor signaling (i.e., hedgehog signaling pathway, interleuking [IL]-17 signaling pathway/Th17 differentiation, JAK-STAT signaling pathway, viral carcinogensis). Furthermore, transcription factor (TF) binding prediction analysis suggested that the WD-induced transcriptional skewing of GMPs into the monocyte direction was influenced by the transcription factors GATA1-3, KLF4, and TCF7, certain FOS, JUN, and ATF family members, as well as CREB1 and EGR1 (functional involvement in induction of cell reprogramming, survival, and monocytic differentiation; Figure S2G). Together, these data indicate that the bone marrow compartment strongly responds to WD feeding with an increased myelopoiesis and a deep transcriptional reprogramming of myeloid precursor cells.
WD Causes Functional Reprogramming of Myeloid Precursor Cells
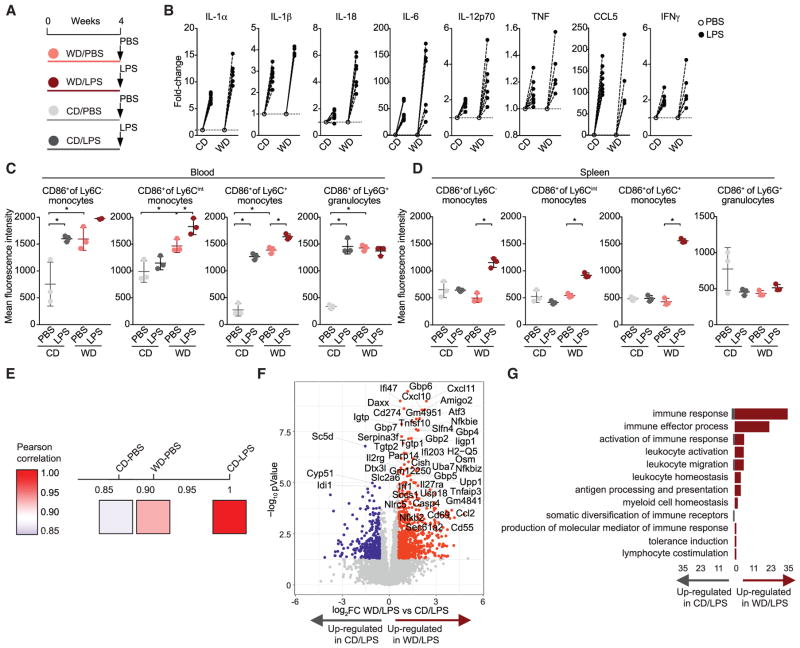
Given that WD feeding induced a global transcriptional reprogramming of GMPs, we next asked whether the immune responses toward a model ligand (i.e., LPS) were altered in vivo. We fed Ldlr−/− mice the CD or WD for 4 weeks and intravenously injected LPS (10 μg) or vehicle control (PBS) 6 hr before sacrifice and cell harvest (Figure 3A). LPS treatment of WD-fed mice resulted in higher serum levels of most inflammatory cytokines and chemokines when compared to LPS-treated mice fed CD (Figure 3B). Furthermore, the activation status of circulating and splenic monocyte subsets as measured by CD86 surface expression was amplified in LPS-challenged WD-fed mice when compared to LPS-challenged CD-fed mice (Figures 3C and 3D). In contrast, CD86 surface expression in circulating and splenic granulocytes remained unchanged upon LPS treatment in WD-fed mice (Figures 3C and 3D). These data suggest that WD feeding primes the immune system for systemic and local innate immune responses toward an inflammatory trigger.
Figure 3. WD Primes for Inflammatory Responses to LPS.
(A) Schematic representation of the WD and LPS manipulations. Female Ldlr−/− mice were fed CD or WD for 4 weeks. 6 hr prior to sacrifice mice were intravenously challenged by LPS or PBS.
(B) Serum cytokine levels in LPS-treated CD- or WD-fed mice were normalized to levels in PBS control groups and represented as fold-change.
(C and D) Activation status of circulating (C) and splenic (D) myeloid subsets from female Ldlr−/− mice treated as indicated.
(E) Pearson correlation analysis of top 1,000 DE genes of transcriptomes from GMPs isolated from mice treated as indicated.
(F) Volcano plot indicating transcriptomic changes between CD/LPS and WD/LPS. Significantly upregulated (FC > 1.5, FDR-adj. p value < 0.05, red) and downregulated (FC < −1.5, FDR-adj. p value < 0.05, blue) genes are shown and the most significantly regulated genes are highlighted.
(G) Number of significantly upregulated (red) and downregulated (gray) GMP genes within immune system associated GO terms in WD/LPS compared to CD-/ LPS-treated mice. n = 3–8 animals per group in (B); n = 3 animals in (C) and (D); ±SEM, p < 0.05 versus CD (B)–(D). Experiments were performed twice independently and data are representative of a single experiment.
See also Figure S3.
To better define how WD-induced reprogramming of myeloid precursor cells influences the responses toward LPS, we next performed RNA-seq analysis of sorted GMPs from mice that received the WD or CD and were exposed to an LPS or PBS challenge 6 hr prior to sacrifice. Pearson correlation matrix analysis of the transcriptional changes in the different treatment conditions showed that WD treatment mimicked the effects of LPS in CD-fed mice, and LPS and WD treatment had synergistic effects on gene transcription (Figure 3E), which was also evident when the gene expression differences are represented as a Volcano plot (Figure 3F). Detailed inspection of the most highly differentially expressed genes revealed that many type I interferon (IFN) response genes, including IRG1, CCL5, CXCL10, GBP4, GBP6, IRF1, and IFI203, were expressed more strongly in WD-fed mice receiving LPS compared to CD-fed mice receiving LPS. Furthermore, functional annotation enrichment analysis for GO terms and KEGG pathways demonstrated that the WD primed GMPs for LPS responses and led to a shift toward increased inflammatory signaling (Figure 3G). As expected, WD feeding was associated with a downregulation of cholesterol biosynthesis pathways; however innate immune defense responses, and in particular anti-viral responses, were greatly enriched (Figures S3B–S3E). These data demonstrate that WD feeding results in GMP priming that triggers enhanced innate immune responses toward LPS.
Long-Lasting Reprogramming of GMPs following WD
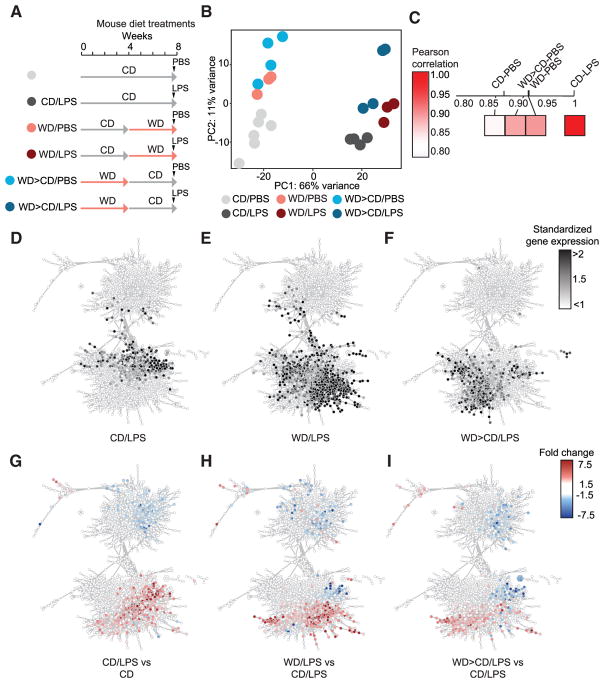
We observed that a 4-week CD resting period normalized the WD-induced systemic cytokine response in the circulation, whereas the innate immune responses of myeloid cells remained enhanced and qualitatively altered (Figures 1C–1E). We next thought to decipher to what extend the transcriptional changes in GMPs induced by WD persisted and how this affected the GMP responsiveness toward LPS. Mice fed CD, 4 weeks WD, or 4 weeks WD followed by 4 weeks CD were intravenously challenged with LPS or PBS 6 hr prior to cell isolation, and then RNA-seq analysis of purified GMPs was performed (Figures 4A and S4A). Principal component analysis (Figure 4B) and Pearson correlation matrix analysis (Figure 4C) demonstrated that the transcriptomic reprogramming of GMPs after WD did not reverse in GMPs isolated from mice that had been switched to CD feeding for 4 weeks after WD feeding. A detailed assessment of genes that were regulated by the different dietary interventions demonstrated that certain gene sets were commonly expressed in GMPs isolated from WD as well as from WD-fed mice that were rested on CD (Figure S4B). Interestingly, other sets of genes were only expressed in GMPs isolated from WD-fed mice or only in GMPs isolated from WD-fed mice that were rested on CD. We therefore analyzed which GO terms were commonly or selectively enriched in GMPs isolated from mice subjected to the different dietary interventions (Figure S4B). We found genes that were commonly upregulated in GMPs from both feeding groups were associated with immune processes and lymphocyte activation (Figure S4B). This shows that even after resting mice from WD, GMPs remain in an activated state and keep a persistent transcriptionally remodeled state that is qualitatively distinct from that of cells isolated from CD-fed mice.
Figure 4. WD Induces Long-Lasting Reprogramming in GMPs.
(A) Schematic of diet and LPS manipulations.
(B) PCA of genes with highest variance (non-adj. p value < 0.05, n = 4672).
(C) Pearson correlation analysis of top 1,000 DE genes of transcriptomes from GMPs isolated from mice treated as indicated.
(D–I) Co-expression network analysis (genes = 4,360, correlation >0.85) based on highly correlated genes among the 11,306 expressed genes. To allow identification of specific gene signatures, Z score transformed average gene expression (D–F) or fold changes in gene expression (G–I) of respective conditions were overlaid onto the co-expression network.
See also Figure S4.
Next, we aimed to better define the effects of the different dietary interventions on the functional responses of GMPs to in vivo LPS challenge. To visualize changes in the transcriptional programs that occur after LPS challenge in mice fed CD, 4 weeks WD, or 4 weeks WD followed by 4 weeks CD, we performed a co-expression network analysis of all genes with changed expression and visualized the changes as standardized gene expression (Figures 4D–4F) or fold changes (Figures 4G–4I). This analysis revealed that LPS challenge induced the expression of many more gene clusters in mice fed a WD than CD (Figures 4D, 4E, 4G, and 4H) and that the LPS response remained altered in GMPs in mice shifted back to CD after WD feeding (Figures 4F and 4I). We found that numerous of the LPS-regulated genes under CD feeding (Figure 4G) were more strongly upregulated or in some cases even counter-regulated after WD feeding (Figure 4H). Of note, the LPS-induced gene expression in GMPs isolated from mice that had been switched to CD after WD feeding remained altered and largely similar to the WD only conditions (Figure 4I). We additionally performed TF motif enrichment analysis for DE genes in GMPs isolated from mice subjected to the different dietary and LPS challenges. Genes that were higher expressed upon LPS challenge in WD-fed compared to CD-fed mice exhibited increased abundance for TF binding sites such as RELA and B, NF-kB1 and 2, STAT1-4, 5A, and B and 6, IKZF1 and 2, as well as IRF1, 3, 7, 8, and 9, JUN and FOS, and KDM4A,B,C,D in the regulatory regions (Figures S4C–S4E). In addition, assessment of TF binding prediction analysis for genes expressed in GMPs that were not regulated upon LPS treatment in CD-fed mice, but upregulated upon WD and LPS treatment revealed a defined set of transcriptional regulators including NRF1, YY1 and YY2, STAT1 and 2, IRF1, 3, 5, 7, 8, and 9, GATA1-3, JUN/FOS, and MEF2A, C, and D (Figure S4D). Notably, TFs that were the most enriched in GMPs from mice treated with WD and LPS remained partly enriched in GMPs from mice fed 4 weeks WD followed by 4 weeks CD and treated with LPS, including among others, YY1 and 2, NRF1/2, MEF2A, C, and D, and ERG (Figure S4E).
Moreover, fold-change analysis of the most strongly LPS-induced genes revealed a preservation of the distinctive type I IFN signature in GMPs (for example GBP6, CXCL10, IFI44, and IFIT3b) from the WD-fed and CD-rested mice compared to those mice fed only CD (Figure 5A). Whereas the marked increase in circulating monocyte quantities seen after WD feeding was not maintained in CD-rested mice (Figure 5B), the quantity of circulating granulocytes was found to be further increased in CD-rested mice compared to WD-fed mice. Additionally, splenic granulocytes displayed a more activated phenotype upon WD feeding and tended to remain more activated in WD- and CD-rested mice compared to CD-fed mice (Figure 5D).
Figure 5. WD Induces Epigenetic Reprogramming of GMPs.
(A) FC-FC plot comparing the LPS response of GMPs with amplitude of difference in LPS responses visualized as color code (red, upregulated genes; blue, downregulated genes).
(B) Total counts of the indicated blood cell populations in female mice fed as shown.
(C and D) Activation status of circulating (C) and splenic (D) myeloid subsets from mice treated as indicated.
(E) Volcano plot displaying open chromatin loci as determined by ATAC-seq in GMPs isolated from mice treated as indicated. Average signal is represented as log2 fold change. Significantly up- (non-adj. p value < 0.05, log2FC > 1) and downregulated (non-adj. p value < 0.05, log2FC < −1) peaks are shown.
(F) Hierarchical clustering, standardized, and visualized as a heatmap show the significant differentially accessible genomic loci (p value < 0.05) in GMPs isolated from mice treated as indicated.
(G) Coverage of ATAC-seq signal for TET2 (top) and TLR4 loci (bottom). n = 3–5 for groups in (B)–(D); ±SEM, p < 0.05 versus CD. Experiments were performed twice independently and data are representative of a single experiment.
See also Figure S4.
To test whether WD changes the overall accessibility of chromatin, we performed global epigenetic profiling of open chromatin by assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq) in GMPs isolated from 4 weeks WD- or CD-fed mice (Figures 5E and 5F). While WD clearly induced changes in the chromatin landscape, these changes were discrete in comparison to LPS stimulation (Figure 5E). Next, differentially regulated peaks were clustered and presented as a heatmap (Figure 5F). We identified 8 clusters of peaks with some genomic loci showing similar chromatin status in GMPs derived from WD-fed or LPS-treated mice but differing from GMPs from CD-fed control mice (Figure 5F, cluster 3). Genes associated with this cluster were linked to GO-terms such as “leukocyte differentiation” or “T cell activation.” Furthermore, peaks particularly enriched in GMPs from WD-fed mice (cluster 4) were associated with genes linked to “IL-6 production” and the “JAK/STAT pathway.” Of interest, enhancer regions including Tet2 and Tlr4 were found to be more open in both GMPs isolated from WD-fed mice and WD followed by CD-fed mice (p < 0.05, |FC| > 2; Figure 5G). In contrast, gene loci for oxysterol-binding protein-related protein 3 (Osbpl3) and Abca1 (p < 0.05, |FC| > 2) were more closed in GMPs isolated from mice fed WD, but reopened in mice fed WD followed by CD and were thus comparable to the CD-fed mice (Figure S4F). These data indicate that WD can effectively reprogram GMPs both on the transcriptional and epigenetic level. Importantly, the broad transcriptional changes in GMPs from LPS-injected mice fed WD and then CD for 4 weeks more closely resembled the altered responses seen in GMPs isolated from WD-fed and LPS-challenged mice suggesting persistence of a long-lasting reprogrammed state after removal of the WD insult.
The IL-1 Pathway Is Part of WD-Induced GMP Reprogramming
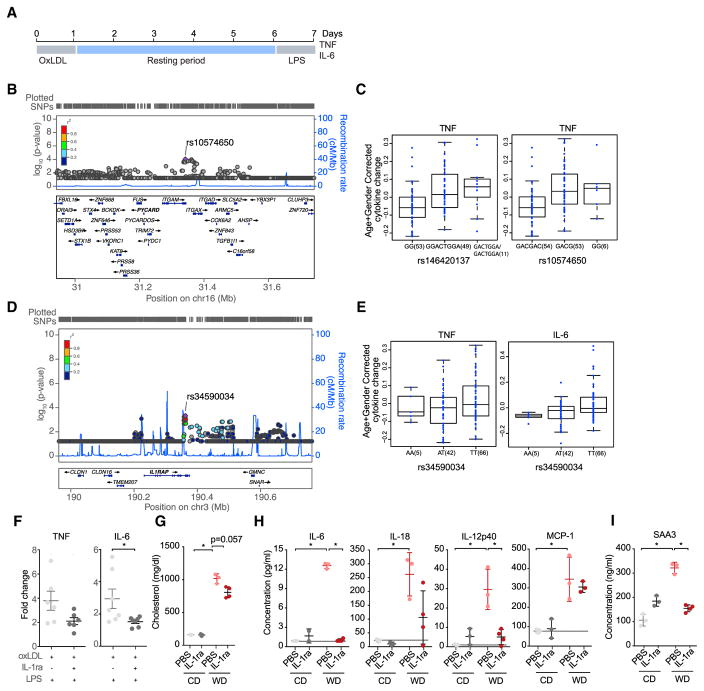
The training effect we observed in our murine model might be further modulated by genetic variation in major pathways involved. To determine such genetic effects and identify those pathways, we performed a functional trained immunity (FTI)-QTL study in human monocytes. We used the pro-inflammatory oxidized form of LDL (oxLDL) that has been associated with WD and is known to induce trained immunity in human monocytes (Bekkering et al., 2014). We subjected adherent monocytes to oxLDL or control, rested cells after a wash step for 5 days, and re-stimulated with LPS using monocytes from 122 healthy individuals (Figure 6A) from the 200 FG cohort (Li et al., 2016). Training of monocytes with oxLDL resulted in an increased responsiveness to LPS re-stimulation, compared to cells pre-incubated with culture medium. The FTI-QTL analysis identified several single nucleotide polymorphisms (SNPs) in the putative regulatory gene regions of (1) PYCARD, the gene encoding the inflammasome adaptor ASC, and (2) IL1RAP, the gene encoding the IL-1 receptor antagonist (IL-1ra). Importantly, several of these genetic variants showed a significant effect on the capacity of oxLDL to induce trained immunity (Figures 6B and 6D: Manhattan plots representing clustered reference SNPs [rs] in the proximity of the gene coding regions for PYCARD or IL1RAP), assessed by the diminished release of TNF (p < 10−4, linear regression model; Figures 6C and 6E) or IL-6 (p < 10−4, linear regression model; Figure 6E). A role of IL-1ra in oxLDL-mediated innate immune training could be confirmed in an independent cohort of humans. Addition of recombinant IL-1ra during the oxLDL-induced training period resulted in lower responses upon LPS challenge after the training period, as analyzed by TNF and IL-6 levels in the culture supernatants (Figure 6F). Given the important role of inflammasomes in regulating IL-1, and the suggestion by the companion papers that IL-1 is involved in cellular reprogramming, we further tested the effects of IL-1 blockage on the systemic WD-induced inflammatory response in vivo. We subjected Ldlr−/− mice to recombinant IL-1ra or PBS vehicle control during a 4-week course of CD or WD feeding. There was a trend towards decreased serum cholesterol levels in IL-1ra treated mice (Figure 6G). Additionally, IL-1 blockade reduced the WD-induced systemic inflammation, demonstrated by the decrease of inflammatory cytokines (Figure 6H), as well as the acute phase protein SAA3 (Figure 6I). Together, these data provide evidence for the importance of the inflammasome and the downstream IL-1R signaling pathway for the induction of trained immunity by oxLDL in vitro or by WD in vivo.
Figure 6. PYCARD and IL-1RAP SNPs Influence oxLDL-Induced Training Effects.
(A) Schematic overview of the in vitro FTI-QTL protocol.
(B–D) Manhattan plots representing reference single nucleotide polymorphisms (rs) in the PYCARD gene locus (B) or in the IL1RAP gene locus (D), (C) TNF production capacity in different genotyping groups from (B).
(E) TNF and IL-6 production capacity between different genotyping groups from (D).
(F) Monocyte oxLDL training was performed in presence or absence of recombinant IL-1ra followed by LPS stimulation. Shift in cytokine levels represented as fold-change (oxLDL/+/− IL-1ra).
(G–I) Systemic serum cholesterol levels (G), cytokines (H), and acute phase response (I) in Ldlr−/− mice treated as indicated. n = 3–4 mice per group (G–I); means ± SEM, p < 0.05 versus non-IL-1ra treatment (RPMI only) (F); versus CD and PBS treatment (G–I).
The NLRP3 Inflammasome Is Required for Sensing WD
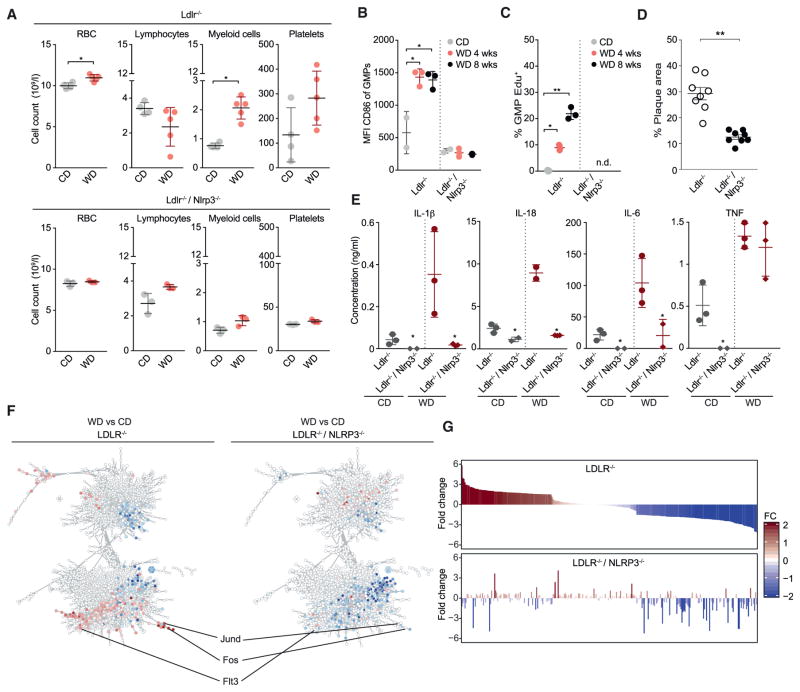
Previous work has identified that oxLDL is capable of activating the NLRP3 inflammasome, and NLRP3 activation has been shown to be part of the pathogenesis of atherosclerosis (Duewell et al., 2010; Sheedy et al., 2013). We hypothesized that triggering of the NLRP3 inflammasome by pro-inflammatory factors arising after WD could contribute to trained immunity, as observed by the hematopoietic response in mice. We therefore generated Nlrp3−/−/Ldlr−/− mice and analyzed their systemic and cellular responses to WD. Strikingly, WD did not induce peripheral monocytosis in Nlrp3−/−/Ldlr−/− mice compared to WD-fed Ldlr−/− mice (Figure 7A). WD furthermore failed to induce GMP activation as assessed by CD86 surface expression (Figure 7B), and did not induce increased GMP proliferation in Nlrp3−/−/Ldlr−/− mice (Figure 7C). Additionally, the WD-induced augmentation of the LPS-induced systemic inflammatory cytokine responses observed in Ldlr−/− were largely blunted in Nlrp3−/−/Ldlr−/− mice, especially in inflammasome-dependent cytokines (Figure 7E). Transcriptional profiling of GMPs isolated from WD- or CD-fed Ldlr−/− or Nlrp3−/−/Ldlr−/− mice combined with computational network analysis (Figure 7F) and rank plot visualization (Figure 7G) demonstrated that the WD-induced transcriptional reprogramming of GMPs was mostly dependent on NLRP3. In agreement with the lack of inflammatory effects of WD, Nlrp3−/−/Ldlr−/− mice showed markedly reduced atherosclerotic plaque size after 8 weeks of WD feeding (Figure 7D).
Figure 7. NLRP3-Dependent Myeloid Progenitor Priming.
(A) Total counts of circulating cell subsets from female Ldlr−/− and Nlrp3−/−/Ldlr−/− mice treated as indicated.
(B and C) Activation status (B) and proliferative capacity (C) of GMPs isolated from Ldlr−/− or Nlrp3−/−/Ldlr−/− mice fed as indicated.
(D) Atherosclerotic plaque lesion size in WD-fed (8 weeks) female Ldlr−/− or Nlrp3−/−/Ldlr−/− mice.
(E) Serum cytokine response 6 hr post LPS injection in female Ldlr−/− and Nlrp3−/−/Ldlr−/− mice fed as indicated.
(F) Co-expression network analysis of transcriptional changes induced by WD in female Ldlr−/− and Nlrp3−/−/Ldlr−/− mice. Fold changes are overlaid onto co-expression networks.
(G) Rank plot visualization of fold changes of genes represented in (F). n = 2–5 for groups in (A)–(C) and (E) (experiments were performed twice independently, and data are representative of a single experiment), n = 5–9 animals in (D); ±SEM, p < 0.05 versus CD (A–C and E) versus Ldlr−/− (D).
Together, our data strongly support a key function of the NLRP3 inflammasome in myeloid cell reprogramming in the context of WD feeding.
DISCUSSION
Genetic and lifestyle factors are key drivers of chronic non-communicable inflammatory diseases, which currently represent the vast majority of death burden in Western societies (Mega et al., 2015). Importantly, an unhealthy lifestyle, which includes factors such as noxious diets, little exercise and sleep, and unfavorable environmental triggers, can independently of the genetic risk strongly increase the rate of incident cardiovascular events and increase the susceptibility for other chronic inflammatory diseases (Khera et al., 2016). Therefore, it is of great importance to better define how these factors mechanistically influence inflammatory processes.
Alterations in diets can have deleterious effects on immune responses and drive the development of a range of inflammatory diseases (Thorburn et al., 2014). Yet, whether and how “inflammatory” diets can provoke trained immunity leading to potentially long-lasting alterations of immune responses was unknown. Early studies documented that WD feeding of rabbits induced enhanced cytokine production toward LPS stimulation in cells within the aortic tissue suggesting that WD could prime for augmented innate immune responses (Clinton et al., 1991; Fleet et al., 1992). Here, we have investigated whether WD consumption can trigger trained immunity and whether this results in modified secondary immune responses in a mouse model for atherosclerosis, because wild-type mice do not spontaneously develop atherosclerosis, unless serum cholesterol levels are raised experimentally by genetic deletion of LDLR or APOE. The Ldlr−/− mouse model is relevant to human atherosclerosis as patients with LDLR mutations are highly susceptible for the development of early atherosclerosis. In our studies we have focused on deciphering inflammatory effects of WD in the early, initiation phase of atherosclerosis before significant amounts of plaque have deposited in the vessel walls in Ldlr−/− mice. This minimized the potential contribution of plaque-derived immune cells and also putative changes that might occur due to plaque regression that can occur after changing the diet from WD to CD. Our studies revealed that while WD provoked only transient systemic inflammatory responses, it led to long-lasting alterations of myeloid cell responses toward different innate immune stimuli. These studies thus establish that not only microbes but also complex sterile inflammatory triggers, such as a WD, can induce trained immunity.
To better understand how WD induces trained immunity, and because other studies have linked hypercholesterolemia to reprogramming of hematopoietic stem cells (Murphy et al., 2011; Seijkens et al., 2014; van Kampen et al., 2014; Yvan-Charvet et al., 2010), we performed functional and transcriptional analyses of GMPs, the proximate precursors of circulating myeloid subsets. Notably, blood leukocyte counts and in particular the numbers of circulating monocytes were shown to be tightly associated with hypercholesterolemia and incidence of cardiovascular events (Friedman et al., 1974; Swirski and Nahrendorf, 2013). Furthermore, inflammatory blood monocytes that arise from activated hematopoietic precursors during WD feeding develop into disease promoting macrophages in atherosclerotic plaques and likely other organs (Swirski et al., 2007). Our studies revealed that WD induces GMP proliferation and skewing toward the development of activated and potentially harmful monocytes, which was accompanied by long-lived transcriptional and epigenetic reprogramming. While the ATAC sequencing results suggest that epigenetic changes are induced by WD feeding and might contribute to the transcriptomic reprogramming, other mechanisms of epigenetic regulation may also be induced. Indeed, opening of chromatin is only one aspect of the epigenetic processes that control transcription, and it is likely that there are further mechanisms that drive the complex long-term transcriptomic changes we have demonstrated. Nevertheless, our studies suggest that the immune system misinterprets a WD as a threat to the host as it sets in motion powerful anti-infectious mechanisms such as hematopoiesis and maintenance of a hyper-responsive state with the generation of myeloid cells that are programmed to respond to secondary inflammatory triggers more potently.
Recent studies support the idea that cell-fate decisions and a potential reprogramming already happens at the level of non-committed HSPCs, particularly under conditions of systemic stress induced by bacterial or viral infections (Boettcher and Manz, 2017) but also during hyperlipidemia and hyperglycemia (Brasacchio et al., 2009; Nagareddy et al., 2014; Singer et al., 2014). The work by the International Trained Immunity Consortium (INTRIM), reported in this issue of Cell now demonstrates that trained immunity can be induced on the level of HSPCs. Mitroulis et al. (2018 [this issue of Cell]) show that the trained immunity model ligand β-glucan induces metabolic and transcriptional rewiring in HSPCs through IL-1R signaling, which protects the hematopoietic system from chemotherapy-induced myeloablation.
By showing that WD induces a “memory” in myeloid progenitors our work raised the question of which mechanisms are involved in sensing the complex inflammatory trigger. LDL cholesterol overload, oxidation of LDL (oxLDL), as well as fatty acids can all induce a continuous state of intracellular stress and activate innate immune pathways (Dorrestein et al., 2014; Sheedy et al., 2013). Additionally, WD-induced microbial alterations could also drive for innate immune cell reprogramming, which could be tested using germ-free mice in future studies. However, the metabolic, neuroendocrine, and molecular control mechanisms that mediate the biological effects of WD are wide-ranging and their individual contributions are difficult to decipher in vivo. We thus performed a reductionist in vitro trained immunity QTL study in human monocytes to exploit human genetic variability as a discovery approach. Intriguingly, this analysis revealed that oxLDL-induced trained immunity was influenced by several independent genetic polymorphisms in the putative regulatory region of PYCARD, the gene encoding the principal inflammasome adaptor protein ASC, and also in the gene for the endogenous IL-1 inhibitory molecule IL-1RAP. Together with a large array of studies linking NLRP3 to the inflammatory response in metabolic diseases (Duewell et al., 2010; Rhoads et al., 2017; Sheedy et al., 2013), this provided evidence for a potential key role of the NLRP3 inflammasome pathway in the induction of trained immunity under conditions of WD feeding. Strongly supporting this hypothesis, we observed an almost complete protection from WD-induced changes in systemic inflammation, induction of hematopoiesis and functional and transcriptomic reprogramming of GMPs isolated from Ldlr−/−/NLRP3−/− mice. Future studies using cell-specific deletions will have to be performed to clarify the important question as to which cell-type activation of the inflammasome is required to induce the observed epigenetic reprogramming effects we have demonstrated.
Nevertheless, NLRP3 qualifies as a principal WD insult sensor that mediates most aspects of the WD-induced inflammation and trained immunity we have demonstrated in bone marrow precursor cells. In support of this hypothesis, patients with primary hypercholesterolemia reportedly have increased levels of inflammasome-dependent IL-18 and evidence for systemic inflammation (El Messal et al., 2006; Narverud et al., 2011). Additionally, cholesterol overload in hematopoietic progenitors could also lead to a sustained state of intracellular stress, linked to increased inflammatory signaling and a potential long-term reprogramming. Supportively, the studies by Bekkering et al. (2018 [this issue of Cell]) reported in this issue of Cell show that dysbalanced cholesterol biosynthesis with an accumulation of mevalonate can drive trained immunity induction. On the other hand, it appears plausible that NLRP3 could also instigate inflammatory pathologies toward a range of sterile danger signals not limited to WD, as numerous triggers of relevance in human chronic inflammatory pathologies have been linked to NLRP3 activation (Heneka et al., 2013; Hornung et al., 2008; Martinon et al., 2006).
Our study, together with work by Mitroulis et al. (2018 [this issue of Cell]) in this issue of Cell, implies that an inflammasome-mediated product, such as IL-1β, is likely the central endogenous mediator of the mechanisms resulting in the induction of trained immunity. Indeed, early work had already established that injections of IL-1β before experimental infection can prevent lethality from infections (van der Meer et al., 1988). Additionally, IL-1β, produced by activated adipose tissue macrophages in obesity, was shown to interact with the IL-1R expressed on CMPs, which stimulated the proliferation of GMPs (Nagareddy et al., 2014). Hence, it is likely that IL-1β is the key messenger generated by NLRP3 following WD-mediated activation. It is further tempting to speculate that these mechanisms are part of the clinical benefit observed in the recent CANTOS trial, which tested IL-1β blockade in humans at cardiovascular risk (Ridker et al., 2017).
Curiously, functional annotation analysis of the transcriptomic changes demonstrated that GMPs responded in a fashion that shares features of a viral infection characterized by the appearance of a strong type I IFN signature. It would be interesting to better understand the complex interplay between type I IFNs signaling and the IL-1β signaling after WD exposure, given that type I IFNs can regulate inflammasome activation and IL-1β synthesis (Labzin et al., 2016). Furthermore, a causal link between WD-induced type I IFN production and the risk of virus infections and auto-immunity is conceivable and should prospectively be investigated in rodent models of infections or in human populations. In addition, we detected signaling pathways in GMPs associated with cell proliferation, anti-apoptosis, loss of stem cell quiescence, and favored lineage differentiation into monocytes (Olsson et al., 2016), which potentially raises GMP lifespan and facilitates memory induction. These cell phenotypes were largely maintained even 4 weeks after shifting mice to CD feeding.
In summary, WD feeding can result in a remodeled cellular compartment in the bone marrow leading to increased proliferation, skewing of hematopoiesis, and generating exaggerated responses to exogenous and endogenous triggers of inflammation. By integrating the inflammatory effects of WD, the NLRP3 inflammasome appears to be critical for this response. Hence, this may pave the road for new therapeutic CVD interventions, such as the use of small molecule inhibitors that block the NLRP3 signaling pathway.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD45 (clone 30-F11) | BioLegend | Cat# 103131 |
| anti-mouse CD11b (clone M1/70) | BioLegend | Cat# 101245 |
| anti-mouse CD11c (clone N418) | BioLegend | Cat# 117317 |
| anti-mouse Ly6C (clone HK1.4) | BioLegend | Cat# 128015 |
| anti-mouse Ly6G (clone 1A8) | BioLegend | Cat# 127605 |
| anti-mouse CD8b (clone H35-17.2) | eBioscience | Cat# 11-0083-85 |
| anti-mouse CD4 (clone RM4-5) | eBioscience | Cat# 11-0042-85 |
| anti-mouse Ly6G (clone 1A8) | BD PharMingen | Cat# 553127 |
| anti-mouse Sca-1 (clone D7) | BioLegend | Cat# 108120 |
| anti-mouse cKit (clone ACK2) | eBioscience | Cat# 47-1172-82 |
| anti-mouse CD16/32 (FcyRII/III; clone2.4 G2) | BD PharMingen | Cat# 560540 |
| anti-mouse CD34 (clone HM34) | BioLegend | Cat# 128606 |
| anti-mouse F4/80 (clone BM8) | BioLegend | Cat# 123117 |
| anti-mouse MHCII (clone M5/114.15.2) | BioLegend | Cat# 107605 |
| anti-mouse CD86 (clone GL-1) | BioLegend | Cat# 105021 |
| anti-mouse CD45R (B220; clone RA3-6B2) | eBioscience | Cat# 11-0452-85 |
| anti-mouse CD11b (clone M1/70) | eBioscience | Cat# 11-0112-82 |
| anti-mouse CD3e (clone 500A2) | eBioscience | Cat# 11-0033-82 |
| anti-mouse TER-119 (clone TER-119) | eBioscience | Cat# 11-5921-85 |
| anti-mouse CD2 (clone RM2-5) | eBioscience | Cat# 11-0021-85 |
| anti-mouse CD19 (clone 1D3) | eBioscience | Cat# 11-0193-85 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| E.coli derived ultrapure LPS (serotype 0111:B4) | Sigma-Aldrich | Cat# L-4391 |
| Anakinra (Kineret, IL-1ra) | in house order via pharmacy | N/A |
| E. coli derived ultrapure LPS (serotype K12) | Invivogen | Tlrl-eklps |
| Pam3CSK4 | Invivogen | Tlrl-pms |
| Resiquimod (R848) | Invivogen | Tlrl-r848 |
| CpG-ODN 1826 | Invitrogen | Tlrl-1826 |
| Trizol | ThermoFisher Scientific | Cat# 15596026 |
| Oxidized LDL | Alfa Aesar | Cat# J65591 |
| E. coli derived LPS (serotype 055:B5) | Sigma-Aldrich | Cat# L2880 |
| Critical Commercial Assays | ||
| Infinity Cholesterol Reagents kit | ThermoFisher Scientific | Cat# TR13421 |
| Mouse Cytokine single/23-plex assay | BioRad Laboratories | Cat# 10014905 |
| Mouse CRP ELISA | R&D | Cat# MCRP00 |
| Mouse SAA3 ELISA | Invitrogen/ThermoFisher Scientific | Cat# KMA0021 |
| Human IL-6 DuoSet ELISA kit | R&D | Cat#DY206 |
| Human TNF DuoSet ELISA kit | R&D | Cat# DY210 |
| ProCarta Beads (IL-10, CCL5, IL-18, IL-6, TNF, MCP-1, IL- 12p40, IL-1a, IL-1b) | ProCarta Bioplex | Cat# EPX01A-20614/26009/20618/20603/20607/26005/26033/20611/26002-901 |
| CD11b magnetic MicroBeads | Miltenyi | Cat# 130-049-601 |
| ClickiT EDU ALexaFluor647 Flow Cytometry Assay Kit | ThermoFisher Scientific | Cat# C10634 |
| miRNeasy Kit | Quiagen | Cat# 217004 |
| TruSeq RNA Sample Preparation Kit (v.2) | Illumina | Cat# RS-122-2001 |
| KAPA Library Quantification Kit (v8.17) | Kapa Biosystems | Cat# KK4824 |
| Nextera DNA Sample Prep. Kit | Illumina | Cat# FC-121-1030 |
| PCR Purification MinELute Kit | Quiagen | Cat# 28004 |
| Deposited Data | ||
| Raw data files for RNA and ATAC sequencing | NCBI Gene Expression Omnibus | GEO: GSE97926 |
| Experimental Models: Cell Lines | ||
| Human: primary healthy PBMCs | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock No.: 00664 |
| Mouse: B6.129S7-Ldlr tm1Her/J | The Jackson Laboratory; In house breeding | Stock No. 002201 |
| Mouse: Bl6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | Stock No. 002014 |
| Mouse: Nlrp3−/−/Ldlr−/− | In house breeding | N/A |
| Software and Algorithms | ||
| FACS DIVA Software | BD | In house license |
| FlowJo Software(v.10) | TreeStar | In house license |
| Luminex xMAP Technology Software | Luminex xPONENT; Milliplex Analyst | In house license |
| CASAVA (v1.8.2); bcl2fastq Conversion Software | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| TopHat (Hisat2-v2.0.6) | Johns Hopkins University, Center for Computational Biology | http://ccb.jhu.edu/software/tophat/index.shtml |
| Docker System (RNA-seq pre-processing) | kathrinklee/rna-seq-pipeline-hisat2 | https://hub.docker.com/r/kathrinklee/rna-seq-pipeline-hisat2/ |
| Partek Genomic Suite (PGS) Software (v6.6) | Partek | In house license |
| DESeq2 R package (v1.14.1) | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ggplot2 R package | ggplot2 | http://ggplot2.tidyverse.org |
| cluster profile R package (v3.2.11) | Bioconductor | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Cytoscape software (v3.3) | Cytoscape | http://www.cytoscape.org/download.php |
| BiNGO Cytoscape plugin (v 2.44) | VIB Department of Plant Systems Biology, UGent | http://apps.cytoscape.org/apps/bingo |
| enrichment map Cytoscape plugin (v1.2) | Cytoscape | http://apps.cytoscape.org/apps/enrichmentmap |
| BioLayout Express 3D(v3.3) | BioLayoutExpress3D Team | https://kajeka.com/download-graphia-pro/ |
| iRegulon (v1.3) | KU Leuven, Stein Aerts Lab | http://iregulon.aertslab.org/download.html#download |
| Monocle 2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/monocle.html |
| Seurat | NYU Center for Genomics and Systems Biology, Satija Lab | http://satijalab.org/seurat/install.html |
| Cibersort | Stanford University, Newman Lab | https://cibersort.stanford.edu |
| Bowtie (v1.1.1) | Johns Hopkins University | http://bowtie-bio.sourceforge.net/index.shtml |
| PICARD (v1.134) | Broad Institute | http://broadinstitute.github.io/picard |
| MACS2 (v2.1.0.20140616) | Harvard University, Xiaole S. Liu’s Lab | https://github.com/taoliu/MACS |
| GenomicRanges R package (v1.26.3) | Bioconductor | GenomicRanges R package |
| Bedtools (v2.25.0) | University of Utah, Quinlan Lab | http://bedtools.readthedocs.io/en/latest/ |
| ChIPseeker R package (v1.10.3) | Bioconductor | https://bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| HOMER (v 4.6) | University of California San Diego, Glass Lab | http://homer.ucsd.edu/homer/download.html |
| Gviz R package (v1.18.1) | Bioconductor | https://bioconductor.org/packages/release/bioc/html/Gviz.html |
| HumanOmniExpressExome-8 (v1.0) | Illumina | https://support.illumina.com/downloads/humanomniexpressexome_product_files.html |
| LeicaQwin 3.5.1 (computerized morphometric image analysis) | Leica Microsystems | in house license |
| GraphPad Prism7 | GraphPad Software | in house license |
| Other | ||
| Western Diet | Harlan Teklad | Cat#TD.88137 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact, Eicke Latz (eicke.latz@uni-bonn.de).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Experiments were approved by the Institutional Animal Care and Use Committees of the University of Massachusetts Medical School and performed according to local ethics regulations (IACUC 1945, UMass Medical School), and in accordance with the NIH guidelines. C57BL/6J were purchased from The Jackson Laboratory. Ldlr−/− and Nlrp3−/− were originally purchased from The Jackson Laboratory and kept in house. Nlrp3−/−/Ldlr−/− were bred in house. All mice were previously backcrossed over ten generations to the C57Bl6/J background. For all in vivo animal studies (Western Diet feeding studies, LPS challenge, in vivo cell proliferation) age (8 weeks of age) and sex-matched female wild-type, Ldlr−/−, Nlrp3−/−/Ldlr−/− mice were used with at least 3 mice per genotype. If mouse numbers per genotype were < 3, experiments were at least performed twice independently. During experimental settings mice had ad libitum access to food and water, and were housed under a 12 hour light-dark cycle.
Human subjects
Experiments were conducted according to the principles expressed in the Declaration of Helsinki as Ethics Statement. Healthy individuals of Western European descent gave written informed consent to donate venous blood to use for research (Li et al., 2016). 122 volunteers were between 23–73 years old, and consisted of 77% males and 23% females. The data shown in Figure 6 represent the combined analysis for female and male subjects. For the SNPs identified in the pycard locus there was no gender-specific difference identified (rs146420137, oxLDL+LPSTNF, female: 0.0004, male: 0.00768; rs10574650, oxLDL+LPSTNF, female: 0.0036, male: 0.0048), whereas for the SNP identified in the IL-1ra locus only male subjects trended (rs34590034, oxLDL+LPSTNF, female: 0.2550, male: 0.085) or reached (rs34590034, oxLDL+LPSIL-6, female: 0.1885, male: 0.0010) significance, when analyzed individually. During all in vitro culture assays human isolated monocytes were kept in RPMI 1640 culture medium supplemented with 50 μg/mL gentamycin, 2 mM glutamax and 1 mM pyruvate at 37°C in a 5%CO2 atmosphere.
METHOD DETAILS
Mouse in vivo studies
To induce hyperlipidemia/hypercholesterolemia female mice were fed a Western diet (Teklad 88137) consiting of 17.3% protein, 21.2% fat (saturated fat 12.8%, monounsaturated fat 5.6%, polyunsaturated fat 1%) and 48.5% carbohydrates. Chow diet (Prolab Isopro RMH 30; LabDiet) consisted of 25% protein, 14% fat (ether extract) and 60% carbohydrates. To study diet effects on long-term reprogramming of myeloid (progenitor) subsets, female mice were fed a WD for 4 weeks, and subsequently subjected to regular chow diet for additional 4 weeks. To study diet-induced innate immune cell priming in vivo female mice received an intravenous injection of PBS (vehicle control) or E. coli derived ultrapure LPS (0111:B4; 10μg/mouse) six hours prior to sacrifice. To study the involvement of the IL-1/IL-1R signaling pathway in innate immune cell reprogramming in vivo female mice were daily i.p. injected with PBS (vehicle control) or Anakinra (human recominant IL-1ra; 10 mg/kg BW) during a 4 weeks course of CD or WD feeding. Blood, spleen and bone marrow (BM) were collected for single cell isolation and additional flow cytometry analysis, for immunohistochemical tissue preparation, for RNA isolation and further gene expression analysis, or for additional in vitro stimulation experiments.
Isolation of mouse plasma lipoproteins by FPLC
For lipoprotein separation by FPLC, total sera cholesterol levels were first determined with standard enzymatic assays (see below for kit description). Then a 50 μL sera aliquot was pre-warmed to 37°C for 5 min. followed by filtration through a PVDF 0.45 μm membrane filter. The filtered samples (20 μl) were subsequently fractionated by fast performance liquid chromatography (FPLC) gel filtration on a Superose 6 PC 3.2 /30 column at 4°C (GE Healthcare, Uppsala, Sweden). The elution fractions were monitored using absorbance at 280 nm, with a constant flow of 40 μl/min and fractions (40 μl) were collected beginning 18 min. after sample injection. Cholesterol in each fraction was measured by the Infinity cholesterol reagents (Thermo Fisher Scientific, VA, USA), and the area under the curve for vLDL, LDL and HDL was determined in comparison to a standard of known amounts of human vLDL, LDL and HDL run in parallel.
Mouse serum MultiPlex Cytokine Measurements
Multi-cytokines in mouse sera were measured with the BioPlex Mouse Cytokine 23-plex assay together with BioPlex mouse cytokine single-plexes of IL-18 and VEGF (Bio-Rad Laboratories, Hercules, CA) on a Bio-Plex 200 system powered by Luminex xMAP Technology. In some experiments a Procarta Plex 10-plex bead assay from ThermoFisher and measured on a MAGPIX instrument using Luminex xMAP Technology. The analysis was performed according to the manufacturer’s protocol with the following modifications: use of only half of the volumes, adapted incubation times (serum: 60-30-30), serum centrifugation for 10 min. at 13200 × rpm before analysis.
Measurement of mouse serum acute phase proteins
Acute phase proteins in mouse sera were measured using commercial ELISA kits for SAA3 (Invitrogen/ThermoFisher Scientific) and CRP (R&D) in accordance with the manufacturer’s instructions.
Murine White Blood Cell (WBC) counts
Total WBC counts in freshly isolated murine blood were performed by collecting blood in EDTA-coated tubes and analyzing them using a hematology cell counter (Abaxis HM5).
Flow cytometry of circulating and splenic leukocyte subsets in mouse
Total number of myeloid cells, monocyte subsets and neutrophils were identified as depicted in Figure S2A. Blood was collected via cardiac puncture into ethylene-diaminetetraacetate (EDTA) lined tubes and immediately placed on ice. Mice were perfused wth 10ml of sterile phosphate buffered saline (PBS) via the left ventricle before harvesting and further processing of organs. Spleens were collected into RPMI1640/0.1% fetal calf serum (FCS) containing vials and stored on ice until further processing. All following steps were performed on ice. Red blood cells (RBCs) were lysed (3 times for 5 minutes) and leukocytes were centrifuged, washed and re-suspended in flow cytometry buffer for staining (PBS containing 0.5% bovine serum albumine (BSA) and 2mM EDTA). Spleens were minced into small pieces, gently crushed and filtered twice with PBS through a 70μm cell strainer. RBC lysis was performed once for 3 minutes before resuspending splenic cells in flow cytometry buffer and antibody staining with a cocktail of antibodies against CD45 PerCp-Cy5.5, CD11b BV510, CD11c BV421, Ly6C-APC, Ly6G-FITC, F4/80 PE, CD64 PE-Cy7, MHCII APC-Cy7, CD86 Pacific Blue (all from BioLegend). Monocytes were identified as CD45hi CD11b+ Ly6G− and further separated into Ly6Chi, Ly6Cint and Ly6C−; neutrophils were identified as CD45hi CD11b+ Ly6G+ Ly6Clo. Cells were analyzed by LSRII using the FACS Diva software (BD). All flow cytometry data were analyzed using FlowJo Software (Tree Star Inc.).
In vivo proliferation assay
For in vivo proliferation studies, mice were injected intraperitoneally with 1 mg of 5-(ethynyl-2′-deoxyuridine) (EdU) 24 hours prior to sacrifice. In preparation for subsequent flow cytometry analysis, cell populations were immunostained as described above, fixed in 4% paraformaldehyde (PFA) and further treated for EdU staining following the manufacturer’s instructions of the Click-iT System (ThermoScientific). AlexaFluor647-conjugated azide was used to detect incorporated EdU. Stained cells were analyzed by LSRII using the BD Diva software and analyzed by FlowJo. Proliferation was quantified and expressed as percentage of EdU-positive cells.
Ex vivo stimulation of bone marrow cells and splenic monocytes
Bone marrow and splenic single cell suspensions were prepared as described above. Splenic CD11b+ monocytes were further purified by positive selection using a cocktail of CD11b magnetic beads (Miltenyi), reaching a purity of 95%. Cells in 96-well flat-bottom wells (100.000 cells/well) were cultured in RPMI1640 supplemented with L-Glut, 10%FCS, 1% Ciprofloxacin at 37°C in a 5% CO2 atmosphere. After a 2 hour resting period in the incubator (37°C, 5%CO2), cells were stimulated with TLR-ligands for 6 hours: Pam3CysK4 (10ng/ml; TLR2 ligand), LPS (10ng/ml; TLR4 ligand), R848 (10μg/ml; TLR7/8 ligand), and CpG (10μM; TLR9 ligand). Subsequently, supernatants were collected for cytokines measurement by MultiPlex bead array (ThermoFisher).
Flow cytometric cell sorting of murine bone marrow hematopoietic stem cells
Hematopoietic stem cell precursor cells (HSPCs) and myeloid progenitor cells including common myeloid progenitor cells (CMPs) and granulocyte-monocyte progenitor cells (GMPs) from bone marrow were analyzed by flow cytometry as described in Figure S2D. Bone marrow was harvested from femurs and tibias by flushing bones with cold Hank’s balanced salt solution (HBSS) containing 0.1% BSA and 2mM EDTA. RBC lysis was performed once for 5 minutes, followed by antibody staining with a cocktail of antibodies against lineage-committed cells (CD3e, CD19, CD45R (B220), CD11b, TER-119, CD2, CD8b, CD4, Ly6G; all FITC-labeled and from eBioscience), Sca-1 Pacific Blue, c-kit (CD117) APC-Cy7, CD34 APC, CD16/CD32 (FcyRII/III) PerCp-Cy5.5 (all from BioLegend). HSPCs were identified as lin- Sca-1+ c-kit+; CMPs as lin− Sca1− c-kit+ CD34int FcγRII/IIIint; GMPs as lin− Sca1− c-kit+ CD34int FcγRII/IIIhi. Isolated and stained bone marrow cells were either directly analyzed by LSRII (BD), or flow sorted by FACS Aria II for further RNA isolation and RNA/ATAC-sequencing, or in vitro assays.
RNA Isolation
1.7 × 105–16 × 105 GMPs were sorted and subsequently lysed in TRIZOL (Invitrogen) and total RNA was extracted using the miRNAeasy kit (Quiagen) according to the manufacturer’s protocol. The precipitated RNA was solved in RNase-free water. The quality of the RNA was assessed by measuring the ratio of absorbance at 260 nm and 280 nm using a Nanodrop 2000 Spectrometer (Thermo Scientific) and by visualization of 28S and 18S band integrity on a Tapestation 2200 (Agilent).
Generation of cDNA Libraries and Sequencing
Total RNA was converted into libraries of double-stranded cDNA molecules as a template for high-throughput sequencing using the Illumina TruSeq RNA Sample Preparation Kit v2. Briefly, mRNA was purified from 100–500 ng of total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in Illumina proprietary fragmentation buffer. First strand cDNA was synthesized using random oligonucleotides and SuperScript II. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities and enzymes were removed. After adenylation of 3′ends of DNA fragments, Illumina adaptor oligonucleotides were ligated to prepare for hybridization. DNA fragments with ligated adaptor molecules were selectively enriched using Illumina PCR primers in a 15 cycles PCR reaction. Size-selection and purification of cDNA fragments with preferentially 200 bp in insert length was performed using SPRIBeads (Beckman-Coulter). Size distribution of cDNA libraries was measured using the Agilent high sensitivity DNA assay on a Tapestation 2200 (Agilent). cDNA libraries were quantified using KAPA Library Quantification Kits (Kapa Biosystems). After cluster generation on a cBot, 75 bp single read sequencing was performed on a HiSeq1500 and de-multiplexed using CASAVA v1.8.2.
RNaseq Data Analysis
Pre-processing of RNaseq data was performed by a standardized and reproducible pipeline based on the Docker system (Docker image is available via docker hub, limesbonn/hisat2). Briefly, alignment to the mouse reference genome mm10 from UCSC was conducted by Hisat2 (Hisat2, 2.0.6) (Kim et al., 2015) using standard settings Aligned BAM files were imported into Partek Genomics Suite (PGS) software (version 6.6, Partek) for further analysis, and mRNA quantification was performed using mm10 RefSeq Transcripts (2015-11-03) as annotation file. Afterward, raw gene counts were normalized using the DESeq2 algorithm in R (package version 1.14.1). After DESeq normalization, the normalized read counts were imported back into PGS and floored by setting all read counts less than 1 to a value of exactly 1. Subsequently, we removed all genes that had a maximum overall group mean below 10. After filtering the data comprised 11306 present genes. Correlation was calculated as Pearson correlation of normalized read counts. Differentially expressed genes were identified by a two-way ANOVA model taking the diet and treatment as factors, with genes with highest variance (n = 4672) being defined as genes with a significant p value (p value < 0.05) for the interaction of diet and treatment. Principal component analysis was performed using PGS. Heatmaps and several plots were generated using the ggplot2 plotting library. Functional annotation of gene sets was performed using the clusterProfiler R package (version 3.2.11). RNa-seq data from Nlrp3−/− /Ldlr−/− mice were normalized and analyzed separately from data obtained from Ldlr−/− mice following the same workflow as described above. Network visualization of Gene Ontology Enrichment Analysis (GOEA) was performed with Cytoscape (version 3.3) and the BiNGO plugin (Version 2.44) with a FDR-corrected hypergeometric p-value of 0.001. The Cytoscape plugin enrichment map (Version 1.2) displayed the GOEA results as a network of GO-terms, and for assistance of annotation we used the word cloud cytoscape app (version 3.1.0).
Co-expression network analysis
In order to perform a topological mapping of global changes in gene expression we used a co-expression network approach based on correlation. The gene expression matrices containing the normalized read counts for the 11306 present genes were imported into BioLayout (Version 3.3) and co-regulation networks were generated with a Pearson correlation cutoff of 0.85. Co-expressed genes were visualized as a network (n = 4,360 genes) by Cytoscape applying a Force-directed layout. To identify the topology of the co-expression network the positive standardized gene expression (scaled and centered average expression) was calculated and mapped onto the co-expression network. In addition, we mapped the expression differences in a LPS response under CD as fold change to the network. To visualize the diet dependent effects on a LPS response we overlaid the color-coded fold changes between WD/LPS and CD/LPS or WD > CD/LPS and CD/LPS, respectively.
Transcription factor prediction analysis
In order to predict potential regulators of the observed gene expression changes we performed transcription factor binding prediction (TFBP) in the region 20kb around the transcriptional start site (TSS). To identify transcriptional regulators of genes upregulated in a WD versus CD LPS response, genes with a significant difference in gene expression (FC > 1.5, non-adj. p value < 0.05) between WD/LPS and CD/LPS were used for TFBP. In addition, we examined transcriptional regulators of genes only regulated in a WD/LPS or an WD > CD/LPS but not in a LPS response under standard chow diet. Therefore, genes were used for TFBP with no considerable gene expression changes between CD/LPS and CD (−1.3 < FC < 1.3) and a significant difference between WD/LPS and CD/LPS or WD > CD/LPS and CD/LPS, respectively. For the gene-based motif enrichment analysis, we used iRegulon (version 1.3) in combination with the provided 10K position weight matrices (PWMs) motif collection. To identify only highly enriched transcription factor predictions we filtered the results by an enrichment score of 3 and only reported transcription factors with a maximum false discovery rate (FDR-adj. p value) of less than 0.001. Resulting transcriptional regulators were grouped into clusters with high motif similarity using the STAMP algorithm with iRegulon standard parameters. The transcriptional regulators in their respective clusters were ordered by decreasing fold change and their gene expression was visualized as log2FC.
Assessment of GMP-commitment using single cell RNaseq data combined with support vector regression
To investigate the effect of nutrition on GMP commitment toward either the monocytic or the granulocytic developmental branch, we utilized a publicly available dataset, which comprised single cell transcriptome data of various murine hematopoietic progenitor cells (GEO: GSE70235, GEO: GSE70240). Here, we used the available normalized count data of only GMPs that were found either in the monocytic or granulocytic cluster (109 cells). Next, we constructed a developmental trajectory using Monocle 2. For this purpose, we transformed the expression levels in transcripts per million mapped reads (TPM) to estimate mRNA counts per cell (RPC) using the Census implementation of Monocle 2. We set the lower detection limit to an RPC of 1 and defined variable genes by taking advantage of a dispersion plot for which the mean expression cutoff was set to 1 and the dispersion cutoff was set to 1.75. Based on these variable genes, we computed a trajectory, which consisted of three branches with two of them were either monocytic (enriched for Csfr1) or granulocytic (enriched for S100A8) committed. Next, we calculated differentially expressed (DE) genes between the GMPs either committed toward the monocytic or the granulocytic lineage using Seurat. As tests for differential expression, we utilized a likelihood-ratio test for single cell gene expression, a standard AUC classifier and Tobit-test. When limiting the evaluation to genes which showed on average at least 1-fold difference (natural log-scale) between the two populations and which were detected in at least 25% of the cells in either of the two populations, all three tests resulted in the same number of DE genes (195 genes). For support vector regression, we defined the calculated DE genes as support vectors. The signature file was generated by population-wise calculation of the total TPM count per gene and filtering for the defined support vectors. The mixture file was based on variable genes (ANOVA, non-adj. p value < 0.05) across all conditions in the present dataset. Support vector regression was performed with 100 permutations and by disabling quantile normalization.
Cell preparation for ATAC-sequencing
For ATAC-sequencing, 5.000 – 50.000 flow sorted GMPs were collected in ice-cold HBSS containing 0.1%BSA and 2mM EDTA, and immediately processed following previously published protocols. In brief, sorted cells were spun down at 500 × g for 5 minutes at 4°C, washed once in cold PBS, and spun down in 50 μl cold lysis buffer at 500 × g for 10 minutes at 4°C. Immediately thereafter the transposition reaction was started by adding Nextera’s Tn5 Transposase (TDE1) in reaction buffer. The transposition reaction mix was incubated for 30 minutes at 37°C, DNA was purified using a Quiagen MinElute PCR purification kit. ATAC-seq libraries were purified using a PCR purification MinElute kit (Quiagen) and quantified using KAPA library quantification kits (Kapa biosystems) and a D1000 assay on a Tapestation 2200 (Agilent). Libraries were sequenced in a 50 bp single read run on a HiSeq 1500 (Illumina).
Analysis of ATACseq data
For Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) data analysis, short reads were aligned to the mouse genome version mm10 with Bowtie v1.1.1. Duplicate reads were flagged with Picard v1.134 prior to peak calling with MACS2 v2.1.0.20140616. Consensus peak regions across all samples were generated with the ‘reduce’ function of the Bioconductor GenomicRanges v1.26.3 package, followed by read counting per sample, which was done with bedtools multiBamCov v2.25.0. Blacklisted analysis regions with coordinate adjustments by the LiftOver tool to mm10, as well as regions that were not consistently present across samples of the same analysis group were excluded from the analysis. Differentially accessible regions were determined with the DESeq2 v1.14.1 package and mapped to nearest genes with the ChIPseeker v1.10.3 package. To read coverage tracks of ATAC signal we combined replicate samples (2 biological replicates, for WD > CD/PBS) and created normalized (to 10,000,000 reads) bigWig files using the HOMER (version 4.6) command makeUCSCfile. For visualization, the read coverage tracks were smoothed by averaging over a sliding window of 200 bases and all tracks for a given region were scaled to the highest overall peak and visualized by the Gviz package (version 1.18.1). Global changes in chromatin accessibility are visualized as heatmap and similarly accessible regions are clustered by hierarchical clustering using pheatmap package v 1.0.8 in R.
Histology of mouse atherosclerotic lesions
Mice were anesthesized by an overdose isoflurane and perfused through the left cardiac ventricle with PBS containing 1% PFA. Hearts were dissected, fixed overnight in 4%PFA frozen in OCT Tissue Tek Compound (Sakura), and cut into 4 μM thick serial sections. To measure plaque volume in the aortic root, plaque area was measured for each valve for consecutive sections at 20 μm intervals that covered the entire lesion. H&E staining was performed as described previously (Duewell et al., 2010). Slides were analyzed in a blinded manner using a Leica DM3000 light microscope (Leica Microsystems) coupled to a computerized morphometric system (Leica Qwin 3.5.1).
Isolation of human PBMCs and in vitro trained immunity model
Venous blood was drawn from the cubital vein of healthy volunteers into 10 mL EDTA tubes (Monoject). PBMCs were isolated by density centrifugation of blood diluted 1:1 in pyrogen-free saline over Ficoll-Paque (Pharmacia Biotech). Cells were washed twice in saline and resuspended in RPMI 1640 culture medium (Invitrogen) supplemented with 50 μg/mL gentamycin, 2 mM glutamax (GIBCO), and 1 mM pyruvate (GIBCO). Cells were counted in a Coulter counter (Coulter Electronics) and adjusted to 5 × 106 cells per milliliter. A 100 μL volume was added into flat-bottom 96-well plates (Corning) and cells were incubated at 37°C. After 1 hour, cells were washed three times with 200 μL warm PBS to remove non-adherent cells. Subsequently, monocytes were incubated with culture medium only (negative control) or 10 μg/mL oxLDL for 24 hours in 10% pooled human serum. In an additional experimental setup Anakinra (recombinant IL-1ra; 10ug/ml) was added during oxLDL incubation for 24 hours. Cells were washed once with 200 μL warm PBS and incubated for 5 days in culture medium with 10% human pooled serum. The medium was changed once on day 3 of incubation. On day 6, the supernatant was discarded and the cells were re-stimulated with 200 μL culture medium or 10 ng/mL E. coli LPS (serotype 055:B5, Sigma-Aldrich). After 24 hours, supernatants were collected and stored at −20°C until cytokine measurement. Cytokine production was measured in supernatants using commercial ELISA kits for TNF and IL-6 (R&D systems) in accordance with the manufacturer’s instructions.
Mapping of genetic variants in human PBMCs
Raw cytokine levels were first log-transformed. Then, the ratio of cytokine measurements between the second and the first stimulation was computed to capture the enhanced trained response or tolerant response. Individuals of 200FG cohort were genotyped using the Illumina HumanOmniExpressExome-8 v1.0 and the data was imputed, as previously described. In total there were 122 samples with both DNA and cytokine measurements. We focused on the genetic effect of polymorphisms within a window of 250kb around PYCARD, NLRP3, CASPASE1 and IL-1RAP genes on the oxLDL-induced trained immunity. The ratio of log-transformed cytokine data was mapped to genotype data using a linear model with age and gender as covariates.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including the exact value of n, the definition of center, dispersion and precision measures (mean ± SEM) and statistical significance are reported in the Figures and Figure Legends. All data are expressed as the mean ± SEM. Data is judged to be statistically significant when p < 0.05 by two-tailed Student’s t test (if normally distributed); nonparametric data are analyzed using a Mann-Whitney U-test. To compare several groups, a one-way ANOVA (with Dunnet’s post-test) or repeated-measures ANOVA with multiple testing correction are used. In figures asterisks denote statistical significance (*, p < 0.05; **, p < 0.01; ***). Statistical analysis was performed in GraphPad PRISM 7 (Graph Pad Software Inc.).
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw data files for the RNA sequencing and ATAC sequencing analysis reported in this paper is NCBI GEO: GSE97926.
Supplementary Material
Figure S1. Systemic and Cellular Responsiveness Are Altered upon WD Feeding, Related to Figure 1
(A) Systemic lipoprotein levels in female Ldlr−/− mice fed as shown.
(B) Body weights of female Ldlr−/− mice treated as indicated.
(C) Acute phase reactant levels in female Ldlr−/− mice treated as indicated.
(D and E) Cytokine and chemokine response of (D) bone marrow cells or (E) splenic CD11b+ monocytes isolated from female Ldlr−/− mice following diet manipulation and treated ex vivo with vehicle or different TLR stimuli for 6h as indicated.
n = 5 mice per group in (B) and n = 6–9 mice per group in (C); ± SEM, p < 0.05 versus CD (C), versus un-stimulated cells (D and E).
Figure S2. WD Feeding Induces a Transcriptional Reprogramming in Bone Marrow Myeloid Precursor Cells, Related to Figure 2
(A) FACS gating strategy.
(B and C) Relative numbers (B) and activation status (C) of splenic myeloid subsets isolated from female Ldlr−/− mice treated as indicated.
(D) FACS sorting gating strategy for the different myeloid progenitor subsets.
(E) Schematic representation of the computational analysis as indicated.
(F) Functional enrichment analysis showing overrepresentation of genes upregulated in GMPs isolated from female Ldlr−/− mice treated as indicated in Gene Ontology terms or KEGG pathways.
(G) Transcription factor binding prediction analysis of significantly upregulated genes in GMPs isolated from female Ldlr−/− mice fed WD or CD.
Motif enrichment analysis was performed on 20kb area centered around the transcriptionally start site of upregulated genes. Log2FC in genes expressed in GMPs and visualized for predicted transcriptional regulators. Boxes represent TFs with high motif similarity (FDR-adj. p value < 0.001). Colors indicate the normalized enrichment score of motif enrichment. n = 3–5 animals per group; ± SEM, p < 0.05 (B, C).
Figure S3. WD Priming Induces a Cellular Hyper-responsiveness upon LPS Challenge, Related to Figure 3
(A) Representative flow cytometry histogram analysis from myeloid cell subsets from CD and WD fed female Ldlr−/− mice.
(B–E) Functional enrichment analysis showing overrepresentation of genes upregulated (B and C) or downregulated (D and E) in GMPs isolated from mice treated as indicated.
Figure S4. WD Feeding Induces a Sustained Altered Transcriptional and Epigenetic Phenotype in Bone Marrow Myeloid Progenitors, Related to Figures 4 and 5
(A) Schematic describing the procedure of computational analysis of RNaseq data for Figure 4 and S4.
(B) Functional analysis of genes commonly upregulated in GMPs from mice treated as indicated. Top five enriched gene ontology terms of these gene lists are shown. DE genes: non-adjust. p value < 0.05, FC > 1.5.
(C–E) Transcription factor binding prediction of regulated genes in GMPs isolated from mice treated as indicated and selected genes as depicted in the schematics. Selection of input genes for analysis: (C) Genes upregulated in WD/LPS versus CD/LPS (FC > 1.5, non-adj. p value < 0.05), (D) Genes not regulated in LPS response under CD (−1.3 < FC < 1.3), but significantly upregulated in LPS response upon WD feeding in GMPs (FC > 1.3, non-adj. p value < 0.05) and (E) Genes not regulated in LPS response under CD (−1.3 < FC < 1.3), but significantly upregulated in LPS response upon WD > CD feeding in GMPs (FC > 1.3, non-adj. p value < 0.05). Motif enrichment analysis was performed on a 20kb area centered around the transcription start site of upregulated genes. Log2 FC is visualized for predicted transcriptional regulators from conditions as indicated. Boxes represent TFs with high motif similarity (FDR-adj. p value < 0.001). Color indicates the normalized enrichment score of motif enrichment. Selected clusters are shown.
(F) Coverage of ATACseq signal for OSBPL3 (left panel) and ABCA1 loci (right panel).
Highlights.
Western diet triggers innate immunity, NOD-like, and IFN signaling pathways
Western diet alters in vivo LPS responses of GMPs
Western diet induces long-lasting trained immunity in myeloid cells
NLRP3 recognizes Western diet and mediates trained immunity
Acknowledgments
E.L., M.L.F., and A.C. are supported by grants from the NIH (1R01HL112661 and R01HL101274). E.L. is supported by the DFG (SFB670 and TRR 83, 57) and an ERC Consolidator grant (InflammAct). E.L. and P.D. are supported by the DFG (SFB1123). J.L.S. is supported by the DFG (SFB645 and SFB704), the European Union’s Horizon 2020 research and innovation program under grant agreement 733100 (SYSCID), and a grant from the Federal Ministry for Economic Affairs and Energy (BMWi Project FASTGENOMICS). E.L. is a co-founder of IFM-Therapeutics and a member of its scientific advisory board. M.G.N. is supported by an ERC Consolidator grant (310372) and a Spinoza grant of the Netherlands Organization for Scientific Research (NWO). N.P.R. is supported by a Dr. Dekker grant from the Netherlands Heart Foundation (2012T051). N.P.R., M.G.N., and L.A.B.J. received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement 667837. M.G.N., L.A.B.J., and N.P.R. are supported by the IN-CONTROL CVON grant from the Netherlands Heart Foundation (CVON2012-03). M.G.N., J.L.S., and E.L. are members of the Excellence Cluster ImmunoSensation.
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at https://doi.org/10.1016/j.cell.2017.12.013.
DECLARATION OF INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Conceptualization, A.C. and E.L.; Methodology, A.C., P.G., and E.L.; Software, P.G., K.H., K.B., C.J.S., K.K., T.U., J.S.-S., and J.L.S.; Investigation, A.C., M.A.R.L., K.P., P.D., D.B., M.H.P., M.O., S.J.C.F.M.M., V.K., T.E., A.S., L.A.B.J., N.P.R., Y.L., L.A.G., and M.G.N.; Resources, M.G.N., J.L.S., and E.L.; Writing – Original Draft, A.C., P.G., M.G.N., and E.L; Writing – Review & Editing, A.C., P.G., J.L.S., M.L.F., and E.L.; Visualization, A.C. and E.L.; Supervision, E.L.; Project Administration, E.L.; Funding Acquisition, A.C., M.L.F., P.D., J.L.S., M.G.N., and E.L.
References
- Bekkering S, Quintin J, Joosten LAB, van der Meer JW, Netea MG, Riksen NP. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler Thromb Vasc Biol. 2014;34:1731–1738. doi: 10.1161/ATVBAHA.114.303887. [DOI] [PubMed] [Google Scholar]
- Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden CDCC, Li Y, Popa CD, ter Horst R, van Tuijl J, Netea-Maier RT, et al. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell. 2018:172. doi: 10.1016/j.cell.2017.11.025. Published online January 11, 2018. [DOI] [PubMed]
- Boettcher S, Manz MG. Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 2017;38:345–357. doi: 10.1016/j.it.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol. 2016;16:35–50. doi: 10.1038/nri.2015.8. [DOI] [PubMed] [Google Scholar]
- Clinton SK, Fleet JC, Loppnow H, Salomon RN, Clark BD, Cannon JG, Shaw AR, Dinarello CA, Libby P. Interleukin-1 gene expression in rabbit vascular tissue in vivo. Am J Pathol. 1991;138:1005–1014. [PMC free article] [PubMed] [Google Scholar]
- Dorrestein PC, Mazmanian SK, Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nuñez G, Schnurr M, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messal M, Beaudeux JL, Drissi A, Giral P, Chater R, Bruckert E, Adlouni A, Chapman MJ. Elevated serum levels of proinflammatory cytokines and biomarkers of matrix remodeling in never-treated patients with familial hypercholesterolemia. Clinica Chimica Acta. 2006;366:185–189. doi: 10.1016/j.cca.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Fleet JC, Clinton SK, Salomon RN, Loppnow H, Libby P. Atherogenic diets enhance endotoxin-stimulated interleukin-1 and tumor necrosis factor gene expression in rabbit aortae. J Nutr. 1992;122:294–305. doi: 10.1093/jn/122.2.294. [DOI] [PubMed] [Google Scholar]
- Friedman GD, Klatsky AL, Siegelaub AB. The leukocyte count as a predictor of myocardial infarction. N Engl J Med. 1974;290:1275–1278. doi: 10.1056/NEJM197406062902302. [DOI] [PubMed] [Google Scholar]
- GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12:204–212. doi: 10.1038/ni.2001. [DOI] [PubMed] [Google Scholar]
- Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, Griep A, Axt D, Remus A, Tzeng TC, et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labzin LI, Lauterbach MAR, Latz E. Interferons and inflammasomes: cooperation and counterregulation in disease. J Allergy Clin Immunol. 2016;138:37–46. doi: 10.1016/j.jaci.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Li Y, Oosting M, Deelen P, Ricaño-Ponce I, Smeekens S, Jaeger M, Matzaraki V, Swertz MA, Xavier RJ, Franke L, et al. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat Med. 2016;22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield M, Devlin JJ, Nordio F, Hyde C, Cannon CP, Sacks F, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I, Ruppova K, Wang B, Chen LS, Grzybek M, Grinenko T, Eugster A, Troullinaki M, Palladini A, Kourtzelis I, et al. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell. 2018;172:147–161. doi: 10.1016/j.cell.2017.11.034. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AJ, Akhtari M, Tolani S, Pagler T, Bijl N, Kuo CL, Wang M, Sanson M, Abramowicz S, Welch C, et al. ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J Clin Invest. 2011;121:4138–4149. doi: 10.1172/JCI57559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagareddy PR, Kraakman M, Masters SL, Stirzaker RA, Gorman DJ, Grant RW, Dragoljevic D, Hong ES, Abdel-Latif A, Smyth SS, et al. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 2014;19:821–835. doi: 10.1016/j.cmet.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narverud I, Ueland T, Nenseter MS, Retterstøl K, Telle-Hansen VH, Halvorsen B, Ose L, Aukrust P, Holven KB. Children with familial hypercholesterolemia are characterized by an inflammatory imbalance between the tumor necrosis factor α system and interleukin-10. Atherosclerosis. 2011;214:163–168. doi: 10.1016/j.atherosclerosis.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9:355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Netea MG, Joosten LAB, Latz E, Mills KHG, Natoli G, Stunnenberg HG, O’Neill LAJ, Xavier RJ. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads JP, Lukens JR, Wilhelm AJ, Moore JL, Mendez-Fernandez Y, Kanneganti TD, Major AS. Oxidized low-density lipoprotein immune complex priming of the Nlrp3 inflammasome involves TLR and FcγR cooperation and is dependent on CARD9. J Immunol. 2017;198:2105–2114. doi: 10.4049/jimmunol.1601563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, et al. CANTOS Trial Group. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- Seijkens T, Hoeksema MA, Beckers L, Smeets E, Meiler S, Levels J, Tjwa M, de Winther MPJ, Lutgens E. Hypercholesterolemia-induced priming of hematopoietic stem and progenitor cells aggravates atherosclerosis. FASEB J. 2014;28:2202–2213. doi: 10.1096/fj.13-243105. [DOI] [PubMed] [Google Scholar]
- Sheedy FJ, Grebe A, Rayner KJ, Kalantari P, Ramkhelawon B, Carpenter SB, Becker CE, Ediriweera HN, Mullick AE, Golenbock DT, et al. CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat Immunol. 2013;14:812–820. doi: 10.1038/ni.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer K, DelProposto J, Morris DL, Zamarron B, Mergian T, Maley N, Cho KW, Geletka L, Subbaiah P, Muir L, et al. Diet-induced obesity promotes myelopoiesis in hematopoietic stem cells. Mol Metab. 2014;3:664–675. doi: 10.1016/j.molmet.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski FK, Nahrendorf M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science. 2013;339:161–166. doi: 10.1126/science.1230719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski FK, Libby P, Aikawa E, Alcaide P, Luscinskas FW, Weissleder R, Pittet MJ. Ly-6Chi monocytes dominate hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. J Clin Invest. 2007;117:195–205. doi: 10.1172/JCI29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- van der Meer JW, Barza M, Wolff SM, Dinarello CA. A low dose of recombinant interleukin 1 protects granulocytopenic mice from lethal gram-negative infection. Proc Natl Acad Sci USA. 1988;85:1620–1623. doi: 10.1073/pnas.85.5.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kampen E, Jaminon A, van Berkel TJC, Van Eck M. Diet-induced (epigenetic) changes in bone marrow augment atherosclerosis. J Leukoc Biol. 2014;96:833–841. doi: 10.1189/jlb.1A0114-017R. [DOI] [PubMed] [Google Scholar]
- Yvan-Charvet L, Pagler T, Gautier EL, Avagyan S, Siry RL, Han S, Welch CL, Wang N, Randolph GJ, Snoeck HW, Tall AR. ATP-binding cassette transporters and HDL suppress hematopoietic stem cell proliferation. Science. 2010;328:1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer S, Grebe A, Latz E. Danger signaling in atherosclerosis. Circ Res. 2015;116:323–340. doi: 10.1161/CIRCRESAHA.116.301135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Systemic and Cellular Responsiveness Are Altered upon WD Feeding, Related to Figure 1
(A) Systemic lipoprotein levels in female Ldlr−/− mice fed as shown.
(B) Body weights of female Ldlr−/− mice treated as indicated.
(C) Acute phase reactant levels in female Ldlr−/− mice treated as indicated.
(D and E) Cytokine and chemokine response of (D) bone marrow cells or (E) splenic CD11b+ monocytes isolated from female Ldlr−/− mice following diet manipulation and treated ex vivo with vehicle or different TLR stimuli for 6h as indicated.
n = 5 mice per group in (B) and n = 6–9 mice per group in (C); ± SEM, p < 0.05 versus CD (C), versus un-stimulated cells (D and E).
Figure S2. WD Feeding Induces a Transcriptional Reprogramming in Bone Marrow Myeloid Precursor Cells, Related to Figure 2
(A) FACS gating strategy.
(B and C) Relative numbers (B) and activation status (C) of splenic myeloid subsets isolated from female Ldlr−/− mice treated as indicated.
(D) FACS sorting gating strategy for the different myeloid progenitor subsets.
(E) Schematic representation of the computational analysis as indicated.
(F) Functional enrichment analysis showing overrepresentation of genes upregulated in GMPs isolated from female Ldlr−/− mice treated as indicated in Gene Ontology terms or KEGG pathways.
(G) Transcription factor binding prediction analysis of significantly upregulated genes in GMPs isolated from female Ldlr−/− mice fed WD or CD.
Motif enrichment analysis was performed on 20kb area centered around the transcriptionally start site of upregulated genes. Log2FC in genes expressed in GMPs and visualized for predicted transcriptional regulators. Boxes represent TFs with high motif similarity (FDR-adj. p value < 0.001). Colors indicate the normalized enrichment score of motif enrichment. n = 3–5 animals per group; ± SEM, p < 0.05 (B, C).
Figure S3. WD Priming Induces a Cellular Hyper-responsiveness upon LPS Challenge, Related to Figure 3
(A) Representative flow cytometry histogram analysis from myeloid cell subsets from CD and WD fed female Ldlr−/− mice.
(B–E) Functional enrichment analysis showing overrepresentation of genes upregulated (B and C) or downregulated (D and E) in GMPs isolated from mice treated as indicated.
Figure S4. WD Feeding Induces a Sustained Altered Transcriptional and Epigenetic Phenotype in Bone Marrow Myeloid Progenitors, Related to Figures 4 and 5
(A) Schematic describing the procedure of computational analysis of RNaseq data for Figure 4 and S4.
(B) Functional analysis of genes commonly upregulated in GMPs from mice treated as indicated. Top five enriched gene ontology terms of these gene lists are shown. DE genes: non-adjust. p value < 0.05, FC > 1.5.
(C–E) Transcription factor binding prediction of regulated genes in GMPs isolated from mice treated as indicated and selected genes as depicted in the schematics. Selection of input genes for analysis: (C) Genes upregulated in WD/LPS versus CD/LPS (FC > 1.5, non-adj. p value < 0.05), (D) Genes not regulated in LPS response under CD (−1.3 < FC < 1.3), but significantly upregulated in LPS response upon WD feeding in GMPs (FC > 1.3, non-adj. p value < 0.05) and (E) Genes not regulated in LPS response under CD (−1.3 < FC < 1.3), but significantly upregulated in LPS response upon WD > CD feeding in GMPs (FC > 1.3, non-adj. p value < 0.05). Motif enrichment analysis was performed on a 20kb area centered around the transcription start site of upregulated genes. Log2 FC is visualized for predicted transcriptional regulators from conditions as indicated. Boxes represent TFs with high motif similarity (FDR-adj. p value < 0.001). Color indicates the normalized enrichment score of motif enrichment. Selected clusters are shown.
(F) Coverage of ATACseq signal for OSBPL3 (left panel) and ABCA1 loci (right panel).