Abstract
Amphiphilic alpha-helices are common motifs used in numerous biological systems including membrane channels/pores and antimicrobial peptides (AMPs), and binding proteins, and a variety of synthetic biomaterials. Previously, an amphiphilic peptide with lysine-containing motifs was shown to reversibly bind the anionic porphyrin meso-Tetra(4-sulfonatophenyl)porphyrin (TPPS42−) and promote the formation of excitonically coupled conductive J-aggregates. The work presented here focuses on the use of this amphiphilic peptide and derivatives as a potential antimicrobial agent. AMPs are naturally occurring components of the innate immune system, which selectively target and kill bacteria. Sequence derivatives were synthesized in which the position of the Trp, used as a fluorescence reporter, was changed. Additional variants were synthesized where the hydrophobic amino acids were replaced with Ala to reduce net hydrophobicity or where the cationic Lys residues were replaced with diaminopropionic acid (Dap). All peptide sequences retained the ability to bind TPPS42− and promote the formation of J-aggregates. The peptides all exhibited a preference for binding anionic lipid vesicles compared to zwitterionic bilayers. The Trp position did not impact antimicrobial activity, but the substituted peptides exhibited markedly lower efficacy. The Dap-containing peptide was only active against E. coli and P. aeruginosa, while the Ala-substituted peptide was inactive at the concentrations tested. This trend was also evident in bacterial membrane permeabilization. The results indicate that the amphiphilic porphyrin binding peptides can also be used as antimicrobial peptides. The cationic nature is a driver in binding to lipid bilayers, but the overall hydrophobicity is important for antimicrobial activity and membrane disruption.
Keywords: Antimicrobial peptides, fluorescence, lipid binding, porphyrin binding, membrane permeabilization
Graphical Abstract
1. Introduction
In recent years, significant interest has been paid to the development of new antibacterials due to the rapid and widespread development of antibiotic resistance. Resistance development in bacteria has been attributed to the overuse and misuse of antibiotics which allows surviving bacteria, that is those with an evolutionarily programmed increased tolerance to the drug, to propagate (1). The World Health Organization recently released a report evaluating the clinical pipeline of antibiotics and determined this pipeline is inadequate to address the coming needs for new and better antimicrobials (2). In addition to the traditional small molecule approaches, natural products (3), metals (4, 5), polymers (6, 7), bacteriophage (8), and peptides have all been areas of development to address the need for new therapies.
Antimicrobial peptides (AMPs) are a class of naturally occurring peptides, which are found in the innate immune system of all higher organisms and exhibit high selectivity for targeting bacteria over host cells. This class of molecules has diverse structural and amino acid sequence characteristics, making prediction and design of AMPs an area of great interest. Generally, AMPs exhibit an amphiphilic chemical / amino acid composition, a net positive charge, and often adopt very simple 3D structures, in many cases as simple as a single, facially amphiphilic α-helix. The mechanism of action is widely believed to include bacterial membrane disruption (9), but more studies are identifying additional or alternate mechanisms by which the AMPs may bring about bacterial cell death. Some AMPs are also known to be involved in immunomodulatory processes (10) or the disruption of bacterial signaling pathways (11).
While peptides are naturally occurring molecules, the chemical flexibility and the relative ease of synthesis and purification have fostered the development of peptides for applications in non-biological settings such as biomaterials development (12, 13). The literature contains numerous examples of peptides as the functional building blocks in hydrogels (13), nanoparticles (14), structural scaffolds (15), and in energy capture/storage (16, 17). Energy capture, transfer, and storage applications have often built on naturally occurring motifs, such as iron-sulfur clusters, chelated metal ions, or porphyrins and porphyrin aggregates, as the foundation of these designed materials (18–20). Specifically, proteins and peptides have been used as scaffolds to orient the photon absorbing or electron transfer components in these materials, similar to the naturally occurring photosynthetic or electron transfer machinery in cells.
Previously, our group reported the development of peptide based scaffolds for the binding and orientation of an anionic porphyrin, meso-tetra(4-sulfonatophenyl)porphyrin (TPPS42−), into an excitonically coupled J-aggregate structure(21). This peptide scaffold was originally synthesized based on the amphiphilic AMP ISM-78 also referred to as r-pexiganan(22). This peptide was shown to promote J-aggregate formation over a wider range of pH values than previously reported (21). The J-aggregate is defined as a dye or other absorptive species that exhibits an absorption peak which red-shifts upon supramolecular organization (23). These J-aggregates are characterized by excitonically coupled oligomers in which contain multiple coupled transition dipoles(23). This peptide-induced stabilization of the J-aggregate allowed for the measurement of the inter-porphyrin electron transfer dynamics (24). The peptide scaffold used in our study, and similar designed scaffolds by other groups, revolved around developing soluble, cationic, facially amphiphilic helices which use cationic side chains to bind the anionic sulfonate groups of the TPPS42− porphyrin (21, 25). At low pH in aqueous solutions, TPPS42− adopts the J-aggregate formation, the Soret band shifts ~55 nm from 434 nm to ~490 nm, while the Q-bands shift and collapse into a single peak at ~705 nm (21). Notably, the peptides used in our studies did not form helices under pH conditions where TPPS42− formed a J-aggregate, but did adopt a helical conformation at neutral pH. While TPPS42− has been studied by many groups as a possible photosensitizer in photodynamic therapy, the peptide-scaffolded TPPS42− aggregates were not developed for this application (26, 27).
Due to the structural parallels to AMPs and previous reported activity of the parent sequence, the porphyrin binding peptide and sequence derivatives (referred to as PBP herein) were tested for antimicrobial activity against a variety of Gram-positive and Gram-negative strains. Two variants of PBP in which the position of the Trp residue was altered were also investigated. A series of fluorescence based binding assays were performed to characterize the interactions with bilayers and the structure of peptides were monitored using circular dichroism. PBP contains nine lysine residues imparting a +9 charge, which should promote binding to bacterial cell surfaces, and 7 hydrophobic residues (Leu, Ile, Val, and Phe) which imparts hydrophobic character and can drive insertion of the peptide into lipid bilayers. Investigation of two additional sequences in which the hydrophobic amino acids were replaced with Ala (PBP-A) or the Lysine residues were replaced with a lysine analog containing a shortened side chain (PBP-X), were used to probe the role of membrane penetration and perturbation on activity. The antimicrobial and biophysical characterization were performed on the free peptides, in the absence of TPPS42−. However, all peptides retained the ability to bind and promote TPPS42− J-aggregate formation, but only PBP, PBP-11W, and PBP-18W had broad spectrum antimicrobial activity while PBP-X exhibited selective activity against selected Gram-negative strains.
2. Materials and Methods
2.1 Materials
Lipids were obtained from Avanti Polar Lipids Inc. (Alabaster, AL). All other chemicals were from Sigma-Aldrich (St. Louis, MO) or VWR (Radnor, PA). Meso-tetra(4-sulfonatophenyl)porphyrin dihydrochloride was from Frontier Scientific (Logan, UT). PBP-A and PBP-X were synthesized by Anaspec, Inc (Fremont, CA). PBP and Trp variant peptides were synthesized in-house using FMOC-based solid phase peptide synthesis followed by purification. Peptides were cleaved from the resin using a mixture of 92.5:2.5:2.5:2.5 trifluoroacetic acid (TFA)/H2O/triisopropyl silane/ethanedithiol. Crude peptide samples were precipitated from the cleavage mixture in cold diethyl ether, followed by centrifugation. Pellets containing the peptide was dissolved in 9:1 H2O:Acetonitrile and were purified by reversed phase HPLC using a linear gradient of water and acetonitrile supplemented with 0.1 % TFA. All peptides were purified using a Jupiter 300 C4 column (Phenomenex, Torrance, CA) at a flow rate of 10ml/min. Peptide identity and purity were confirmed by ESI-MS. Eluted HPLC fractions containing peptide were pooled and lyophilized for storage. Working stock solutions were made in 3:1 H2O:Ethanol in the range of 100–200μM and stored at −20° C. Lipids: (16:0–18:1) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphotidylcholine (POPC), (16:0–18:1) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphotidylglycerol (POPG), and 10-doxyl nonadecane (10-DN) were used without further purification and stored as stocks in chloroform at −20°C. Buffers used in all assays were PBS (150 mM NaCl, 50 mM sodium phosphate; pH 7.0), 10X diluted PBS (for CD measurements), Z-Buffer (0.1 M Na2HPO4/NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.05 M β-mercapthoethanol, pH 7.0), or 20mM sodium acetate (C2H3NaO2) pH 3.5.
2.2 Methods
2.2.1 Absorbance Spectroscopy
Absorbance spectra of TPPS42− were collected as reported previously (21). Briefly, TPPS42− was dissolved in 6mM KOH as a stock solution. An aliquot of the stock was added to 1mL of sodium acetate buffer for a final TPPS42− concentration of 3μM. Absorbance spectra were recorded on the TPPS42− samples before and after the addition of 1μM peptide using a Genesys 10S UV-Vis spectrophotometer.
2.2.2 Lipid Vesicle Preparation
Appropriate volumes of lipid in chloroform were transferred to test tubes with desired volume and ratio of lipid, being either 100% POPC or 75/25 POPC/POPG. Lipid mixtures were dried under flowing N2 gas followed by vacuum dessication of the lipid film for 60 minutes to remove any remaining solvent. The lipid film was resuspended in PBS for a total volume of 1400 μL at 1000 μM. Resuspended lipid solutions were sonicated using Special Ultrasonic Cleaning Sonicator for 20 minutes. For CD and quenching experiments, SUVs were created using the ethanol dilution method (28). Briefly, lipid films were dissolved in 10 μL of ethanol to which the appropriate volume PBS were added while vortexing to a total volume of 800 μL accounting for the future addition of peptide. In the case of 10-DN quenching experiments, the final concentration of lipids was 250 μM, with 10% of the POPC replaced with 10-DN in those samples.
2.2.3 Lipid Binding Assay
Samples containing 2μM peptide in PBS at a final volume of 800μL were prepared. Background samples lacking peptide were also prepared. Fluorescence emission spectra were collected using a Horiba FluoroMax-4 Spectrofluorometer with λex = 280 nm and emission over the range 300–400 nm. Sonicated lipid vesicles were titrated into each cuvette and remeasured after each titration step. The barycenter and ΔBarycenter were calculated as previously described (29). All samples were performed at least in duplicate. All spectra were recorded in semi-micro style quartz cuvettes.
2.2.4 Fluorescence Quenching
Samples of 800μL containing 5μM peptide in PBS or a blank lacking peptide were prepared. All intensity measurements were measured with a Horiba FluoroMax-4 Spectrofluorometer. Initial intensity values (F0) were recorded prior to addition of acrylamide using λex = 295 and λem = 340 to reduce inner filter effects of the acrylamide. Samples were titrated with 10μL aliquots from a stock of 4 M acrylamide in water and the fluorescence was remeasured after each addition. Intensity values were corrected for dilution and inner filter effects as reported previously (30). Ksv was calculated from averaging the slope of the best fit line for each replicate and is reported as the average and standard deviation of these linear fits.
Quenching with 10-DN was performed by preparing vesicles with and without 10-DN imbedded in the bilayer. For these experiments, λex = 280 and λem = 340. The average F0 intensity was used in the calculation of the F0 / F values. The Q-ratio was calculated as described previously (31). Briefly, the Q-ratio is calculated by the following formula:
where the acrylamide quenching ratio is calculated from samples containing 0.235 M acrylamide.
2.2.5 Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra were recorded on a Jasco J-810 Spectropolarimeter over the range 190–260 nm at 50 nm/min. Samples were prepared under four different conditions: 10x diluted PBS, 10x diluted PBS with 100 mM SDS, and 1:1 10x diluted PBS:trifluoroethanol, or 250 μM PC:PG vesicles in 10x diluted PBS. In all cases containing no vesicles, peptide concentration was 5 μM. Scans containing vesicles possessed 3 μM peptide. Samples lacking peptide were measured as background scans. All samples were scanned 64 times and averaged before background subtraction.
2.2.6 Minimum Inhibitory Concentration
Bacterial strains E. coli D31 (the chromosomal penicillin V-resistant isolate in the 1968 study by Burman et al (32), K. pneumonia (ATCC: 700603), P. aeruginosa (ATCC:10145), and S. aureus (ATCC: 27660) were incubated in separate Mueller Hinton broth for ~18 hours at 37°C. Aliquots of the overnight cultures were diluted 1:200 in 25 mL fresh MH broth. The diluted cultures were incubated with shaking at 37°C until the optical density at 600nm (OD600) was within the range 0.2–0.6. After reaching optimal absorbance, the cultures were diluted into more fresh MH broth to a final density of ~ 105 CFU/mL. 90 μL the diluted cultures were pipetted into wells of a 96-well plate containing serial dilutions of peptides for a final sample volume of 100 μL. No peptide controls were included where 10μL of distilled was supplemented. The plates were covered and incubated for 18 hours at 37°C. After incubation, OD600 was measured for each plate and MIC determined by the lowest concentration with no visible growth. Data reported are the average of 3 independent samples.
2.2.7 Outer Membrane Permeabilization Assay
A single colony of E.coli D31was transferred into LB broth with 100 μg/mL ampicillin (LB-Amp) and incubated at 37°C with shaking for 18 hours. The culture was diluted in fresh LB-Amp at a 1:240 ratio. The diluted culture was placed back in incubation at 37°C with shaking until OD600 reached 0.2–0.6. Once the appropriate OD600 was reached, the culture was centrifuged at 2500 rpm for 15 minutes in a benchtop clinical centrifuge. The supernatant was discarded, followed by resuspension of the bacterial pellet in an identical volume of PBS. Nitrocefin solution was prepared through dissolving 1 mg nitrocefin in 100μL DMSO and subsequently diluting with 1.9 mL PBS to achieve a final stock concentration of 500 μg/mL nitrocefin. The nitrocefin solution was covered in aluminum foil and stored at 4°C before dispensing into plates.
Solutions were dispensed into a 96 well plate in the subsequent order: 10 μL of peptide with serial dilutions starting from 15 μM (excluding the last row which had 10 μL distilled H2O as a negative control), 80 μL E.coli D31 in all wells, and finally 10 μL nitrocefin stock solution. Following the addition of nitrocefin to the wells, the absorbance was immediately recorded at 486 nm and was recorded every 5 minutes over the next 90 minutes. Data reported are the average of 3 independent samples.
2.2.8 Inner Membrane Permeabilization Assay
A single colony of E.coli D31 was inoculated into 3 mL of LB broth. The culture was incubated for 18 hours at 37°C with shaking, followed by a 240-fold dilution into fresh LB broth supplemented with 100 μL of 100 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) to induce expression of the –galactosidase gene. The diluted culture was incubated with shaking until an OD600 of 0.2–0.5 was reached.
The following solutions were transferred into each well of a 96 well plate in the order listed: 56 μL of Z-buffer, 10 μL of serial diluted peptide starting from 15 μM (except the last row containing 10μL distilled H2O), 19 μL E.coli D31, and 15 μL of 4 mg/mL ortho-Nitrophenyl-β-galactoside (ONPG) in Z-buffer. Immediately after adding ONPG absorbance was recorded at 420nm and was recorded every 5 minutes over the next 90 minutes. Data reported are the average of 3 independent samples.
2.2.9 Flow Cytometry
Flow cytometry was used to identify cell permeability by incorporation of propidium iodide (PI) into S. aureus under various concentrations of CTAB detergent, PBP, PBP-11, PBP-18, PBP-X, PBP-A peptides using a BD FACSCelesta flow cytometer. Approximately 10,000 cells per sample in 100 μL of PBS, permeabilization reagent, and 5 μg/mL PI solution, were incubated for 30 min at 23°C counted in duplicate, and fluorescent signal was evaluated Ex 488nm laser, Em 575 filter. Percentage of cells found to be permeable for all samples was established by gating around cells on histogram illustrating PI signal with known permeabilization CTAB 102 μM (Sup Fig 1).
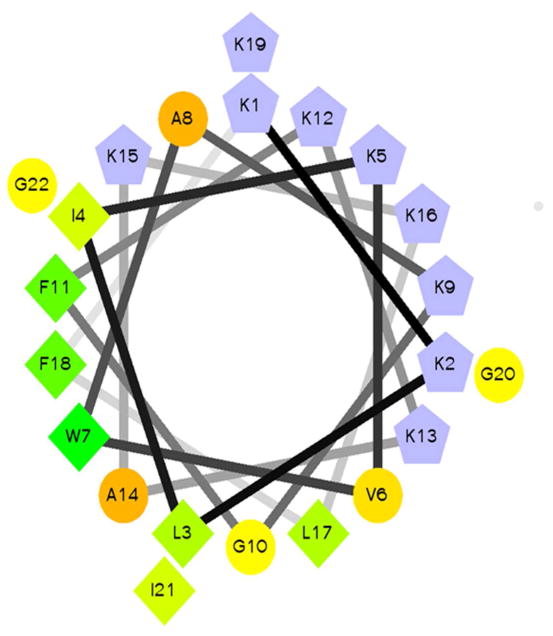
Figure 1. Helical wheel of PBP.
A helical wheel diagram of PBP is shown. Residue shapes correspond to hydrophobic (diamonds), polar (circles), and cationic (pentagons). The color pattern is blue for cationic residues and green to yellow for hydrophobics according to the internal scale of the HWP program. Helical wheel projection was made using Helical Wheel Projection Creator by Don Armstrong and Raphael Zidovetzki (33).
2.2.10 Hemolysis
Hemolysis of sheep red blood cells (RBCs) was used to identify cell permeabilization by leakage of hemoglobin at various concentrations of CTAB detergent, PBP, PBP-W11, PBP-W18, PBP-X, or PBP-A. A 5ml aliquot of defibrinated sheep blood (HemoStat Laboratories) was mixed with 5ml sterile PBS. The cells were isolated via centrifugation for 5min in a benchtop clinical centrifuge. The upper layer was aspirated and the cell pellet was resuspended to a final volume of 10ml. This was repeated for a total of 3 washes/isolations. Next, 135 μL of RBCs was added to wells of a 96-well plate containing 15 μL of serially diluted peptide or CTAB. The plate was covered and allowed to incubate at 37°C with gentle shaking for 60 min. The plate was then subjected to low speed centrifugation for 10 min to pellet the RBCs. The analysis was performed by carefully mixing 6 μL of the supernatant with 94 μL of fresh PBS and measuring the absorbance at 420 nm using a Molecular Devices M5 plate reader. Percent hemolysis was calculated based on the absorbance of each well compared to those wells with no peptide/CTAB and those with the highest concentration (2mM) CTAB.
2.2.11 Cytotoxicity of peptides toward HeLa Cells
Cytotoxicity of PBP peptide variants on HeLa cells was assayed by CellTiter-Blue reagent from Promega. In this assay, active cellular metabolism is measured by the enzymatic conversion of resarzurin to resorufin (fluorescent at 590 nm). HeLa cells were seeded into a 96-well flat bottom cell culture plate at 100,000 cells per well and grown for 24hrs in 180uL of DMEM 10% FBS, 1% Pen/Strep, 1% L-Glut. After 24 h, cells were exposed to various concentrations (20uL) of PBP peptides beginning at 15um final concentration and serially diluted 2-fold. As a positive control for cytotoxicity, CTAB reagent was used at a starting concentration of 8.2 mM and serially diluted 2-fold. Following 24 h of exposure to peptides, 20uL of CellTiter-Blue reagent was added directly to plate, mixed and incubated at 37°C for 2 h and analyzed on Synergy HT fluorescent plate reader, Ex. 485, Em. 590.
3. Results
3.1 Peptide sequences
Peptides were synthesized using solid-phase methods and purified using reversed phase HPLC. Fractions containing the peptide were isolated and analyzed via ESI-MS to confirm the identity of the peptide in the fractions (Table 1). The peptide sequences are based on the peptide scaffold used previously to promote TPPS42− J-aggregate formation (21). The peptides are highly cationic (+9 charge) which facilitates the 3:1 TPPS42-:peptide stoichiometry observed. When in a helical conformation, the peptides will adopt a facially amphipathic structure (Figure 1). The original scaffold PBP contained a Trp at position 7 which was switched to position 11 (PBP-W11) or 18 (PBP-W18) to serve as a reporter for different positions in the peptide. The PBP-X variant replaced all of the Lys residues with diaminopropionic acid, a non-proteinogenic lysine analog (Figure 1). The PBP-A variant replaced all of the hydrophobic residues with alanine, reducing the net hydrophobicity of the molecule.
Table 1.
Peptide Sequences and Properties
| Name | Sequence | MW calculated | MW found | Net Charge |
|---|---|---|---|---|
| PBP | KKLIKVWAKGFKKAKKLFKGIG | 2517.2 | 2515.5 | +10 |
| PBP-W11 | KKLIKVFAKGWKKAKKLFKGIG | 2517.2 | 2516.4 | +10 |
| PBP-W18 | KKLIKVFAKGFKKAKKLWKGIG | 2517.2 | 2516.4 | +10 |
| PBP-X | XXLIXVWAXGFXXAXXLFXGIG | 2139.2 | 2137.2 | +10 |
| PBP-A | KKAAKAWAKGAKKAKKLAKGAG | 2210.7 | 2209.6 | +10 |
3.2 TPPS42− Binding
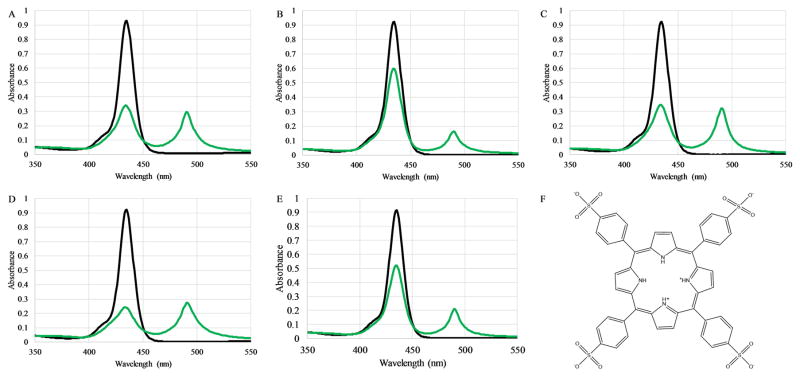
The PBP peptide was originally designed to serve as a scaffold to promote J-aggregate formation of TPPS42− molecules under elevated pH conditions. The scaffold activity at pH 3.6 exhibited a 3:1 TPPS42−:peptide stoichiometry, translating to a 1:3 TPPS42−:Lys stoichiometry (21). The peptides used in the study presented here were first analyzed for the ability to bind TPPS42− and form J-aggregates at pH 3. The formation of J-aggregate structures was monitored by the appearance of the characteristic absorbance peak at 490nm (Figure 2). All five peptides exhibited the ability to promote J-aggregate formation in the expected 1:3 peptide:TPPS42− stoichiometry at pH 1.8. Additional peptide:TPPS42− spectra highlighting the Soret and Q-bands at pH 1.8 and 7.0 are shown in Supplemental Figures 5 and 6.
Figure 2. Peptide-porphyrin binding by absorbance spectroscopy.
The binding of the TPPS42− to the peptides (A) PBP, (B) PBP-W11, (C) PBP-W18, (D) PBP-X, or (E) PBP-A was assessed by absorbance spectroscopy. In all panels the absorbance of 3 μM TPPS42− alone is shown in black while the absorbance spectrum after addition of 1 μM peptide is shown in green. (F) Chemical structure of TPPS42−
3.3 Antibacterial Activity
The antibacterial activity of the peptides was evaluated determining the minimal inhibitory concentration (MIC) by the broth microdilution method. Briefly, a series of serial dilutions of the peptide are prepared and exposed to a culture of bacteria with known density (~105 cfu/ml) and the ability to inhibit bacterial growth is determined after 18h. The peptides were tested against Gram-positive (S. aureus) and Gram-negative (E. coli, K. pneumoniae, and P. aeruginosa) strains in the MIC assay. The results are shown in Table 2. These results show the PBP parent sequence and the two Trp-position variants were similarly active against all strains tested, exhibiting MIC values in the range of 0.46–1.88 μM. Interestingly, the PBP-X containing peptide showed slightly reduced activity against E. coli and P. aeruginosa but exhibited no killing of K. pneumoniae or S. aureus. PBP-A was inactive against all strains tested over the range of concentrations examined (Table 2).
Table 2.
Minimal Inhibitory Concentration of PBP Variants
| PBP | PBP-W11 | PBP-W18 | PBP-X | PBP-A | |
|---|---|---|---|---|---|
| MIC (μM) | |||||
| E. coli | 0.94 | 0.94 | 0.94 | 1.88 | >15 |
| P. aeruginosa | 1.88 | 1.88 | 1.88 | 3.75 | >15 |
| K.Pneumoniae | 1.88 | 1.88 | 1.88 | 15 | >15 |
| S. aureus | 0.94 | 0.47 | 0.94 | >15 | >15 |
3.4 Binding & Interaction with Lipid Bilayers
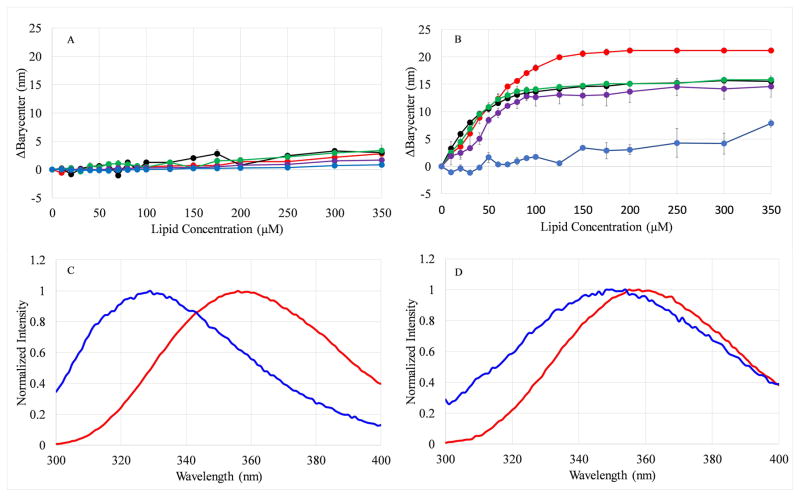
Based on the proposed membrane-active mechanism of action among AMPs, binding or interacting with the bacterial membrane is the first step in this process. However, due to the complexity of bacterial membranes and the abundance of membrane proteins present in them, it is difficult to measure direct binding to bacterial cell surfaces. Additionally, the presence of native membrane proteins prevents the use of Trp fluorescence as a reporter. As such, a model lipid vesicle system was used to directly probe the peptide-lipid interactions. A vesicle composition containing 100% POPC lipids was used as a zwitterionic surface to represent a mammalian or host cell, while vesicles composed of 75%POPC and 25% of the anionic POPG lipid was used to reflect the anionic character of the bacterial cell surface. When the Trp containing peptides bind to the bilayer, the Trp undergoes a change in the polarity of the local environment, resulting in a shift in the fluorescence emission spectrum, which is analyzed using the change in the barycenter (ΔBarycenter) (29). As seen in Figure 3A, there was no shift detected for any of the peptides when titrated with the zwitterionic PC vesicles. On the other hand, there were significant differences in binding to the PC:PG vesicles among the peptides over the range of the titration (Figure 3B). The PBP Trp variants and PBP-X all exhibited dramatic shifts in barycenter >15nm while the PBP-A peptide exhibited ~7nm in barycenter shift at the highest lipid concentration tested (Figure 3, Supplemental Figure 1). The barycenter shifts are consistent with the shifts in λmax of emission (Table 3). The full-width at half-maximum (FWHM) which can inform about population heterogeneity was also calculated for the emission spectra in solution and at 350μM lipid (Table 3). If two populations of peptides existed, such as bound and unbound or deeply inserted in the bilayer and shallowly inserted, the FWHM would dramatically increase as the steady state fluorescence emission would be a composite of emission spectra from two differently emitting populations, one blue shifted and one red shifted. However, if the peptides adopt a homogeneous population, the spectrum would be narrower. The values in Table 3 indicate that when bound to PC:PG, all the peptides adopt a relatively uniform population distribution except PBP-A, which displays a somewhat higher FWHM (~75nm). The broader spectrum is consistent with the binding data that exhibited an intermediate barycenter shift for PBP-A, likely indicative of incomplete binding.
Figure 3. Effects of lipid binding on Trp emission barycenter.
The change in the emission barycenter (ΔBc) of the Trp emission spectra were calculated after each step in a titration of 2μM peptide with (A) 100 % PC vesicles or (B) 75:25 PC:PG vesicles. In both panels colors represent PBP (red), PBP-W11 (black), PBP-W18 (green), PBP-X (purple), and PBP-A (blue). All data represent the average of 2–6 samples. Error bars representing the standard deviation are included for all data points but are obscured by the symbols in some cases. Representative spectra of PBP (C) and PBP-A (D) are also shown. In both (C) and (D), red curves represent emission from the peptide in the absence of lipid, while blue curves represent emission in the presence of 350μM lipid.
Table 3.
Spectroscopic and Fluorescence Quenching Data
| λ max (nm) | FWHM (nm) | Ksv (M−1) | Q-Ratioa | QIS (nm)b | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||
| Buffer | PCPG | Buffer | PCPG | Buffer | PCPG | PCPG | PCPG | |
| PBP | 356 | 329 | 64 | 59 ± 1 | 2.87 ± 0.1 | 1.14 ± 0.57 | 0.84 | 1.9 |
| PBP-W11 | 356 ± 1 | 333 ± 1 | 65 | 66 ± 1 | 10.2 ± 2.9 | 0.89 ± 0.15 | 0.81 | 3.2 |
| PBP-W18 | 357 ± 3 | 333 | 64 | 62 ± 1 | 6.77 ± 1.5 | 1.10 ± 0.14 | 0.78 | 2.0 |
| PBP-X | 357 ± 2 | 334 ± 2 | 64 ± 1 | 71 ± 6 | 3.15 ± 0.5 | 0.45 ± 0.29 | 2.20 | 2.6 |
| PBP-A | 358 ± 1 | 354 ± 1 | 63 ± 1 | 75 ± 1 | 14.4 ± 3.4 | 1.23 ± 0.66 | 6.23 | 3.7 |
In order to gain more insight on the peptide interactions with model membranes, fluorescence quenching approaches were used. Acrylamide was chosen as it effectively quenches Trp residues exposed to the aqueous environment, crosses bilayers, and does not significantly quench Trp residues buried in the bilayer (31). Quenching is analyzed using the Stern-Volmer equation in which the slope of the linear equation (Ksv) is related to the quenching and hence the exposure of the fluorophore to the quencher. The Ksv values for the peptides are shown in Table 3 for both peptide free in solution and when bound to PC:PG vesicles. As expected, the Ksv for the peptide in solution was higher than when bound to vesicles. This confirms that the peptide is indeed binding to the bilayer and adopting a conformation that shields the Trp from the acrylamide quencher.
The experiments were extended with the application of the Dual-Quencher Analysis (DQA) method. Briefly, this approach relies on the combination of quenching by the aqueous quencher acrylamide and the membrane-imbedded quencher 10-doxyl nonadecane (10-DN) (31). The ratio of quenching by these molecules (Q-ratio) is directly related to the depth of the Trp in the bilayer (28, 31). A Trp located near the center of the bilayer would exhibit a Q-ratio near 0.1, while a Trp at the surface of the bilayer can exhibit Q-ratios >2 (28, 34, 35). Additionally, monitoring the λmax or barycenter of the quenched spectra in comparison to the unquenched can yield similar information to the FWHM, in this case referred to as quencher induced shift (QIS). These results are shown in Table 3. The Trp variants exhibit remarkably similar Q-ratios, indicating these three peptides likely adopt a similar orientation in the bilayer and the Trp is located at an intermediate depth. However, the PBP-X and PBP-A peptides exhibit much higher Q-ratios, indicative of shallow, surface conformations. Again, the PBP-X exhibits the highest QIS, indicative of some extent population heterogeneity, although the net value is relatively small.
3.5 Secondary structure analysis
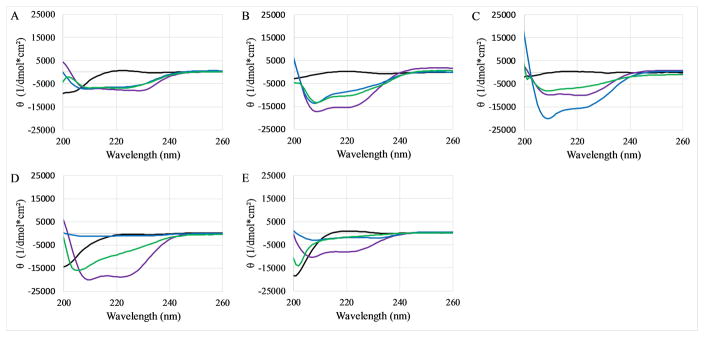
Many antimicrobial peptides adopt an α-helical conformation upon binding to lipid bilayers (36). Considering the small size of the peptides and thus the inability to form higher order folded conformations, circular dichroism (CD) spectroscopy can be used to gain a good understanding of the secondary structural elements present in these peptide. CD spectra were recorded of the peptides dissolved in dilute PBS, in the presence of SDS micelles (a membrane mimetic), in the presence of PC:PG vesicles, or dispersed in 50:50 PBS:TFE (a helix promoting solvent). In all cases, the peptides exhibited CD spectra corresponding to disordered or random coil conformations when dissolved in PBS buffer (Figure 4, black lines). Similarly, all peptides exhibited α-helical spectral signatures dispersed in PBS:TFE (Figure 4, purple lines). In the SDS micelles (Figure 4, blue lines) and PC:PG vesicles (Figure 4, green lines), only the PBP and Trp variants exhibited the canonical helical spectral signature. Both PBP-X and PBP-A exhibited very weak helical spectra in SDS, but appear to be generally lacking structure when bound to vesicles.
Figure 4. CD spectra of peptides.
CD spectra of peptides were collected in 10x diluted PBS (black), in the presence of 10mM SDS (blue), in the presence of 250μM PC:PG vesicles (green), or dispersed in 50:50 PBS:TFE (purple). The graphs represent (A) PBP, (B) PBP-W11, (C) PBP-W18, (D) PBP-X, (E) PBP-A. In all cases peptide concentration was 5 μM. All data represent the average of 64 scans after subtraction of background spectra lacking peptide.
3.6 Bacterial Membrane Permeabilization
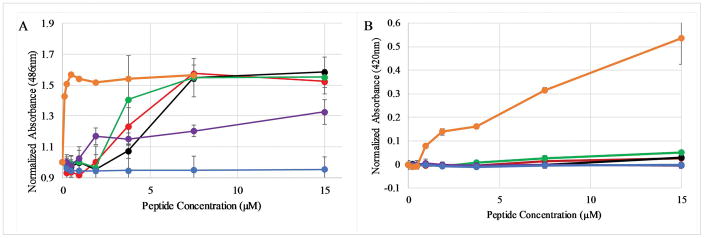
The binding assays indicate the ability of 4 of 5 PBP peptides to interact with bilayers, however as mentioned previously, the natural target membranes are significantly more complex than simple lipid vesicles. The ability of these peptides to permeabilize intact bacterial membranes was examined next. Permeability of E.coli was measured using two different enzyme-chromophore pairs: β-lactamase and nitrocefin for the outer membrane or β–galactosidase and ortho-Nitrophenyl-β-galactoside (ONPG) for the inner membrane. Briefly, the assay relies on the limited permeability of the substrates across the intact bacterial membrane resulting in a low level of enzymatic conversion of the substrate into the chromophoric product. However, if the peptides cause a disruption in the integrity of the bacterial membrane, the small molecule chromophore substrates can more easily transit across the membrane and be converted into the colored product. Assay progression is monitored by absorbance spectroscopy.
The permeabilization of the E.coli outer membrane is shown in Figure 5A. The PBP parent and the Trp variants exhibited similar permeabilization profiles, with maximum levels of permeability reached at 7.5μM peptide(compared to the control molecule polymyxin B). Interestingly, while the PBP-X binds to lipid bilayers in a similar fashion to PBP, this peptide induced less permeabilization of the E.coli outer membrane, even at the highest concentration tested of 15μM. The increase in permeability was dose dependent for these four sequences. Alternatively, the PBP-A peptide induced no detectable permeabilization of the outer membrane. The full time courses of the outer membrane permeabilization assay is shown in supplemental Figure S2. In comparison to the outer membrane activity, none of the peptides induced any measurable membrane permeabilization (Figure 5B). A comparison to the positive control, the cationic detergent CTAB, is also shown. The full time course of the inner membrane permeabilization assay is shown in supplemental Figure S3.
Figure 5. Peptide induced membrane permeabilization.
Peptide induced leakage of substrate across (A) the E.coli outer membrane assayed by β-lactamase conversion of nitrocefin and (B) across the E.coli inner membrane assayed by β-galactosidase conversion of ONPG. The normalized absorbance reflects the absorbance (A) at 486nm (the nitrocefin reaction product) or (B) at 420nm (the ONPG reaction product) normalized by the initial absorbance of the sample. In both panels symbols represent PBP (red), PBP-W11 (black), PBP-W18 (green), PBP-X (purple), and PBP-A (blue), and positive control (orange; polymyxin B in panel A, CTAB in panel B). All data represent the average of 3–6 replicates and error bars represent the standard deviations. In some cases the error bars are obscured by the symbols.
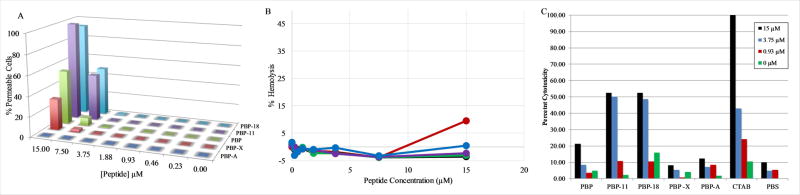
The permeabilization of the Gram-positive S.aureus was investigated using flow cytometry and the DNA-binding dye propidium iodide (PI). Briefly, if the peptides cause a disruption in the S.aureus membrane, the normally impermeant PI can enter the S. aureus cytoplasm and bind to DNA, resulting in fluorescence emission. The results of the S. aureus membrane permeabilization assay is shown in Figure 6A. The peptides followed the same general trend as the E. coli permeabilization with the Trp variants exhibiting significant permeabilization at high peptide concentrations, PBP-X exhibiting a more intermediate profile, and PBP-A inducing no detectable permeabilization of the S. aureus cells. Notably, the PBP peptide induced ~50% permeabilization at 15μM while PBP-W11 and PBP-W18 both exhibited >90% permeabilization at the same concentration. This contrasts with the nearly identical levels of E.coli OM permeabilization for these sequences across the range of concentrations tested.
Figure 6. Membrane permeabilzation and cytoxicity assays.
(A) Flow cytometry analysis of PI leakage into S. aureus after treatment with peptides. The percent permeabilization was determined from PI intensity. (B) Hemolysis of sheep RBCs by peptides PBP (red), PBP-W11 (black), PBP-W18 (green), PBP-X (purple), and PBP-A (blue). The percent permeabilization was determined by comparing the absorbance at 415nm in samples treated with peptides to that of samples treated with 8mM CTAB (set at 100% leakage). Error bars are included but are obscured by the symbols in all cases. Standard deviations were <1% in all cases. (C) Cytoxicity against HeLa cells measured by CTB assay. Cells were incubated with varying concentrations of peptide for 24 h and cell viability was assessed. Representative data from one of two biologicial replicates shown, all samples normalized to cytotoxicity of positive control CTAB.
3.7 Hemolysis and Cytoxicity Assays
The high selectivity of AMPs for bacterial over host membranes is a critical functional property of these molecules and is a discriminating factor between potentially therapeutic AMPs and those that are simply broad spectrum membrane permeabilizing peptides. As a means of testing the PBPs to disrupt mammalian membranes, we used a well characterized hemolysis assay using sheep red blood cells. The results of this assay are shown in Figure 6B. The percent hemolysis was calculated in comparison to leakage from untreated RBCs and that from RBCs treated with a detergent. In all cases, the PBP peptides induced very little hemolysis at all concentrations tested. Only the PBP sequence exhibited any meaningful hemolytic activity, ~10%, at the highest concentration tested of 15μM.
The cytoxicty of the peptides was directly measured against the commonly utilized HeLa model system which is derived from human cervical cancer cells. Cells were treated for 24 h with varying concentrations of the PBP peptides and cell viability was subsequently assessed by the CellTiter-Blue (CTB) method, which measures the conversion of resarzurin to resorufin (fluorescent at 590nm) by metabolically active and thus viable cells. The results of this assay are shown in Figure 6C. In contrast to the hemolysis assay, PBP-11 and PBP-18 exhibited significant (~50%) toxicity toward HeLa cells at 15 μM and 3.75 μM while PBP exhibited much less toxic effect (~20%) at 15 μM. PBP-X and PBP-A exhibited little to no toxicity, very similar to PBS control. Interestingly, these results mimic those seen in PI permeability flow cytometry assays with PBP-11 and PBP-18 having the most significant effect followed by PBP, PBP-X, and PBP-A having no effect.
4. Discussion
The viability of AMPs as potential therapeutics has been an area of investigation since the wider study of host-defense peptides took off in the late 1980’s. The relative ease of synthesis, natural variation, controlled sequence as compared to random copolymers, and selectivity for bacteria over host cells are all beneficial traits that would be ideal for development into a therapeutic. Generally, naturally isolated and designed AMPs share the common traits of being net cationic, possessing some hydrophobic side chains, and are thought to act through a membrane-active mechanism. However, despite significant effort, no consensus sequences have been identified, nor have any AMPs been successfully piloted through clinical trials. While MSI-78 (aka pexiganan) and a variety of “MSI” peptides have been studied(37), we could only find two reports on the revisimer efficacy(22, 38), and only one of these has any biophysical characterization of the peptide(22)
Focusing on the Trp position in the peptide, it is clear that the replacement of Phe with Trp does not have a significant effect on any of the measured properties of the peptide. The binding to lipid vesicles was almost identical for all three variants (PBP, PBP-W11, PBP-W18), with the exception that the PBP exhibited a slightly higher Δbarycenter (21 nm vs. 16 nm). These three peptides showed similar profiles in antibacterial activity (Table 2), spectral properties (Table 3), secondary structure (Figure 4), and induction of bacterial membrane permeabilization (Figures 5, S2, and S3). This is not surprising as both Trp and Phe are bulky, aromatic amino acids and are both very nonpolar (39, 40). It is well known that Trp exhibits a strong preference for locating at or near the headgroup interface region in helical proteins (41), however the data indicate this preference does not impact the behavior of the PBP peptide when the Trp is relocated within the sequence. In all cases, the Trp appears to locate at an intermediate depth, consistent with a facially amphiphilic helix with the hydrophobic face buried in the lipid bilayer core (28, 42). The data also indicate that the peptide is relatively uniform in penetrating the bilayer. The Trp placement near the N-terminus, center of the peptide, and near the C-terminus yielded very similar depth profiles, indicating the peptide is likely adopting a uniform orientation, near parallel to the plane of the bilayer. Interestingly, despite the similar properties, the PBP displayed a total barycenter shift ~5nm greater than PBP-W11 and PBP-W18. It is not clear why the Trp emission would be shifted if the overall depth in the bilayer and access to aqueous and membrane imbedded quenchers is so similar. Nonetheless, the similarity in bilayer topography is also consistent with the very similar antmicrobial activity, supporting the hypothesis that the peptides exert the antimicrobial effect by disrupting bacterial membranes. However, the PBP sequence did show both a decreased permeability in PI assays in S. aureus as well as less cytotoxic activity, so there appears to be a sequence dependent effect.
The effects of the more significant sequence changes in PBP-X and PBP-A are much more dramatic than the Trp positional effects. The PBP-A lost all ability to kill bacteria at the highest concentrations tested (15μM), which is consistent with the weaker binding to lipid bilayers (Figure 3). The simplest explanation is because of the overall reduced hydrophobicity in the PBP-A, the peptide cannot effectively partition into the bilayer core, thus it cannot induce a significant disruption in the bilayer integrity. This is consistent with the reduced barycenter shift exhibited at the highest lipid concentrations, and the very shallow depth according to the Q-ratio. This is consistent with previous findings from our laboratory using AMPs (29) and from other groups using polymeric mimics of peptides (43, 44). However, caution must be taken to prevent over interpretation of the Q-ratio. Very large Q-ratios lose quantitative meaning relating to depth as the formula relies on a ratio of acrylamide quenching to 10-DN quenching. As the quenching of Trp by 10-DN is distance dependent, a very shallow or peptide not bound to the bilayer may experience little to no quenching by the 10-DN, drastically reducing the denominator in the Q-ratio and thus resulting in a very large Q-ratio value (28, 31). In the case of PBP-A, the broadened emission spectra and weak quenching by 10-DN indicate there may be some small fraction of peptides bound to the bilayer at the highest lipid concentrations. It should also be noted that during the experimental analysis, the PBP-A peptide appeared to show a propensity to aggregate. This was evidenced by a biphasic Stern volmer plot, with minimal quenching at low acrylamide concentrations (data not shown). Freshly dissolved PBP-A eliminated this behavior and was used for all future experiments.
The PBP-X exhibited the most varied set of characteristics. The binding ad acrylamide quenching alone indicate the peptides are binding with strong affinity for the anionic lipid bilayers, shielding the Trp from quencher and exhibiting the expected shift in barycenter. However, the Q-ratio indicated a more shallow depth profile compared to PBP, PBP-W11, and PBP-W18. This may indicate a somewhat different topography or orientation in the bilayer that induces the large shift in barycenter while still being more protected from the membrane bound quencher 10-DN. This altered topography may result from the shortened side chains in Dap causing the peptide backbone to orient more closely to the bilayer surface. A shallower topography is consistent with the Dap side chains being unable to “snorkel” and the side chain amide remain in the polar environment (34, 43, 45–49). The shortened side chains reduce the rotational flexibility of the helix as a whole, thus limiting the orientations it can adopt on the bilayer surface to satisfy both the cationic groups and the hydrophobic groups. Similar effects have been previously demonstrated with surface associated peptides containing Asp (42). Most interestingly, the antimicrobial activity of the PBP-X was mixed, with activity near PBP against E. coli and P. aeruginosa, but showing markedly less activity against K. pneumoniae and S. aureus compared to PBP. The activity against S. aureus is especially interesting as the PBP-X showed very low levels of S. aureus membrane permeabilization, but still prevented bacterial growth. This may indicate the mechanism does not require large-scale permeabilization to accommodate large molecule diffusion across the membrane to induce bacterial cell death. Alternatively, the difference in bacterial cell density and therefore the number of peptides per cell, differs in the MIC and flow cytometry experiments which may give rise to the differences. Additionally, PBP-X peptide exhibited weak to no helical characteristics in the CD spectra when bound to bilayers. This is not unexpected as previous reports have shown inclusion of Dap reduces helical content(50). However, this will directly impact the way the peptide orients on/in the bilayer, potentially reducing the role of the facial amphiphilicity of the AMP structure.
The results show that the balance of hydrophobicity and cationic charge is clearly important for the antimicrobial activity of these peptides. These findings are consistent with previously shown trends in antimicrobial polymers and peptide mimetics (51). The cationic charge is generally thought to provide AMPs the selectivity between bacterial and host membranes, while the hydrophobicity drives membrane disruption. Additionally, the hydrophobicity of membrane active antimicrobials is directly linked to the cytotoxic effects against host cells, with increased hydrophobicity causing increased cytotoxicity (7, 51, 52). The PBP peptides follow this trend, with the most hydrophobic peptides causing the most, although minimal in these studies, RBC lysis. Similarly, the least hydrophobic peptide had no antibacterial activity, likely because it cannot partition into the bilayer core. However, more than the balance of hydrophobics and cationic charge, the actual structure of these functional groups may play a significant role in the activity. Because the peptides adopt an amphipathic α-helix when bound to the lipid surface, the depth the peptide partitions into the bilayer core is influenced by the side chain length, both hydrophobic and cationic. The ability of cationic side chains to snorkel and the ability of the hydrophobics to partition into the nonpolar core, as well as how the side chains orient when bound, are clearly important factors in the activity of AMPs.
Overall, the PBP peptides yield several interesting observations regarding AMPs. First, the binding and quenching studies support the interfacial orientation of these peptides. The leakage studies coupled with the MIC assays support the membrane disruption model for the mechanism of action of these peptides, in both Gram positive and Gram negative strains. Finally, the dual functionality of the PBPs, antimicrobial and porphyrin binding, demonstrates the potential for the development of multifunctional peptides as the basis for materials and therapeutics. Taken together these observations can help guide the development of new peptides with optimal efficacy and toxicity profiles for antimicrobial applications but also have the potential to add functionality based on the specific demands of the application.
Supplementary Material
Acknowledgments
The authors would like to thank the College of Science and Mathematics for the purchase of the flow cytometer. This work was funded by NIH 1R15GM094330 to G.A.C. and by junior faculty SEED funding to B.R.C. The authors would also like to thank Kenichi Kuroda for helpful discussions.
Footnotes
Conflict of interest
All authors declare no potential financial or personal conflict of interest.
Authors’ contribution
Caputo and Carone participated in research design; Shirley and Chrom conducted the experiments and performed data analysis; and Caputo, Shirley, and Carone contributed to the writing of the manuscript. Richards performed cytotoxicity assays.
References
- 1.Prevention CfDCa. Antibiotic Resistance Threats in the United States, 2013 2013 [Google Scholar]
- 2.Organization, W. H. Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline, including tuberculosis. Geneva: Sep, 2017. p. 48. [Google Scholar]
- 3.Wright GD. Something old, something new: revisiting natural products in antibiotic drug discovery. Canadian journal of microbiology. 2014;60:147–154. doi: 10.1139/cjm-2014-0063. [DOI] [PubMed] [Google Scholar]
- 4.Villapun VM, Dover LG, Cross A, Gonzalez S. Antibacterial Metallic Touch Surfaces. Materials. 2016;9 doi: 10.3390/ma9090736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goderecci SS, Kaiser E, Yanakas M, Norris Z, Scaturro J, Oszust R, Medina CD, Waechter F, Heon M, Krchnavek RR, Yu L, Lofland SE, Demarest RM, Caputo GA, Hettinger JD. Silver Oxide Coatings with High Silver-Ion Elution Rates and Characterization of Bactericidal Activity. Molecules. 2017;22 doi: 10.3390/molecules22091487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda K, Caputo GA. Antimicrobial polymers as synthetic mimics of host-defense peptides, Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology. 2013;5:49–66. doi: 10.1002/wnan.1199. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi H, Caputo GA, Vemparala S, Kuroda K. Synthetic Random Copolymers as a Molecular Platform To Mimic Host-Defense Antimicrobial Peptides. Bioconjugate chemistry. 2017;28:1340–1350. doi: 10.1021/acs.bioconjchem.7b00114. [DOI] [PubMed] [Google Scholar]
- 8.Young R, Gill JJ. MICROBIOLOGY. Phage therapy redux--What is to be done? Science. 2015;350:1163–1164. doi: 10.1126/science.aad6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Koh JJ, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Frontiers in neuroscience. 2017;11:73. doi: 10.3389/fnins.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock RE, Haney EF, Gill EE. The immunology of host defence peptides: beyond antimicrobial activity. Nature reviews Immunology. 2016;16:321–334. doi: 10.1038/nri.2016.29. [DOI] [PubMed] [Google Scholar]
- 11.Choi J, Groisman EA. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Molecular microbiology. 2016;101:1024–1038. doi: 10.1111/mmi.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier JH, Segura T. Evolving the use of peptides as components of biomaterials. Biomaterials. 2011;32:4198–4204. doi: 10.1016/j.biomaterials.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branco MC, Sigano DM, Schneider JP. Materials from peptide assembly: towards the treatment of cancer and transmittable disease. Current opinion in chemical biology. 2011;15:427–434. doi: 10.1016/j.cbpa.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doll TAPF, Dey R, Burkhard P. Design and optimization of peptide nanoparticles. Journal of Nanobiotechnology. 2015;13:73. doi: 10.1186/s12951-015-0119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KH, Ko DK, Kim YT, Kim NH, Paul J, Zhang SQ, Murray CB, Acharya R, DeGrado WF, Kim YH, Grigoryan G. Protein-directed self-assembly of a fullerene crystal. Nature communications. 2016;7:11429. doi: 10.1038/ncomms11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JH, Lee JH, Lee YJ, Nam KT. Protein/peptide based nanomaterials for energy application. Current opinion in biotechnology. 2013;24:599–605. doi: 10.1016/j.copbio.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Solomon LA, Sykes ME, Wu YA, Schaller RD, Wiederrecht GP, Fry HC. Tailorable Exciton Transport in Doped Peptide-Amphiphile Assemblies. ACS nano. 2017;11:9112–9118. doi: 10.1021/acsnano.7b03867. [DOI] [PubMed] [Google Scholar]
- 18.Reig AJ, Pires MM, Snyder RA, Wu Y, Jo H, Kulp DW, Butch SE, Calhoun JR, Szyperski T, Solomon EI, DeGrado WF. Alteration of the oxygen-dependent reactivity of de novo Due Ferri proteins. Nature chemistry. 2012;4:900–906. doi: 10.1038/nchem.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fry HC, Lehmann A, Saven JG, DeGrado WF, Therien MJ. Computational design and elaboration of a de novo heterotetrameric alpha-helical protein that selectively binds an emissive abiological (porphinato)zinc chromophore. Journal of the American Chemical Society. 2010;132:3997–4005. doi: 10.1021/ja907407m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder RA, Butch SE, Reig AJ, DeGrado WF, Solomon EI. Molecular-Level Insight into the Differential Oxidase and Oxygenase Reactivities of de Novo Due Ferri Proteins. Journal of the American Chemical Society. 2015;137:9302–9314. doi: 10.1021/jacs.5b03524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuciauskas D, Caputo GA. Self-assembly of peptide-porphyrin complexes leads to pH-dependent excitonic coupling. The journal of physical chemistry B. 2009;113:14439–14447. doi: 10.1021/jp905468y. [DOI] [PubMed] [Google Scholar]
- 22.Neubauer D, Jaskiewicz M, Migon D, Bauer M, Sikora K, Sikorska E, Kamysz E, Kamysz W. Retro analog concept: comparative study on physico-chemical and biological properties of selected antimicrobial peptides. Amino acids. 2017;49:1755–1771. doi: 10.1007/s00726-017-2473-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurthner F, Kaiser TE, Saha-Moller CR. J-aggregates: from serendipitous discovery to supramolecular engineering of functional dye materials. Angewandte Chemie. 2011;50:3376–3410. doi: 10.1002/anie.201002307. [DOI] [PubMed] [Google Scholar]
- 24.Kuciauskas D, Kiskis J, Caputo GA, Gulbinas V. Exciton annihilation and energy transfer in self-assembled peptide-porphyrin complexes depends on peptide secondary structure. The journal of physical chemistry B. 2010;114:16029–16035. doi: 10.1021/jp108685n. [DOI] [PubMed] [Google Scholar]
- 25.Kokona B, Kim AM, Roden RC, Daniels JP, Pepe-Mooney BJ, Kovaric BC, de Paula JC, Johnson KA, Fairman R. Self assembly of coiled-coil peptide-porphyrin complexes. Biomacromolecules. 2009;10:1454–1459. doi: 10.1021/bm9000553. [DOI] [PubMed] [Google Scholar]
- 26.Nardo L, Kristensen S, Tonnesen HH, Hogset A, Lilletvedt M. Solubilization of the photosensitizers TPCS(2a) and TPPS(2a) in aqueous media evaluated by time-resolved fluorescence analysis. Die Pharmazie. 2012;67:598–600. [PubMed] [Google Scholar]
- 27.Peng Q, Moan J, Farrants G, Danielsen HE, Rimington C. Localization of potent photosensitizers in human tumor LOX by means of laser scanning microscopy. Cancer letters. 1991;58:17–27. doi: 10.1016/0304-3835(91)90019-e. [DOI] [PubMed] [Google Scholar]
- 28.Caputo GA, London E. Analyzing transmembrane protein and hydrophobic helix topography by dual fluorescence quenching. Methods in molecular biology. 2013;974:279–295. doi: 10.1007/978-1-62703-275-9_13. [DOI] [PubMed] [Google Scholar]
- 29.Saint Jean KD, Henderson KD, Chrom CL, Abiuso LE, Renn LM, Caputo GA. Effects of Hydrophobic Amino Acid Substitutions on Antimicrobial Peptide Behavior. Probiotics and antimicrobial proteins. 2017 doi: 10.1007/s12602-017-9345-z. [DOI] [PubMed] [Google Scholar]
- 30.Ridgway Z, Picciano AL, Gosavi PM, Moroz YS, Angevine CE, Chavis AE, Reiner JE, Korendovych IV, Caputo GA. Functional characterization of a melittin analog containing a non-natural tryptophan analog. Biopolymers. 2015;104:384–394. doi: 10.1002/bip.22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caputo GA, London E. Using a novel dual fluorescence quenching assay for measurement of tryptophan depth within lipid bilayers to determine hydrophobic alpha-helix locations within membranes. Biochemistry. 2003;42:3265–3274. doi: 10.1021/bi026696l. [DOI] [PubMed] [Google Scholar]
- 32.Burman LG, Nordstrom K, Boman HG. Resistance of Escherichia coli to penicillins. V. Physiological comparison of two isogenic strains, one with chromosomally and one with episomally mediated ampicillin resistance. Journal of bacteriology. 1968;96:438–446. doi: 10.1128/jb.96.2.438-446.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Don Armstrong RZ. Helical Wheel Projectinos. Id: wheel.pl, v 1.4, 2009-10-20 21:23, 4 2009-10-20 21: 23:36 don Exp ed. [Google Scholar]
- 34.Caputo GA, London E. Cumulative effects of amino acid substitutions and hydrophobic mismatch upon the transmembrane stability and conformation of hydrophobic alpha-helices. Biochemistry. 2003;42:3275–3285. doi: 10.1021/bi026697d. [DOI] [PubMed] [Google Scholar]
- 35.Moroz YS, Binder W, Nygren P, Caputo GA, Korendovych IV. Painting proteins blue: beta-(1-azulenyl)-L-alanine as a probe for studying protein-protein interactions. Chemical communications. 2013;49:490–492. doi: 10.1039/c2cc37550h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyansky AA, Chugunov AO, Vassilevski AA, Grishin EV, Efremov RG. Recent advances in computational modeling of alpha-helical membrane-active peptides. Current protein & peptide science. 2012;13:644–657. doi: 10.2174/138920312804142147. [DOI] [PubMed] [Google Scholar]
- 37.Lee DK, Bhunia A, Kotler SA, Ramamoorthy A. Detergent-type membrane fragmentation by MSI-78, MSI-367, MSI-594, and MSI-843 antimicrobial peptides and inhibition by cholesterol: a solid-state nuclear magnetic resonance study. Biochemistry. 2015;54:1897–1907. doi: 10.1021/bi501418m. [DOI] [PubMed] [Google Scholar]
- 38.Iwahori A, Hirota Y, Sampe R, Miyano S, Numao N. Synthesis of reversed magainin 2 analogs enhanced antibacterial activity. Biological & pharmaceutical bulletin. 1997;20:267–270. doi: 10.1248/bpb.20.267. [DOI] [PubMed] [Google Scholar]
- 39.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. Journal of molecular biology. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 40.White SH, Heijne Gv. How Translocons Select Transmembrane Helices. Annual review of biophysics. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 41.Senes A, Chadi DC, Law PB, Walters RF, Nanda V, Degrado WF. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. Journal of molecular biology. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 42.Caputo GA, London E. Position and ionization state of Asp in the core of membrane-inserted alpha helices control both the equilibrium between transmembrane and nontransmembrane helix topography and transmembrane helix positioning. Biochemistry. 2004;43:8794–8806. doi: 10.1021/bi049696p. [DOI] [PubMed] [Google Scholar]
- 43.Palermo EF, Vemparala S, Kuroda K. Cationic spacer arm design strategy for control of antimicrobial activity and conformation of amphiphilic methacrylate random copolymers. Biomacromolecules. 2012;13:1632–1641. doi: 10.1021/bm300342u. [DOI] [PubMed] [Google Scholar]
- 44.Palermo EF, Lee DK, Ramamoorthy A, Kuroda K. Role of cationic group structure in membrane binding and disruption by amphiphilic copolymers. The journal of physical chemistry B. 2011;115:366–375. doi: 10.1021/jp1083357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zelezetsky I, Tossi A. Alpha-helical antimicrobial peptides—Using a sequence template to guide structure–activity relationship studies. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2006;1758:1436–1449. doi: 10.1016/j.bbamem.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 46.Ojemalm K, Higuchi T, Lara P, Lindahl E, Suga H, von Heijne G. Energetics of side-chain snorkeling in transmembrane helices probed by nonproteinogenic amino acids. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:10559–10564. doi: 10.1073/pnas.1606776113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gebhardt M, Henkes LM, Tayefeh S, Hertel B, Greiner T, Van Etten JL, Baumeister D, Cosentino C, Moroni A, Kast SM, Thiel G. Relevance of lysine snorkeling in the outer transmembrane domain of small viral potassium ion channels. Biochemistry. 2012;51:5571–5579. doi: 10.1021/bi3006016. [DOI] [PubMed] [Google Scholar]
- 48.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of molecular biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 49.Chamberlain AK, Lee Y, Kim S, Bowie JU. Snorkeling preferences foster an amino acid composition bias in transmembrane helices. Journal of molecular biology. 2004;339:471–479. doi: 10.1016/j.jmb.2004.03.072. [DOI] [PubMed] [Google Scholar]
- 50.Cheng RP, Girinath P, Ahmad R. Effect of lysine side chain length on intra-helical glutamate--lysine ion pairing interactions. Biochemistry. 2007;46:10528–10537. doi: 10.1021/bi700701z. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda K, Caputo GA, DeGrado WF. The role of hydrophobicity in the antimicrobial and hemolytic activities of polymethacrylate derivatives. Chemistry. 2009;15:1123–1133. doi: 10.1002/chem.200801523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S, Hyun S, Lee Y, Lee Y, Yu J. Nonhemolytic Cell-Penetrating Peptides: Site Specific Introduction of Glutamine and Lysine Residues into the alpha-Helical Peptide Causes Deletion of Its Direct Membrane Disrupting Ability but Retention of Its Cell Penetrating Ability. Biomacromolecules. 2016;17:3007–3015. doi: 10.1021/acs.biomac.6b00874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.